Introduction
Esophageal carcinoma is one of the most common
malignancies of the digestive system and the sixth leading cause of
cancer-associated mortality worldwide (1). Esophageal carcinoma includes
esophageal squamous cell carcinoma (ESCC) and esophageal
adenocarcinoma, of which ESCC is the most common pathological type
in Asia (2,3). Currently, the treatment strategies for
ESCC include surgery, radiotherapy, and chemotherapy (4,5).
Despite advancements in these techniques, the 5-year survival rate
of patients with ESCC was <20% in 2012 (6,7). Thus,
it is important to identify effective biomarkers for the diagnosis
and treatment of patients with ESCC (8,9). In
recent years, research on non-coding RNAs in the field of molecular
biology has attracted great interest due to its association with
tumors (10,11).
MicroRNAs (miRNAs/miRs) are a class of highly
conserved non-coding RNA molecules, which negatively regulate gene
expression by binding to the 3'-untranslated region of target
mRNAs, resulting in mRNA degradation and inhibition of translation
(12,13). Increasing evidence suggest that
miRNAs play key roles in gene expression regulation and signal
transduction pathways in cancer (14,15).
It has been reported that downregulated miR-378 expression exerts a
tumor suppressive role in non-small cell lung cancer (NSCLC), which
provides an innovative and candidate target for the diagnosis and
treatment of patients with NSCLC (16). Yang et al (17) comprehensively analyzed the role of
miRNAs and mRNAs in ESCC, and microarray analysis demonstrated that
miR-378 expression was abnormally downregulated in ESCC. However,
the prognostic value of miR-378 and its regulatory effect on tumor
progression in ESCC remain unclear.
With the aim of exploring effects of miR-378 on
ESCC, the present study determined the expression levels of miR-378
in ESCC tissues and cell lines, and then analyzed the correlation
between its expression and clinical parameters as well as the
prognosis value for ESCC. Preliminarily, the molecular mechanism of
action of miR-378 on ESCC was investigated.
Materials and methods
Patients and tissue samples
A total of 135 patients with ESCC at Yidu Central
Hospital were enrolled in the present study between June 2013 and
June 2015. The mean age for all patients was 62 years old with a
range of 38-72 years; males slightly predominated (54%). Included
cases were histopathologically confirmed to have ESCC and all
patients agreed to participate in the study. Patients who had
previously received any radiation or chemotherapy for ESCC prior to
surgery were excluded from the present study. Other eligibility
criteria were as following: ≥18 years of age; and no history of
concurrent cancer in other organs or history of previous cancer in
any organ. Pathological tissue samples and paired normal tissue
samples (5 cm away from the tumor tissues) were collected via
surgical resection. The fresh specimens were immediately frozen in
liquid nitrogen and stored at -80˚C until subsequent
experimentation. Patients were classified according to the seventh
edition of TNM staging criteria for malignant tumors, as revised by
the International Union against Cancer and the American Cancer
Federation in 2009(18). The
present study was approved by the Medical Ethics Committee of Yidu
Central Hospital of Weifang (Weifang, China; approval no. 201210)
and written informed consent was provided by all patients prior to
the study start. Survival analysis was performed via a 5-year
telephone follow-up study (from the date of surgery treatment to
death or the last observation).
Cell culture and transfection
The ESCC cell lines TE-1, KYSE-150, Eca-109 and TE-8
were purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences, while the human esophageal epithelial
cell line, Het-1A, was purchased from the American Type Culture
Collection. All cells were maintained in RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), at 37˚C
with 5% CO2. miR-378 mimic (50 nM; 5'-CUC
CUGACUCCAGGUCCUGUGU-3'), mimic negative control (NC, 50 nM;
5'-UCACAACCUCCUAGAAAGAGUAGA-3'), miR-378 inhibitor (50 nM;
5'-ACACAGGACCUGGAGUCA GGAG -3') and inhibitor NC (50 nM;
5'-UUCUCCGAACGU GUCACGUTT-3') were synthesized by Guangzhou RiboBio
Co., Ltd. 20 nM of miR-378 mimic or inhibitor transfection mix was
prepared in Lipofectamine® 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. The transfection mix and 1.5x105 cells
were seeded in medium in the same 6-well plates at 37˚C. Following
24 h, the medium along with the transfection reagent was replaced
with fresh medium. After another 24 h of medium replacement at
37˚C, the cells were harvested for the subsequent
experimentation.
RNA reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from ESCC tissues and cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and the RNA quality and quantity were verified
using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.). Total RNA
was reverse transcribed into cDNA using the TaqMan miRNA reverse
transcription kit (Thermo Fisher Scientific, Inc.) using the
following temperature protocol: 16˚C holding for 30 min, 42˚C for
30 min, 85˚C for 5 min and then 4˚C. qPCR was subsequently
performed using the SYBR-Green I Master mix kit (Invitrogen; Thermo
Fisher Scientific, Inc.) on an ABI 7500 system. The amplification
of U6 small nuclear RNA was used for the normalization. The primers
used in the present study were as follows: U6 forwards,
5'-CGCTTCACGAATTTGCGTGTCAT-3' and reverse
5'-GCTTCGGCAGCACATATACTAAAAT-3'; and miR-378-5p forwards,
CAAACCTCCTCCTGACTCCAG and reverse, TATGCTTGTTCTCGTCTCTGTGTC. The
PCR conditions were as follows: 95˚C for 30 sec; and 40 cycles of
95˚C for 5 sec and 60˚C for 30 sec and 72˚C for 20 sec. Relative
expression levels were calculated using the 2-ΔΔCq
method (19). All experiments were
performed in triplicate.
Cell proliferation assay
Of the four ESCC cell lines, miR-378 expression was
significantly lower in TE-1 and KYSE-150 cells compared with
Eca-109 and TE-8 cells. Thus, the TE-1 and KYSE-150 cell lines were
selected for subsequent experimentation. Transfected TE-1 and
KYSE-150 cells were centrifuged at 100 x g for 5 min at 4˚C.
Thereafter, the cell suspension was prepared in RPMI-1640 medium
containing 10% FBS and then seeded into 96-well plates at a density
of 2x103 cells per well. Subsequently, 10 µl Cell
Counting Kit-8 (CCK-8) reagent (Dojindo Molecular Technologies,
Inc.) was added to each well and incubated for 1-2 h at 37˚C, with
5% CO2. Cell proliferation was analyzed at a wavelength
of 450 nm, using a microplate reader system (Molecular Devices
LLC).
Cell migration and invasion
assays
TE-1 and KYSE-150 cells were starved for 24 hours in
a serum-free medium before invasion or migration experiments. A
total of 2x105 transfected ESCC cells were plated in the
upper chambers of Transwell plates (8 µm pores; BD Biosciences),
and finally, 200 µl serum-free RPMI-1640 was added to the upper
chamber. At 37˚C, the cells were cultured for 24 h in an incubator
with 5% CO2. For the invasion assay, Transwell membranes
were precoated with Matrigel. RPMI-1640 medium (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS was plated in
the lower chambers. Cells were fixed with 4% paraformaldehyde
solution for 20 min at room temperature and subsequently stained
with 0.1% crystal violet for 20 min at room temperature. Stained
cells were counted in five randomly selected fields using an
Olympus IX-70 fluorescence microscope at 200x magnification.
Statistical analysis
Statistical analysis was performed using SPSS 21.0
software (IBM Corp.) and GraphPad Prism 5.0 software (GraphPad
Software, Inc.). All experiments were performed in triplicate and
data are presented as the mean ± standard deviation. Paired
Student's t-test was used to compare differences between two
groups, while one-way ANOVA and Tukey's post-hoc tests were used to
compare differences between multiple groups. χ2 test was
used to compare the association between miR-378 expression and
clinic data of patients. Survival analysis was performed using the
Kaplan-Meier method and compared with the use of log-rank test to
determine the statistical significance. Univariate and multivariate
Cox regression analysis was performed to determine the prognostic
value of miR-378 in ESCC. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-378 expression in ESCC tissues and
cell lines
RT-qPCR analysis was performed to detect miR-378
expression in ESCC tissues and cell lines. The results demonstrated
that miR-378 expression was significantly lower in ESCC tissues
(0.567±0.321) compared with adjacent normal tissues (1.02±0.326;
P<0.001; Fig. 1A). Similarly,
miR-378 expression was significantly lower in the four ESCC cell
lines compared with the normal cell line, particularly in KYSE-150
and TE-1 cells (P<0.01, P<0.001, Fig. 1B). Notably, miR-378 expression was
three times higher in Het-1A cells compared with KYSE-150 cells,
and twice as high compared with TE-1 cells.
Association between miR-378 expression
and the clinicopathological characteristics of patients with
ESCC
The median relative expression of miR-378 was used
as the threshold (20,21). All patients were divided into two
groups, the miR-378 high expression group (n=67) and the miR-378
low expression group (n=68). The results demonstrated that miR-378
expression was significantly associated with the TNM stage
(P=0.001) and lymph node metastasis (P=0.01) of patients with ESCC.
However, no significant associations were observed between miR-378
expression and age, sex, smoking, drinking or tumor differentiation
degree among patients with ESCC (P>0.05, Table I).
 | Table IAssociation between miR-378 expression
and the clinicopathological characteristics of patients with
esophageal squamous cell carcinoma (n=135). |
Table I
Association between miR-378 expression
and the clinicopathological characteristics of patients with
esophageal squamous cell carcinoma (n=135).
| | miR-378
expression | |
|---|
| Characteristic | Number of
patients | Low (n=68) | High (n=67) | P-value |
|---|
| Age, years | | | | 0.793 |
|
<65 | 64 | 33 | 31 | |
|
≥65 | 71 | 35 | 36 | |
| Sex | | | | 0.790 |
|
Male | 73 | 36 | 37 | |
|
Female | 62 | 32 | 30 | |
| Smoking | | | | 0.550 |
|
No | 82 | 43 | 39 | |
|
Yes | 53 | 25 | 28 | |
| Drinking | | | | 0.673 |
|
No | 77 | 40 | 37 | |
|
Yes | 58 | 28 | 30 | |
| Differentiation
degree | | | | 0.788 |
|
Middle/High | 60 | 31 | 29 | |
|
Low | 75 | 37 | 38 | |
| TNM stage | | | | 0.001 |
|
Ⅰ/II | 102 | 43 | 59 | |
|
III | 33 | 25 | 8 | |
| Lymph node
metastasis | | | | 0.010 |
|
Negative | 95 | 41 | 54 | |
|
Positive | 40 | 27 | 13 | |
miR-378 expression is associated with
survival outcomes of patients with ESCC
Survival analysis was performed using the
Kaplan-Meier method and log-rank test, based on miR-378 expression.
The results demonstrated that patients with low miR-378 expression
had a significantly shorter overall survival time (P=0.026;
Fig. 2) than those with high
miR-378 expression. Furthermore, univariate and multivariate Cox
regression analysis indicated that miR-378 expression [P=0.031;
hazard ratio (HR), 2.516], TNM staging (P=0.043; HR, 2.801) and
lymph node metastasis (P=0.036; HR, 0.426) are independent
prognostic factors for overall survival in patients with ESCC
(Table II).
 | Table IIMultivariate Cox regression analysis
for overall survival in patients with esophageal squamous cell
carcinoma. |
Table II
Multivariate Cox regression analysis
for overall survival in patients with esophageal squamous cell
carcinoma.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| MicroRNA-378
expression | 2.124 | 1.053-4.283 | 0.035 | 2.516 | 1.088-5.817 | 0.031 |
| Age | 1.031 | 0.544-1.952 | 0.926 | 1.546 | 0.756-3.160 | 0.232 |
| Sex | 0.892 | 0.468-1.700 | 0.729 | 0.507 | 0.239-1.078 | 0.078 |
| Smoking | 1.089 | 0.568-2.089 | 0.797 | 1.075 | 0.544-2.126 | 0.835 |
| Drinking | 1.373 | 0.710-2.656 | 0.347 | 1.575 | 0.795-3.121 | 0.193 |
| Differentiation
degree | 1.329 | 0.703-2.514 | 0.382 | 1.027 | 0.501-2.107 | 0.942 |
| TNM stage | 1.612 | 0.709-3.662 | 0.254 | 2.801 | 1.031-7.606 | 0.043 |
| Lymph node
metastasis | 0.393 | 0.208-0.745 | 0.004 | 0.426 | 0.191-0.947 | 0.036 |
Downregulation of miR-378 expression
promotes cell proliferation, migration and invasion
Following transfection with miR-378 mimics, mimic
NC, miR-378 inhibitors and inhibitor NC, miR-378 expression in TE-1
and KYSE-150 cells was detected via RT-qPCR analysis. The results
demonstrated that transfection with miR-378 mimic significantly
increased miR-378 expression, the effects of which were reversed
following transfection with miR-378 inhibitor (P<0.01, Fig. 3A). The CCK-8 assay was performed to
assess cell proliferation. The results demonstrated that
overexpression of miR-378 inhibited the proliferation of ESCC
cells, while miR-378 knockdown promoted the proliferation of ESCC
cells compared with the control groups (P<0.05, Fig. 3B).
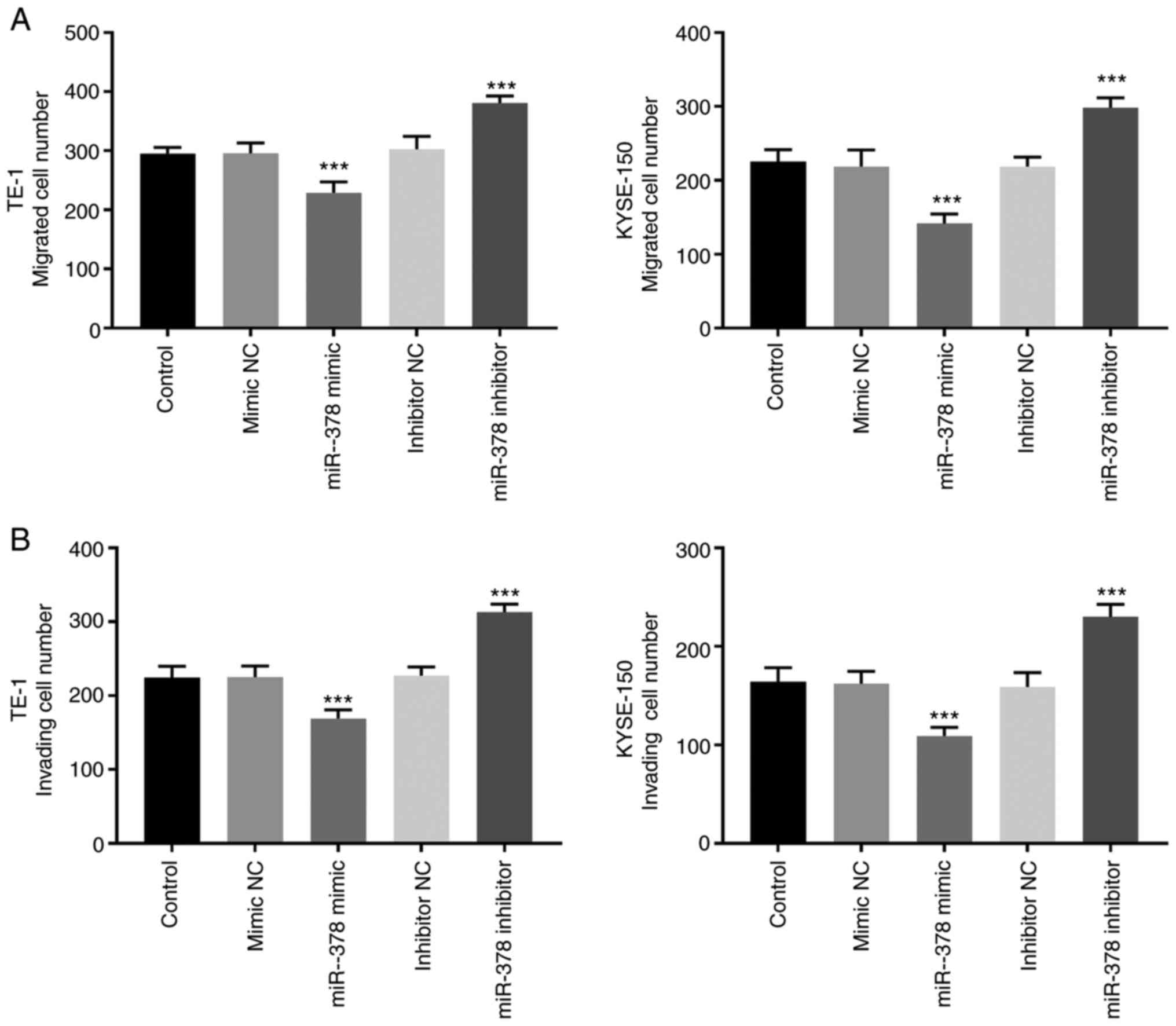
The results of the Transwell assay demonstrated that
the number of migratory and invasive cells significantly decreased
in TE-1 and KYSE-150 cells following transfection with miR-378
mimic, the effects of which were reversed following transfection
with miR-378 inhibitor (P<0.01; Fig.
4A and B).
Discussion
miRNAs can regulate the expression of target genes
at the post-transcriptional level (22-24).
It has been confirmed that approximately one third of genes in the
human genome are regulated by miRNAs (25). Several studies have demonstrated
that miRNAs play important roles in the occurrence and development
of tumors, functioning as either oncogenes or tumor suppressors in
different types of cancer (13,26,27).
ESCC is a common malignancy worldwide, which is a major threat to
human health (5,28). Given that the molecular mechanisms
underlying the occurrence and progression of esophageal carcinoma
are not yet fully understood, the prognosis of patients remains
poor (4,29,30).
Thus, it is important to identify effective molecular targets and
novel therapeutic strategies for patients with esophageal carcinoma
(31,32).
Previous studies have reported that miR-378 plays an
important role in different types of cancer. For example, Li et
al (33) demonstrated that
miR-378 expression is significantly lower in glioma tissues
compared with non-neoplastic brain tissues, and downregulated
miR-378 expression is associated with tumor invasiveness and poor
prognosis of patients with glioma. In colorectal cancer, miR-378
expression is significantly downregulated in colorectal cancer
tissues and cell lines, and low miR-378 expression predicts a
shorter overall survival acting as an independent prognostic factor
(34).
The results of the present study demonstrated an
association between miR-378 expression and the progression of ESCC.
The results indicated that miR-378 expression was significantly
downregulated in ESCC tissues compared with normal adjacent
tissues, suggesting that miR-378 may inhibit ESCC progression. In
tumors, TNM stage represents the degree of tumor development, and
lymph node metastasis is an important factor affecting the survival
of patients. In the present study, patients with low miR-378
expression exhibited an advanced TNM stage and positive lymph node
metastasis. In addition, Kaplan-Meier survival analysis and
multivariate Cox regression analysis demonstrated that patients
with low miR-378 expression had a poor prognosis. Taken together,
these results suggest that miR-378 may serve as a potential
prognostic marker for ESCC.
Zeng et al (35) reported that miR-378 expression is
downregulated in colon cancer tissues and cell lines, and that
overexpression of miR-378 inhibits the proliferation, migration and
invasion of colon cancer cells. Furtherly, cell proliferative,
migratory and invasive abilities are usually associated with the
development of tumors (36). Thus,
the present study assessed the effect of altering miR-378
expression on the biological behaviors of ESCC cells via cell
transfection. The results demonstrated that miR-378 knockdown
promoted the proliferation, migration and invasion of ESCC cells,
while overexpression of miR-378 inhibited these cellular behaviors
compared with untreated ESCC cells. Collectively, these results
suggest that miR-378 plays an inhibitory role in ESCC.
miR-378 has been reported to exert similar
inhibitory effects in other types of cancer, for instance, low
ectopic miR-378 expression inhibits the proliferation, migration
and invasion of MGC-803 gastric cancer cells, suggesting that
miR-378 exerts an anticancer role in gastric cancer (37). This may provide novel diagnostic and
therapeutic options for the future clinical management of human
gastric cancer (37). Taken
together, the results of the present study are consistent with
previous findings (35,38,39).
The present study is not without limitations, such
as, the sample size used was too small or the downstream target
genes of miR-378 have not yet been fully investigated. Thus,
further studies are required to confirm the results presented
here.
In conclusion, the results of the present study
demonstrated that miR-378 expression was downregulated in ESCC
tissues and cell lines, which was associated with a poor prognosis
of patients with ESCC. Notably, miR-378 may act as a tumor
suppressor in the occurrence and development of ESCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WJ, LW and HL conceived and designed the present
study. All authors performed the experiments. WJ, LW and SC
analyzed and interpreted the data. WJ and LW drafted the initial
manuscript. HL critically revised the manuscript for important
intellectual content. All authors have read and approved the final
manuscript. WJ, LW and HL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Yidu Central Hospital of Weifang (Weifang, China,
approval no. 201210) and written informed consent was provided by
all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Alsina M, Moehler M and Lorenzen S:
Immunotherapy of esophageal cancer: Current status, many trials and
innovative strategies. Oncol Res Treat. 41:266–271. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Dong Z, Wang J, Zhan T and Xu S:
Identification of prognostic risk factors for esophageal
adenocarcinoma using bioinformatics analysis. Onco Targets Ther.
11:4327–4337. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hou J, Zou K, Yang C, Leng X and Xu Y:
Clinicopathological and prognostic significance of circulating
tumor cells in patients with esophageal cancer: A meta-analysis.
Onco Targets Ther. 11:8053–8061. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hirano H and Kato K: Systemic treatment of
advanced esophageal squamous cell carcinoma: Chemotherapy,
molecular-targeting therapy and immunotherapy. Jpn J Clin Oncol.
49:412–420. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Reichenbach ZW, Murray MG, Saxena R,
Farkas D, Karassik EG, Klochkova A, Patel K, Tice C, Hall TM, Gang
J, et al: Clinical and translational advances in esophageal
squamous cell carcinoma. Adv Cancer Res. 144:95–135.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dong Z, Wang J, Zhang H, Zhan T, Chen Y
and Xu S: Identification of potential key genes in esophageal
adenocarcinoma using bioinformatics. Exp Ther Med. 18:3291–3298.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jamali L, Tofigh R, Tutunchi S, Panahi G,
Borhani F, Akhavan S, Nourmohammadi P, Ghaderian SMH, Rasouli M and
Mirzaei H: Circulating microRNAs as diagnostic and therapeutic
biomarkers in gastric and esophageal cancers. J Cell Physiol.
233:8538–8550. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li Y, Huang HC, Chen LQ, Xu LY, Li EM and
Zhang JJ: Predictive biomarkers for response of esophageal cancer
to chemo(radio)therapy: A systematic review and meta-analysis. Surg
Oncol. 26:460–472. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang Z, Song Q, Yang S, Zeng R, Li X,
Jiang C, Ding W, Zhang J and Zheng Y: Serum microRNA-218 is a
potential biomarker for esophageal cancer. Cancer Biomark.
15:381–389. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mei LL, Qiu YT, Zhang B and Shi ZZ:
MicroRNAs in esophageal squamous cell carcinoma: Potential
biomarkers and therapeutic targets. Cancer Biomark. 19:1–9.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Correia de Sousa M, Gjorgjieva M, Dolicka
D, Sobolewski C and Foti M: Deciphering miRNAs' action through
miRNA editing. Int J Mol Sci. 20(20)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mishra S, Yadav T and Rani V: Exploring
miRNA based approaches in cancer diagnostics and therapeutics. Crit
Rev Oncol Hematol. 98:12–23. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kang M, Li Y, Zhu S, Zhang S, Guo S and Li
P: MicroRNA-193b acts as a tumor suppressor gene in human
esophageal squamous cell carcinoma via target regulation of KRAS.
Oncol Lett. 17:3965–3973. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Xia D, Tian S, Chen Z, Qin W and Liu Q:
miR-302a inhibits the proliferation of esophageal cancer cells
through the MAPK and PI3K/Akt signaling pathways. Oncol Lett.
15:3937–3943. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ji KX, Cui F, Qu D, Sun RY, Sun P, Chen
FY, Wang SL and Sun HS: miR-378 promotes the cell proliferation of
non-small cell lung cancer by inhibiting FOXG1. Eur Rev Med
Pharmacol Sci. 22:1011–1019. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yang H, Su H, Hu N, Wang C, Wang L, Giffen
C, Goldstein AM, Lee MP and Taylor PR: Integrated analysis of
genome-wide miRNAs and targeted gene expression in esophageal
squamous cell carcinoma (ESCC) and relation to prognosis. BMC
Cancer. 20(388)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Edge S, Byrd DR, Compton CC, Fritz AG,
Greene F and Trotti A (eds): AJCC Cancer Staging Manual. 7th
Edition. Springer-Verlag, New York, NY, 2010.
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Goto T, Fujiya M, Konishi H, Sasajima J,
Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, et
al: An elevated expression of serum exosomal microRNA-191, - 21,
-451a of pancreatic neoplasm is considered to be efficient
diagnostic marker. BMC Cancer. 18(116)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Nie X, Su Z, Yan R, Yan A, Qiu S and Zhou
Y: MicroRNA-562 negatively regulated c-MET/AKT pathway in the
growth of glioblastoma cells. Onco Targets Ther. 12:41–49.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tomaselli S, Panera N, Gallo A and Alisi
A: Circulating miRNA profiling to identify biomarkers of
dysmetabolism. Biomarkers Med. 6:729–742. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang C, Ji Q, Yang Y, Li Q and Wang Z:
Exosome: Function and role in cancer metastasis and drug
resistance. Technol Cancer Res Treat.
17(1533033818763450)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vishnoi A and Rani S: MiRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Codipilly DC, Qin Y, Dawsey SM, Kisiel J,
Topazian M, Ahlquist D and Iyer PG: Screening for esophageal
squamous cell carcinoma: Recent advances. Gastrointest Endosc.
88:413–426. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fu JH: Biomarkers of predicting response
to neoadjuvant chemoradiotherapy in esophageal cancer. Zhonghua Wei
Chang Wai Ke Za Zhi. 16:805–810. 2013.PubMed/NCBI(In Chinese).
|
|
30
|
Hou X, Wen J, Ren Z and Zhang G:
Non-coding RNAs: New biomarkers and therapeutic targets for
esophageal cancer. Oncotarget. 8:43571–43578. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Su X, Gao C, Feng X and Jiang M: miR-613
suppresses migration and invasion in esophageal squamous cell
carcinoma via the targeting of G6PD. Exp Ther Med. 19:3081–3089.
2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yi Y, Lu X, Chen J, Jiao C, Zhong J, Song
Z, Yu X and Lin B: Downregulated miR-486-5p acts as a tumor
suppressor in esophageal squamous cell carcinoma. Exp Ther Med.
12:3411–3416. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li B, Wang Y, Li S, He H, Sun F, Wang C,
Lu Y, Wang X and Tao B: Decreased expression of miR-378 correlates
with tumor invasiveness and poor prognosis of patients with glioma.
Int J Clin Exp Pathol. 8:7016–7021. 2015.PubMed/NCBI
|
|
34
|
Zhang GJ, Zhou H, Xiao HX, Li Y and Zhou
T: miR-378 is an independent prognostic factor and inhibits cell
growth and invasion in colorectal cancer. BMC Cancer.
14(109)2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zeng M, Zhu L, Li L and Kang C: miR-378
suppresses the proliferation, migration and invasion of colon
cancer cells by inhibiting SDAD1. Cell Mol Biol Lett.
22(12)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Nie X, Xia F, Liu Y, Zhou Y, Ye W, Hean P,
Meng J, Liu H, Liu L, Wen J, et al: Downregulation of Wnt3
suppresses colorectal cancer development through inhibiting cell
proliferation and migration. Front Pharmacol.
10(1110)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fei B and Wu H: MiR-378 inhibits
progression of human gastric cancer MGC-803 cells by targeting
MAPK1 in vitro. Oncol Res. 20:557–564. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Guo XB, Zhang XC, Chen P, Ma LM and Shen
ZQ: miR-378a-3p inhibits cellular proliferation and migration in
glioblastoma multiforme by targeting tetraspanin 17. Oncol Rep.
42:1957–1971. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Cui Z, Sun S, Liu Q, Zhou X, Gao S, Peng P
and Li Q: MicroRNA-378-3p/5p suppresses the migration and
invasiveness of oral squamous carcinoma cells by inhibiting KLK4
expression. Biochem Cell Biol. 98:154–163. 2020.PubMed/NCBI View Article : Google Scholar
|