Introduction
There are ~376,300 new colorectal cancer (CRC) cases
according to the data from the Chinese National Cancer Center in
China in 2015 (1,2). The treatment strategies for CRC include
surgery, chemotherapy and radiation therapy. However, the 5-year
overall survival rate for CRC patients at early stages is as high
as 90%, with an extremely low rate 13% for patients at stage IV
(3).
MicroRNAs (miRNAs/miRs) are RNAs with a length of
19-22 nucleotides, which lack the ability to code for proteins
(4). They primarily bind the
3'-untranslated regions (UTRs) of target genes to repress gene
expression (4). miRNAs have been
demonstrated to be aberrantly expressed in CRC, and they have the
potential to be developed as diagnostic or treatment biomarkers.
miR-17 has an elevated expression in both CRC tissues and cell
lines (5). The overexpression of
miR-17 promotes CRC cell metastasis, while knockdown of miR-17
causes the opposite effects via targeting salt inducible kinase 1,
indicating the oncogenic role of miR-17(5). Furthermore, the expression levels of
miR-143-3p have been reported to decrease in CRC tissues, and
functional analyses revealed that miR-143-3p was able to suppress
CRC cell proliferation, migration and invasion capacities via
regulating catenin-δ1(6). Moreover,
abnormal expression levels of miR-1307-3p were identified in human
cancers, including breast cancer and hepatocellular carcinoma
(7,8). In breast cancer, miR-1307-3p was
revealed to be upregulated in cancer tissues and increased
miR-1307-3p expression increased the risk of death (7). Functional analyses revealed that
miR-1307-3p overexpression stimulates breast cancer cell growth
through targeting SET and MYND domain-containing 4 (SMYD4)
(7). In addition, miR-1307-3p was
revealed to be upregulated in hepatocellular carcinoma tissues at
advanced tumor stages, in large tumors and was associated with a
less favorable overall clinical outcome (8). In vitro and in vivo
analyses indicated that the knockdown of miR-1307-3p inhibited
tumor progression via regulating Disabled Homolog 2 interacting
protein (8). To date, it is unclear
whether miR-1307-3p has a role in the development of CRC.
Tumor suppressor candidate 5 (TUSC5) is a protein
that is widely expressed in brown adipocytes and peripheral neurons
(9,10). In recent years, the roles of TUSC5 in
various tumor types have been gradually recognized. For example,
TUSC5 expression was reported to be regulated by miR-3188,
affecting breast cancer cell proliferation, migration and apoptosis
capacity (11). In addition, TUSC5
was also revealed to be downregulated, and served as target for
miR-484, to influence the malignant phenotypes of hepatocellular
carcinoma cells (12).
In the present study, the expression levels of
miR-1307-3p were explored in CRC cells. Additionally, functional
assays were performed to investigate the functions of miR-1307-3p
in CRC progression. Furthermroe, the mechanisms of action that
mediated the functions of miR-1307-3p were explored.
Materials and methods
Cell lines and transfection
The normal human epithelial cell line NCM460 and CRC
cells, including SW480 and SW620, were obtained from the American
Type Culture Collection were grown in DMEM (cat. no. A4192101;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(cat. no. 16000044; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (cat. no. 15140122; Gibco; Thermo Fisher
Scientific, Inc.) in a 37˚C humidified atmosphere containing 95%
atmospheric air and 5% CO2. The miR-1307-3p inhibitor
(5'-CACGACCGACGCCACGCCGAGU-3') and control miRNA (miR-con,
5'-AGGCCAGCCACGGCGCAUCCAC-3') were synthesized by Guangzhou RiboBio
Co., Ltd. Specific small interfering (si)RNA targeting TUSC5
(si-TUSC5; 5'-GGAGAACAAGGAUGACCAATT-3') and the corresponding
control sequence (siR-con; 5'-CAGTCGCGTTTGCGACTGGTT-3') were also
purchased from Guangzhou RiboBio Co., Ltd. Transfections were
conducted using Lipofectamine® 2000 (cat. no. 16168019;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions (final concentrations: miRNA, 200 nM; siRNA, 100 nM).
Cells (SW480 and SW620) were transfected for 48 h before being
collected for subsequent analyses.
Microarray analysis
The GSE123040 dataset (13) was downloaded from the Gene Expression
Omnibus database (https://www.ncbi.nlm.nih.gov/gds/?term=gse123040)
and was used to determine the expression levels of miR-1307-3p in
CRC tissues and normal tissues.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Cultured cells (SW480 and SW620) with or without
oligonucleotides transfection, were lysed to isolate RNA using
TRIzol® reagent (cat. no. 15596018; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instruction. For
miRNA, complementary DNA (cDNA) was synthesized using the miScript
II RT kit (cat. no. 4366596; Thermo Fisher Scientific, Inc.). For
mRNA, cDNA was synthesized using the PrimeScript RT Reagent kit
(cat. no. 6210A, Takara Biotechnology, Inc.). Quantitative PCR was
conducted using an ABI 7500 system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using SYBR Green mix obtained from Takara
Biotechnology Co., Inc. (cat. no. RR820A). Primers sequences used
in the present study were as follows: miR-1307-3p forward,
5'-TGCGGGTCCAGTTTTCCCAGGAA-3' and reverse,
5'-CCAGTGCAGGGTCCGAGGT-3'; U6 snRNA forward,
5'-TGCGGGTGCTCGCTTCGCAGC-3' and reverse, 5'-CCAGTGCAGGGTCCGAGGT-3';
TUSC5 forward, 5'-ATTAGTAAAGTTGTTT-3' and reverse,
5'-CAAAAAACTCTAAAAAAA-3'; GAPDH forward,
5'-AACGTGTCAGTOGTGGACCTG-3' and reverse,
5'-AGTGGGTGTCGCTGTFGAAGT-3'. The procedure used was as follows: 1
cycle at 94˚C for 10 min, 40 cycles at 95˚C for 10 sec, 56˚C for 30
sec and 70˚C for 30 sec. U6 snRNA or GAPDH was regarded as internal
control gene for miR-1307-3p or TUSC5, respectively. Relative
expression levels were calculated using the
2-ΔΔCq method (14).
Cell proliferation assay
Cells (SW480 and SW620) with or without
oligonucleotides transfection, were plated into a 96-well plate at
a density of 3x103 cells/well and then the cell
proliferation rate was detected using the cell counting kit-8 (cat.
no. C0037; CCK-8, Beyotime Institute of Biotechnology) assay
according to the supplier's instruction. At the indicated times, 10
µl CCK-8 was added to the well and further incubated for 2 h at
37˚C. Finally, optical densities for each well were measured at 450
nm.
Cell invasion assay
Matrigel® (cat. no. 40480; BD
Biosciences) was used to pre-coated 8-µm chamber (cat. no. 354480,
Corning, Inc.) at room temperature for 24 h. Subsequently,
1x105 cells suspended in serum-free DMEM were plated in
the upper chamber, while the DMEM containing 10% FBS (cat. no.
16000044; Gibco; Thermo Fisher Scientific, Inc.) was plated in the
lower chamber. After 48 h incubation, invading cells were fixed
with 90% methanol at room temperature for 30 min, stained with 0.1%
crystal violet at room temperature for 15 min, and counted under
inverted light microscope (magnification, x200).
Bioinformatics analysis
TargetScan 7.2 (http://www.targetscan.org/) was utilized to analyze
potential targets for miR-1307-3p.
Dual-luciferase reporter assays
TUSC5 3'-UTR containing binding sites for
miR-1307-3p were inserted into the pmirGLO plasmid obtained from
Promega Corporation to generate wild-type TUSC5 (TUSC5-wt). Mutant
TUSC5 (TUSC5-mt) was constructed using the Site-Directed
Mutagenesis kit (Takara Bio, Inc.). TUSC5-wt or TUSC5-mt were
co-transfected with 100 nM synthetic miRNA into 2x103
CRC cells using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
After transfection for 48 h, the dual-luciferase system (cat. no.
E1910, Promega Corporation) was used to detect relative luciferase
activities with Renilla luciferase activity as internal
control according to the manufacturer's instructions.
Statistical analysis
SPSS version 18.0 (SPSS, Inc.) was used to analyze
data obtained from three independent experiments. Data are
presented as the mean ± SD. Differences in groups were analyzed
using the paired Student's t-test (2 groups) or one-way ANOVAs with
Tukey's post-hoc test (≥3 groups). P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-1307-3p is upregulated in CRC
tissues and cell lines
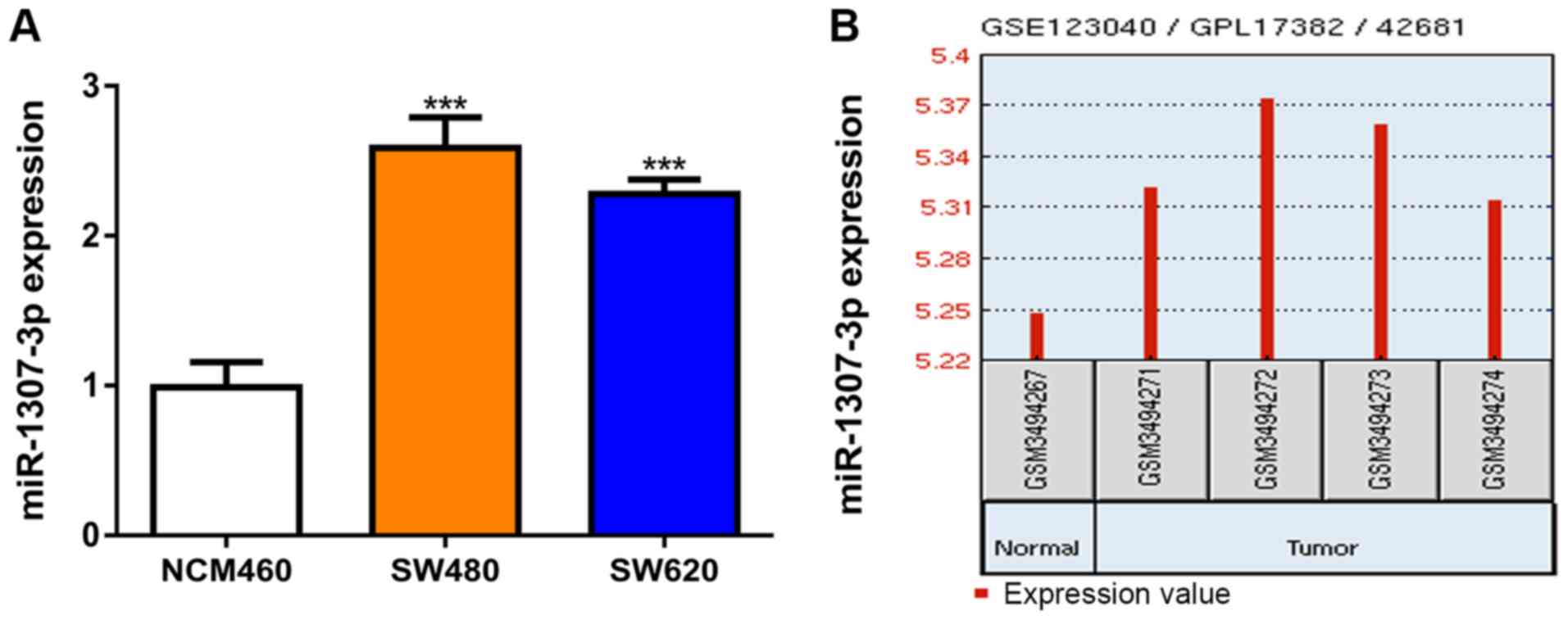
Firstly, miR-1307-3p expression levels were measured
in the CRC and normal cell lines using RT-qPCR. It was revealed
that miR-1307-3p expression was significantly elevated in CRC cells
compared with the normal cell line (Fig.
1A). Moreover, miR-1307-3p expression levels were increased in
CRC tissues compared with normal tissues (Fig. 1B).
miR-1307-3p promotes CRC cell
proliferation and invasion
The effects of miR-1307-3p on CRC cell behavior were
then investigated via loss-of-function experiments. It was
demonstrated that the introduction of a miR-1307-3p inhibitor
significantly decreased miR-1307-3p expression levels in CRC cells
(Fig. 2A). CCK-8 assays revealed
that the optical density value in the miR-1307-3p
inhibitor-transfected group was significantly lower compared with
the miR-con group, as indicated at 72 h (Fig. 2B). Transwell invasion assays were
then performed to evaluate the effect of miR-1307-3p on cell
invasion. It was revealed that there were fewer invasive cells in
the miR-1307-3p inhibitor transfected group compared with the
miR-con group (Fig. 2C). The effect
of miR-1307-3p on proliferation was also tested in the normal cell
line (NCM460). As indicated in Fig.
S1A and B, knockdown of
miR-1307-3p repressed NCM460 cell proliferation.
miR-1307-3p targets TUSC5 in CRC
To explore the mechanisms of action behind
miR-1307-3p-mediated CRC cell behaviors, the targets of miR-1307-3p
were predicted using TargetScan, and it was revealed that TUSC5
represented a potential target (Fig.
3A). Dual-luciferase activity reporter assays revealed that
miR-1307-3p inhibitor transfection increased the luciferase
activity of CRC cells transfected with TUSC5-wt, but not TUSC5-mt
(Fig. 3B). Subsequently, RT-qPCR was
performed to analyze the TUSC5 expression levels in the groups
transfected with miR-1307-3p inhibitor or miR-con. TUSC5 expression
levels were significantly elevated by the miR-1307-3p inhibitor in
comparison with the miR-con (Fig.
3C). Moreover, it was shown that the TUSC5 expression levels
were decreased in the CRC cell lines compared with the normal cell
line (Fig. 3D).
Knockdown of TUSC5 attenuates the
effects of miR-1307-3p on CRC cells
To further confirm the role of TUSC5 in
miR-1307-3p-mediated stimulation of CRC cells, rescue experiments
were performed. It was demonstrated that the transfection of
si-TUSC5 decreased TUSC5 expression levels in CRC cells (Fig. 4A). si-TUSC5 also appeared to
partially reverse the effects of miR-1307-3p inhibitor on TUSC5
expression (Figs. 3C and 4A). It was also found that the knockdown of
TUSC5 appeared to reduce the effects of miR-1307-3p inhibitor on
CRC cell proliferation as indicated at 72 h (Figs. 2B and 4B). Similar results were observed in
Transwell invasion assays. Although not directly compared, TUSC5
downregulation appeared to decrease the impact of the miR-1307-3p
inhibitor on CRC cell invasion capacity (Figs. 2C and 4C).
 | Figure 4Knockdown of TUSC5 restores the
effects of miR-1307-3p on CRC cell behavior. (A) Expression of
TUSC5, (B) Cell proliferation and (C) Cell invasion (magnification,
x200) in CRC cells transfected with si-TUSC5, siR-con or si-TUSC5 +
miR-1307-3p inhibitor. *P<0.05,
**P<0.01, ***P< 0.01 vs. siR-con. con,
control; CRC, colorectal cancer; siR-con, negative control small
interfering RNA; miR, microRNA; OD, optical density; si-TUSC5,
small interfering RNA targeting tumor suppressor candidate 5;
TUSC5, tumor suppressor candidate 5. |
Discussion
The mechanisms of action underlying CRC progression
have been gradually explored in recent years (15). Numerous miRNAs have been identified
to be abnormally expressed in CRC and function as either tumor
drivers or suppressors (5,6). Hence, it is essential to fully
investigate abnormally expressed molecules associated with CRC
carcinogenesis to discover novel biomarkers for prognosis or
treatment (16).
In this present study, miR-1307-3p expression levels
were revealed to be significantly upregulated in CRC tissues and
cell lines compared with normal tissues and cell lines,
respectively. Suppressing miR-1307-3p in CRC cells inhibited CRC
cell proliferation and invasion in vitro. As a tumor-driver
miRNA, miR-1307-3p expression levels were upregulated in several
cancer types, including breast cancer and hepatocellular carcinoma
(7,8). Consistent with previous studies, the
current work indicated that miR-1307-3p functions as an oncogenic
miRNA to promote CRC progression.
Previous studies have indicated that miR-1307-3p
influences cancer progression via regulating the expression of
SMYD4 and DAB2 interacting protein (7,8). In the
present study, it was demonstrated that TUSC5 was a putative target
for miR-1307-3p using TargetScan. In this present study, knockdown
of TUSC5 promoted CRC cell proliferation and invasion; and may have
partially reversed the effects of miR-1307-3p inhibitor on the
malignant phenotypes of CRC cells. However, the mechanisms of
action underlying miR-1307-3p-mediated regulation of TUSC5
expression, and the associated mechanisms in CRC remain unclear and
require further exploration. In addition, it has to be recognized
that the primary limitation of the present work is that the
miR-1307-3p/TUSC5 axis was not validated in a xenograft tumor
model. In the future work, to address this issue, CRC cells could
be inoculated in nude mice and then the synthesized miRNAs injected
into the developing tumors at varying time points. Subsequently,
the tumor volume and tumor weight as well as markers of metastasis,
including ki-67, N-Cadherin and vimentin should be examined to
investigate the effects of miR-654-5p on tumor growth and
metastasis in vivo.
In conclusion, miR-1307-3p was upregulated in CRC
cells and regulated CRC cell proliferation and invasion through
regulating TUSC5. The present study provided evidence that
miR-1307-3p may be therapeutic target for the treatment of CRC.
Supplementary Material
Figure S1. Knockdown of miR.1307-3p
inhibits cell proliferation. (A) miR.1307-3p expression levels and
(B) cell proliferation in NCM460 cells transfected with miR.1307-3p
inhibitor or miR.con. ***P<0.01 vs. miR.con.
miR.1307-3p: microRNA.1307-3p; miR.con: Negative control miRNA; OD,
optical density.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NY, MY, RZ and MW performed the experiments,
analyzed the data and wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Popat S, Hubner R and Houlston RS:
Systematic review of microsatellite instability and colorectal
cancer prognosis. J Clin Oncol. 23:609–618. 2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang C, Liu J, Xu L, Hu W, Wang J, Wang M
and Yao X: MicroRNA-17 promotes cell proliferation and migration in
human colorectal cancer by downregulating SIK1. Cancer Manag Res.
11:3521–3534. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ding X, Du J, Mao K, Wang X, Ding Y and
Wang F: MicroRNA-143-3p suppresses tumorigenesis by targeting
catenin-δ1 in colorectal cancer. Onco Targets Ther. 12:3255–3265.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Han S, Zou H, Lee JW, Han J, Kim HC, Cheol
JJ, Kim LS and Kim H: miR-1307-3p stimulates breast cancer
development and progression by targeting SMYD4. J Cancer.
10:441–448. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen S, Wang L, Yao B, Liu Q and Guo C:
miR-1307-3p promotes tumor growth and metastasis of hepatocellular
carcinoma by repressing DAB2 interacting protein. Biomed
Pharmacother. 117(109055)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Koide H, Shibata T, Yamada N, Asaki T,
Nagao T, Yoshida T, Noguchi Y, Tanaka T, Saito Y and Tatsuno I:
Tumor suppressor candidate 5 (TUSC5) is expressed in brown
adipocytes. Biochem Biophys Res Commun. 360:139–145.
2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Oort PJ, Warden CH, Baumann TK, Knotts TA
and Adams SH: Characterization of Tusc5, an adipocyte gene
co-expressed in peripheral neurons. Mol Cell Endocrinol. 276:24–35.
2007.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen X and Chen J: miR-3188 regulates cell
proliferation, apoptosis, and migration in breast cancer by
targeting TUSC5 and regulating the p38 MAPK signaling pathway.
Oncol Res. 26:363–372. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang S, Wang W, Han X, Wang Y, Ge Y and
Tan Z: Dysregulation of miR484-TUSC5 axis takes part in the
progression of hepatocellular carcinoma. J Biochem. 166:271–279.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Moreno EC, Pascual A, Prieto-Cuadra D,
Laza VF, Molina-Cerrillo J, Ramos-Muñoz ME, Rodríguez-Serrano EM,
Soto JL, Carrato A, García-Bermejo ML and Guillén-Ponce C: Novel
molecular characterization of colorectal primary tumors based on
miRNAs. Cancers (Basel). 11(E346)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak JK and Schmittgen TD: Analysis of
relative gene expression data using quantitative PCR and the
2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Brody H: Colorectal cancer. Nature.
521(S1)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Strubberg AM and Madison BB: MicroRNAs in
the etiology of colorectal cancer: Pathways and clinical
implications. Dis Model Mech. 10:197–214. 2017.PubMed/NCBI View Article : Google Scholar
|