|
1
|
Rits S, Olsen BR and Volloch V:
Protein-encoding RNA to RNA information transfer in mammalian
cells: RNA-dependent mRNA amplification. Identification of chimeric
RNA intermediates and putative RNA end products. Ann Integr Mol
Med. 1:23–47. 2019.PubMed/NCBI
|
|
2
|
Howie H, Rijal CM and Ressler KJ: A review
of epigenetic contributions to post-traumatic stress disorder.
Dialogues Clin Neurosci. 21:417–428. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Holt CE and Schuman EM: The central dogma
decentralized: New perspectives on RNA function and local
translation in neurons. Neuron. 80:648–657. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Maydanovych O and Beal PA: Breaking the
central dogma by RNA editing. Chem Rev. 106:3397–3411.
2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Thakur P, Estevez M, Lobue PA, Limbach PA
and Addepalli B: Improved RNA modification mapping of cellular
non-coding RNAs using C- and U-specific RNases. Analyst.
145:816–827. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu J, Dou X, Chen C, Chen C, Liu C, Xu
MM, Zhao S, Shen B, Gao Y, Han D and He C:
N6-methyladenosine of chromosome-associated regulatory
RNA regulates chromatin state and transcription. Science.
367:580–586. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13(117)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shinoda K, Suda A, Otonari K, Futaki S and
Imanishi M: Programmable RNA methylation and demethylation using
PUF RNA binding proteins. Chem Commun (Camb). 56:1365–1368.
2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen X, Chen X, Zhou Z, Mao Y, Wang Y, Ma
Z, Xu W, Qin A and Zhang S: Nirogacestat suppresses RANKL-Induced
osteoclast formation in vitro and attenuates LPS-Induced bone
resorption in vivo. Exp Cell Res. 382(111470)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
The RNA methyltransferase METTL3 promotes
oncogene translation. Cancer Discov. 6(572)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chen Y, Peng C, Chen J, Chen D, Yang B, He
B, Hu W, Zhang Y, Liu H, Dai L, et al: WTAP facilitates progression
of hepatocellular carcinoma via m6A-HuR-dependent epigenetic
silencing of ETS1. Mol Cancer. 18(127)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li J, Zhu L, Shi Y, Liu J, Lin L and Chen
X: m6A demethylase FTO promotes hepatocellular carcinoma
tumorigenesis via mediating PKM2 demethylation. Am J Transl Res.
11:6084–6092. 2019.PubMed/NCBI
|
|
17
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Parker MT, Knop K, Sherwood AV, Schurch
NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ and Simpson GG:
Nanopore direct RNA sequencing maps the complexity of Arabidopsis
mRNA processing and m6A modification. eLife.
9(e49658)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liao S, Sun H and Xu C: YTH Domain: A
family of N6-methyladenosine (m6A) readers.
Genomics Proteomics Bioinformatics. 16:99–107. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao YL, Liu YH, Wu RF, Bi Z, Yao YX, Liu
Q, Wang YZ and Wang XX: Understanding m6A function
through uncovering the diversity roles of YTH domain-containing
proteins. Mol Biotechnol. 61:355–364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li F, Kennedy S, Hajian T, Gibson E,
Seitova A, Xu C, Arrowsmith CH and Vedadi M: A radioactivity-based
assay for screening human m6A-RNA methyltransferase, METTL3-METTL14
complex, and demethylase ALKBH5. J Biomol Screen. 21:290–297.
2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu Y and Santi DV: m5C RNA and m5C DNA
methyl transferases use different cysteine residues as catalysts.
Proc Natl Acad Sci USA. 97:8263–8265. 2000.PubMed/NCBI View Article : Google Scholar
|
|
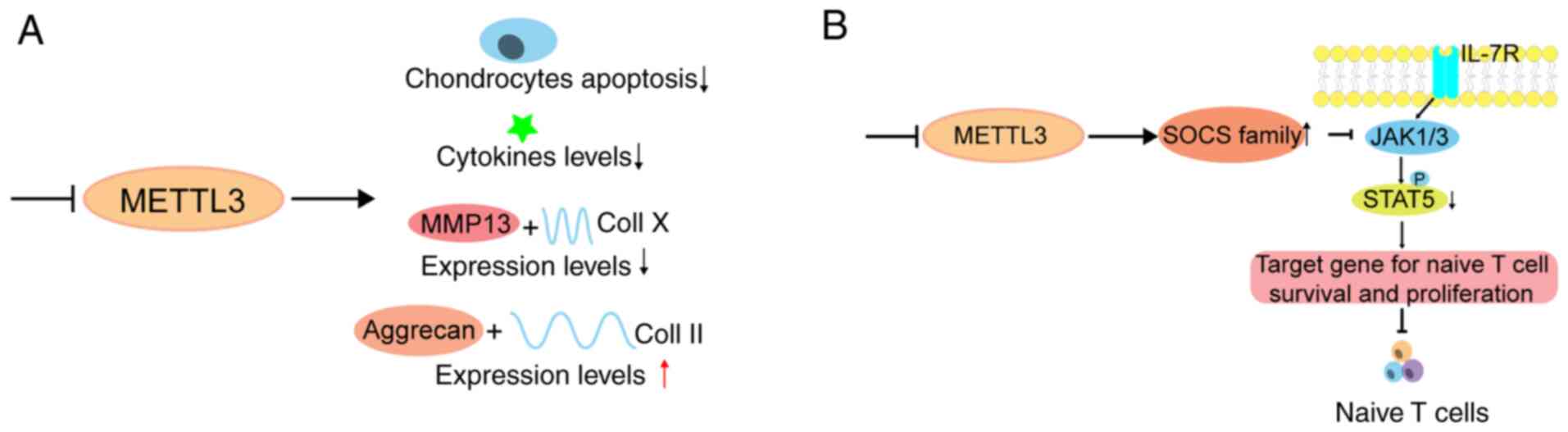
23
|
Roovers M, Wouters J, Bujnicki JM, Tricot
C, Stalon V, Grosjean H and Droogmans L: A primordial RNA
modification enzyme: The case of tRNA (m1A) methyltransferase.
Nucleic Scids Res. 32:465–476. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Grabowski P: Physiology of bone. Endocr
Dev. 16:32–48. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
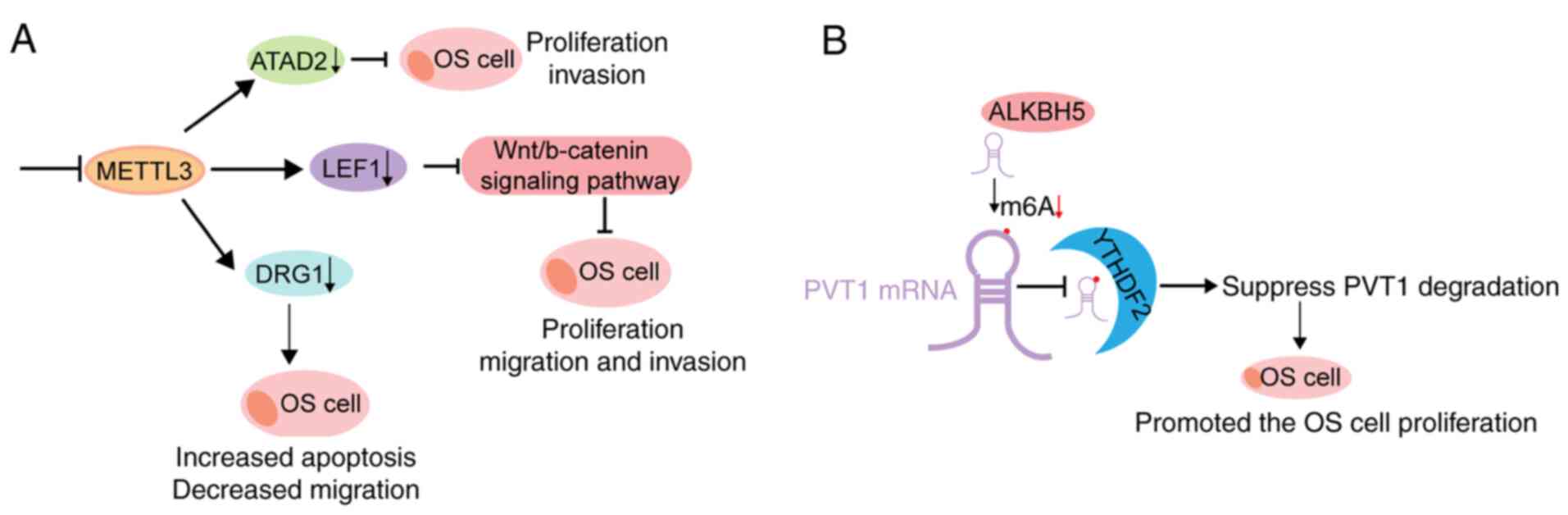
|
Scholtysek C, Kronke G and Schett G:
Inflammation-associated changes in bone homeostasis. Inflamm
Allergy Drug Targets. 11:188–195. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Suominen H: Muscle training for bone
strength. Aging Clin Exp Res. 18:85–93. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Fu R, Lv WC, Xu Y, Gong MY, Chen XJ, Jiang
N, Xu Y, Yao QQ, Di L, Lu T, et al: Endothelial ZEB1 promotes
angiogenesis-dependent bone formation and reverses osteoporosis.
Nat Commun. 11(460)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Landete-Castillejos T, Kierdorf H, Gomez
S, Luna S, García AJ, Cappelli J, Pérez-Serrano M, Pérez-Barbería
J, Gallego L and Kierdorf U: Antlers-Evolution, development,
structure, composition, and biomechanics of an outstanding type of
bone. Bone. 128(115046)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hassan MQ, Tye CE, Stein GS and Lian JB:
Non-coding RNAs: Epigenetic regulators of bone development and
homeostasis. Bone. 81:746–756. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Bocheva G and Boyadjieva N: Epigenetic
regulation of fetal bone development and placental transfer of
nutrients: Progress for osteoporosis. Interdiscip Toxicol.
4:167–172. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Adamik J, Roodman GD and Galson DL:
Epigenetic-based mechanisms of osteoblast suppression in multiple
myeloma bone disease. JBMR Plus. 3(e10183)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Marini F, Cianferotti L and Brandi ML:
Epigenetic mechanisms in bone biology and osteoporosis: Can they
drive therapeutic choices? Int J Mol Sci. 17(1329)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ghayor C and Weber FE: Epigenetic
regulation of bone remodeling and its impacts in osteoporosis. Int
J Mol Sci. 17(1446)2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Montecino M, Stein G, Stein J, Zaidi K and
Aguilar R: Multiple levels of epigenetic control for bone biology
and pathology. Bone. 81:733–738. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kobayashi M, Ohsugi M, Sasako T, Awazawa
M, Umehara T, Iwane A, Kobayashi N, Okazaki Y, Kubota N, Suzuki R,
et al: The RNA methyltransferase complex of WTAP, METTL3, and
METTL14 regulates mitotic clonal expansion in adipogenesis. Mol
Cell Biol. 38:e00116–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou L, Yang C, Zhang N, Zhang X, Zhao T
and Yu J: Silencing METTL3 inhibits the proliferation and invasion
of osteosarcoma by regulating ATAD2. Biomed Pharmacother.
125(109964)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bujnicki JM, Feder M, Radlinska M and
Blumenthal RM: Structure prediction and phylogenetic analysis of a
functionally diverse family of proteins homologous to the MT-A70
subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol.
55:431–444. 2002.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Scholler E, Weichmann F, Treiber T, Ringle
S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G:
Interactions, localization, and phosphorylation of the
m6A generating METTL3-METTL14-WTAP complex. RNA.
24:499–512. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lence T, Paolantoni C, Worpenberg L and
Roignant JY: Mechanistic insights into m6A RNA enzymes.
Biochim Biophys Acta Gene Regul Mech. 1862:222–229. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Haussmann IU, Bodi Z, Sanchez-Moran E,
Mongan NP, Archer N, Fray RG and Soller M: m6A
potentiates Sxl alternative pre-mRNA splicing for robust drosophila
sex determination. Nature. 540:301–304. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Alarcon CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Meyer KD and Jaffrey SR: Rethinking
m6A readers, writers, and erasers. Annu Rev Cell Dev
Biol. 33:319–342. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Geula S, Moshitch-Moshkovitz S,
Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V,
Peer E, Mor N, Manor YS, et al: Stem cells. m6A mRNA methylation
facilitates resolution of naive pluripotency toward
differentiation. Science. 347:1002–1006. 2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hongay CF and Orr-Weaver TL: Drosophila
Inducer of MEiosis 4 (IME4) is required for Notch signaling during
oogenesis. Proc Natl Acad Sci USA. 108:14855–14860. 2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zhong S, Li H, Bodi Z, Button J, Vespa L,
Herzog M and Fray RG: MTA is an Arabidopsis messenger RNA adenosine
methylase and interacts with a homolog of a sex-specific splicing
factor. Plant Cell. 20:1278–1288. 2008.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Clancy MJ, Shambaugh ME, Timpte CS and
Bokar JA: Induction of sporulation in Saccharomyces cerevisiae
leads to the formation of N6-methyladenosine in mRNA: A potential
mechanism for the activity of the IME4 gene. Nucleic Acids Res.
30:4509–4518. 2002.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu
HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor
proliferation of bladder cancer by accelerating pri-miR221/222
maturation in m6A-dependent manner. Mol Cancer.
18(110)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Choe J, Lin S, Zhang W, Liu Q, Wang L,
Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al: mRNA
circularization by METTL3-eIF3h enhances translation and promotes
oncogenesis. Nature. 561:556–560. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X,
Ren F, Cui G and Sun R: Expression patterns and prognostic value of
m6A-related genes in colorectal cancer. Am J Transl Res.
11:3972–3991. 2019.PubMed/NCBI
|
|
54
|
Visvanathan A, Patil V, Arora A, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: Essential role of
METTL3-mediated m6A modification in glioma stem-like
cells maintenance and radioresistance. Oncogene. 37:522–533.
2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wang H, Xu B and Shi J: N6-methyladenosine
METTL3 promotes the breast cancer progression via targeting Bcl-2.
Gene. 722(144076)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme
METTL3 controls myeloid differentiation of normal hematopoietic and
leukemia cells. Nat Med. 23:1369–1376. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Lin S, Liu J, Jiang W, Wang P, Sun C, Wang
X, Chen Y and Wang H: METTL3 promotes the proliferation and
mobility of gastric cancer cells. Open Med (Wars). 14:25–31.
2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dahal U, Le K and Gupta M: RNA m6A
methyltransferase METTL3 regulates invasiveness of melanoma cells
by matrix metallopeptidase 2. Melanoma Res. 29:382–389.
2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zheng W, Dong X, Zhao Y, Wang S, Jiang H,
Zhang M, Zheng X and Gu M: Multiple functions and mechanisms
underlying the role of METTL3 in Human Cancers. Front Oncol.
9(1403)2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wu L, Wu D, Ning J, Liu W and Zhang D:
Changes of N6-methyladenosine modulators promote breast cancer
progression. BMC Cancer. 19(326)2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Li X, Tang J, Huang W, Wang F, Li P, Qin
C, Qin Z, Zou Q, Wei J, Hua L, et al: The M6A methyltransferase
METTL3: Acting as a tumor suppressor in renal cell carcinoma.
Oncotarget. 8:96103–96116. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wei W, Huo B and Shi X: miR-600 inhibits
lung cancer via downregulating the expression of METTL3. Cancer
Manag Res. 11:1177–1187. 2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhang C, Zhang M, Ge S, Huang W, Lin X,
Gao J, Gong J and Shen L: Reduced m6A modification predicts
malignant phenotypes and augmented Wnt/PI3K-Akt signaling in
gastric cancer. Cancer Med. 8:4766–4781. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Iyer LM, Zhang D and Aravind L: Adenine
methylation in eukaryotes: Apprehending the complex evolutionary
history and functional potential of an epigenetic modification.
Bioessays. 38:27–40. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Liu X, Qin J, Gao T, Li C, Chen X, Zeng K,
Xu M, He B, Pan B, Xu X, et al: Analysis of METTL3 and METTL14 in
hepatocellular carcinoma. Aging (Albany NY). 12:21638–21659.
2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Buker SM, Gurard-Levin ZA, Wheeler BD,
Scholle MD, Case AW, Hirsch JL, Ribich S, Copeland RA and
Boriack-Sjodin PA: A mass spectrometric assay of METTL3/METTL14
methyltransferase activity. SLAS Discov. 25:361–371.
2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B,
Li C, Sun L, Qin J, Xu T, et al: METTL14-mediated
N6-methyladenosine modification of SOX4 mRNA inhibits tumor
metastasis in colorectal cancer. Mol Cancer. 19(106)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Gong PJ, Shao YC, Yang Y, Song WJ, He X,
Zeng YF, Huang SR, Wei L and Zhang JW: Analysis of
N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as
tumor suppressor genes in breast cancer. Front Oncol.
10(578963)2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y,
Wang Y, Yang J and Tian F: Mettl14 inhibits bladder TIC
self-renewal and bladder tumorigenesis through
N6-methyladenosine of Notch1. Mol Cancer.
18(168)2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Weng H, Huang H, Wu H, Qin X, Zhao BS,
Dong L, Shi H, Skibbe J, Shen C, Hu C, et al: METTL14 inhibits
hematopoietic stem/progenitor differentiation and promotes
leukemogenesis via mRNA m6A modification. Cell Stem
Cell. 22:191–205 e9. 2018.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5' sites. Cell
Rep. 8:284–296. 2014.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Sorci M, Ianniello Z, Cruciani S, Larivera
S, Ginistrelli LC, Capuano E, Marchioni M, Fazi F and Fatica A:
METTL3 regulates WTAP protein homeostasis. Cell Death Dis.
9(796)2018.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Li H, Su Q, Li B, Lan L, Wang C, Li W,
Wang G, Chen W, He Y and Zhang C: High expression of WTAP leads to
poor prognosis of gastric cancer by influencing tumour-associated T
lymphocyte infiltration. J Cell Mol Med. 24:4452–4465.
2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Bansal H, Yihua Q, Iyer SP, Ganapathy S,
Proia DA, Penalva LO, Uren PJ, Suresh U, Carew JS, Karnad AB, et
al: WTAP is a novel oncogenic protein in acute myeloid leukemia.
Leukemia. 28:1171–1174. 2014.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Horiuchi K, Umetani M, Minami T, Okayama
H, Takada S, Yamamoto M, Aburatani H, Reid PC, Housman DE, Hamakubo
T and Kodama T: Wilms' tumor 1-associating protein regulates G2/M
transition through stabilization of cyclin A2 mRNA. Proc Natl Acad
Sci USA. 103:17278–17283. 2006.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Wen J, Lv R, Ma H, Shen H, He C, Wang J,
Jiao F, Liu H, Yang P, Tan L, et al: Zc3h13 regulates nuclear RNA
m6A methylation and mouse embryonic stem cell
self-renewal. Mol Cell. 69:1028–1038 e6. 2018.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Zhu D, Zhou J, Zhao J, Jiang G, Zhang X,
Zhang Y and Dong M: ZC3H13 suppresses colorectal cancer
proliferation and invasion via inactivating Ras-ERK signaling. J
Cell Physiol. 234:8899–8907. 2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Chen J, Yu K, Zhong G and Shen W:
Identification of a m6A RNA methylation regulators-based
signature for predicting the prognosis of clear cell renal
carcinoma. Cancer Cell Int. 20(157)2020.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Liu T, Li C, Jin L, Li C and Wang L: The
prognostic value of m6A RNA methylation regulators in colon
adenocarcinoma. Med Sci Monit. 25:9435–9445. 2019.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m(6)A mRNA methylation directs translational
control of heat shock response. Nature. 526:591–594.
2015.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Dina C, Meyre D, Gallina S, Durand E,
Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et
al: Variation in FTO contributes to childhood obesity and severe
adult obesity. Nat Genet. 39:724–726. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
84
|
Frayling TM, Timpson NJ, Weedon MN,
Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H,
Rayner NW, et al: A common variant in the FTO gene is associated
with body mass index and predisposes to childhood and adult
obesity. Science. 316:889–894. 2007.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun
L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine
demethylase FTO promotes breast tumor progression through
inhibiting BNIP3. Mol Cancer. 18(46)2019.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C,
Huang H, Nachtergaele S, Dong L, Hu C, et al: FTO plays an
oncogenic role in acute myeloid leukemia as a
N6-Methyladenosine RNA demethylase. Cancer Cell.
31:127–141. 2017.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Yang S, Wei J, Cui YH, Park G, Shah P,
Deng Y, Aplin AE, Lu Z, Hwang S, He C and He YY: m6A
mRNA demethylase FTO regulates melanoma tumorigenicity and response
to anti-PD-1 blockade. Nat Commun. 10(2782)2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Li J, Han Y, Zhang H, Qian Z, Jia W, Gao
Y, Zheng H and Li B: The m6A demethylase FTO promotes the growth of
lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem
Biophys Res Commun. 512:479–485. 2019.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Baltz AG, Munschauer M, Schwanhausser B,
Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew
K, Milek M, et al: The mRNA-bound proteome and its global occupancy
profile on protein-coding transcripts. Mol Cell. 46:674–690.
2012.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Fedeles BI, Singh V, Delaney JC, Li D and
Essigmann JM: The AlkB family of Fe(II)/α-Ketoglutarate-dependent
dioxygenases: Repairing nucleic acid alkylation damage and beyond.
J Biol Chem. 290:20734–20742. 2015.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Pilzys T, Marcinkowski M, Kukwa W, Garbicz
D, Dylewska M, Ferenc K, Mieczkowski A, Kukwa A, Migacz E, Wołosz
D, et al: ALKBH overexpression in head and neck cancer: Potential
target for novel anticancer therapy. Sci Rep.
9(13249)2019.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang
Y, Feng Y, Pan Q and Wan R: RNA demethylase ALKBH5 prevents
pancreatic cancer progression by posttranscriptional activation of
PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer.
19(91)2020.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Shen C, Sheng Y, Zhu AC, Robinson S, Jiang
X, Dong L, Chen H, Su R, Yin Z, Li W, et al: RNA demethylase ALKBH5
selectively promotes tumorigenesis and cancer stem cell
self-renewal in acute myeloid leukemia. Cell Stem Cell.
27:64–80.e9. 2020.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A demethylase
ALKBH5 maintains tumorigenicity of glioblastoma Stem-like cells by
sustaining FOXM1 expression and cell proliferation program. Cancer
Cell. 31:591–606.e6. 2017.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Zhu Z, Qian Q, Zhao X, Ma L and Chen P:
N6-methyladenosine ALKBH5 promotes non-small cell lung
cancer progress by regulating TIMP3 stability. Gene.
731(144348)2020.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Zhang J, Guo S, Piao HY, Wang Y, Wu Y,
Meng XY, Yang D, Zheng ZC and Zhao Y: ALKBH5 promotes invasion and
metastasis of gastric cancer by decreasing methylation of the
lncRNA NEAT1. J Physiol Biochem. 75:379–389. 2019.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Ueda Y, Ooshio I, Fusamae Y, Kitae K,
Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K and Tsujikawa
K: AlkB homolog 3-mediated tRNA demethylation promotes protein
synthesis in cancer cells. Sci Rep. 7(42271)2017.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Mohan M, Akula D, Dhillon A, Goyal A and
Anindya R: Human RAD51 paralogue RAD51C fosters repair of alkylated
DNA by interacting with the ALKBH3 demethylase. Nucleic Acids Res.
47:11729–11745. 2019.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J,
Lu Z, Zheng Z, Dai Q and Wang H: Transfer RNA demethylase ALKBH3
promotes cancer progression via induction of tRNA-derived small
RNAs. Nucleic Acids Res. 47:2533–2545. 2019.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306.
2014.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Yue Y, Liu J and He C: RNA
N6-methyladenosine methylation in post-transcriptional gene
expression regulation. Genes Dev. 29:1343–1355. 2015.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Zhu T, Roundtree IA, Wang P, Wang X, Wang
L, Sun C, Tian Y, Li J, He C and Xu Y: Crystal structure of the YTH
domain of YTHDF2 reveals mechanism for recognition of
N6-methyladenosine. Cell Res. 24:1493–1496. 2014.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Lee Y, Choe J, Park OH and Kim YK:
Molecular mechanisms driving mRNA degradation by m6A
modification. Trends Genet. 36:177–188. 2020.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z,
Perez SP, Suganthan R, He C, Bjørås M and Klungland A:
Ythdf2-mediated m6A mRNA clearance modulates neural
development in mice. Genome Biol. 19(69)2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X,
Liu B, Xiao F, Wang W, Huang G, et al: Loss of YTHDF2-mediated
m6A-dependent mRNA clearance facilitates hematopoietic
stem cell regeneration. Cell Res. 28:1035–1038. 2018.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Huang T, Liu Z, Zheng Y, Feng T, Gao Q and
Zeng W: YTHDF2 promotes spermagonial adhesion through modulating
MMPs decay via m6A/mRNA pathway. Cell Death Dis.
11(37)2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5'UTR
m(6)A promotes cap-independent translation. Cell. 163:999–1010.
2015.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Hu L, Wang J, Huang H, Yu Y, Ding J, Yu Y,
Li K, Wei D, Ye Q, Wang F, et al: YTHDF1 regulates pulmonary
hypertension through translational control of MAGED1. Am J Respir
Crit Care Med. 203:1158–1172. 2021.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu
Z, Li J, Hao P, Zhang Y, Zhang F, et al: m6A facilitates
hippocampus-dependent learning and memory through YTHDF1. Nature.
563:249–253. 2018.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Nishizawa Y, Konno M, Asai A, Koseki J,
Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai
D, et al: Oncogene c-Myc promotes epitranscriptome m6A
reader YTHDF1 expression in colorectal cancer. Oncotarget.
9:7476–7486. 2018.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Zhao X, Chen Y, Mao Q, Jiang X, Jiang W,
Chen J, Xu W, Zhong L and Sun X: Overexpression of YTHDF1 is
associated with poor prognosis in patients with hepatocellular
carcinoma. Cancer Biomark. 21:859–868. 2018.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Zhang Y, Wang X, Zhang X, Wang J, Ma Y,
Zhang L and Cao X: RNA-binding protein YTHDF3 suppresses
interferon-dependent antiviral responses by promoting FOXO3
translation. Proc Natl Acad Sci USA. 116:976–981. 2019.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou
A, Liu J, Che L and Li J: Long noncoding RNA GAS5 inhibits
progression of colorectal cancer by interacting with and triggering
YAP phosphorylation and degradation and is negatively regulated by
the m6A reader YTHDF3. Mol Cancer.
18(143)2019.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Jurczyszak D, Zhang W, Terry SN, Kehrer T,
Bermúdez González MC, McGregor E, Mulder LCF, Eckwahl MJ, Pan T and
Simon V: HIV protease cleaves the antiviral m6A reader protein
YTHDF3 in the viral particle. PLoS Pathog.
16(e1008305)2020.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519.
2016.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Luxton HJ, Simpson BS, Mills IG, Brindle
NR, Ahmed Z, Stavrinides V, Heavey S, Stamm S and Whitaker HC: The
oncogene metadherin interacts with the known splicing proteins
YTHDC1, Sam68 and T-STAR and plays a novel role in alternative mRNA
splicing. Cancers (Basel). 11(1233)2019.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu
Y, Gregory BD, Schultz RM and Wang PJ: Nuclear m6A reader YTHDC1
regulates alternative polyadenylation and splicing during mouse
oocyte development. PLoS Genet. 14(e1007412)2018.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B
and Qian SB: m6A in mRNA coding regions promotes
translation via the RNA helicase-containing YTHDC2. Nat Commun.
10(5332)2019.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Tanabe A, Tanikawa K, Tsunetomi M, Takai
K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, et al:
RNA helicase YTHDC2 promotes cancer metastasis via the enhancement
of the efficiency by which HIF-1α mRNA is translated. Cancer Lett.
376:34–42. 2016.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Kretschmer J, Rao H, Hackert P, Sloan KE,
Hobartner C and Bohnsack MT: The m6A reader protein
YTHDC2 interacts with the small ribosomal subunit and the 5'-3'
exoribonuclease XRN1. RNA. 24:1339–1350. 2018.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Nakano M, Ondo K, Takemoto S, Fukami T and
Nakajima M: Methylation of adenosine at the N6 position
post-transcriptionally regulates hepatic P450s expression. Biochem
Pharmacol. 171(113697)2020.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Bailey AS, Batista PJ, Gold RS, Chen YG,
de Rooij DG, Chang HY and Fuller MT: The conserved RNA helicase
YTHDC2 regulates the transition from proliferation to
differentiation in the germline. eLife. 6(e26116)2017.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Tanabe A, Konno J, Tanikawa K and Sahara
H: Transcriptional machinery of TNF-α-inducible YTH domain
containing 2 (YTHDC2) gene. Gene. 535:24–32. 2014.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Alarcon CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of
m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308.
2015.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295.
2018.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Srivastava M and Deal C: Osteoporosis in
elderly: Prevention and treatment. Clin Geriatr Med. 18:529–555.
2002.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Langdahl BL: Overview of treatment
approaches to osteoporosis. Br J Pharmacol. 178:1891–1906.
2021.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Rosen CJ and Bouxsein ML: Mechanisms of
disease: Is osteoporosis the obesity of bone? Nat Clin Pract
Rheumatol. 2:35–43. 2006.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Raisz LG: Pathogenesis of osteoporosis:
Concepts, conflicts, and prospects. J Clin Invest. 115:3318–3325.
2005.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Palmieri D, Valli M, Viglio S, Ferrari N,
Ledda B, Volta C and Manduca P: Osteoblasts extracellular matrix
induces vessel like structures through glycosylated collagen I. Exp
Cell Res. 316:789–799. 2010.PubMed/NCBI View Article : Google Scholar
|
|
132
|
DeNichilo MO, Shoubridge AJ, Panagopoulos
V, Liapis V, Zysk A, Zinonos I, Hay S, Atkins GJ, Findlay DM and
Evdokiou A: Peroxidase enzymes regulate collagen biosynthesis and
matrix mineralization by cultured human osteoblasts. Calcif Tissue
Int. 98:294–305. 2016.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Long F: Building strong bones: Molecular
regulation of the osteoblast lineage. Nat Rev Mol Cell Biol.
13:27–38. 2011.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Boyle WJ, Simonet WS and Lacey DL:
Osteoclast differentiation and activation. Nature. 423:337–342.
2003.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Villaseñor A, Aedo-Martín D, Obeso D,
Erjavec I, Rodríguez-Coira J, Buendía I, Ardura JA, Barbas C and
Gortazar AR: Metabolomics reveals citric acid secretion in
mechanically-stimulated osteocytes is inhibited by high glucose.
Sci Rep. 9(2295)2019.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Dallas SL, Prideaux M and Bonewald LF: The
osteocyte: An endocrine cell... and more. Endocr Rev. 34:658–690.
2013.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Chen Q, Shou P, Zheng C, Jiang M, Cao G,
Yang Q, Cao J, Xie N, Velletri T, Zhang X, et al: Fate decision of
mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death
Differ. 23:1128–1139. 2016.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Kawai M, Devlin MJ and Rosen CJ: Fat
targets for skeletal health. Nat Rev Rheumatol. 5:365–372.
2009.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Rosen CJ, Ackert-Bicknell C, Rodriguez JP
and Pino AM: Marrow fat and the bone microenvironment:
Developmental, functional, and pathological implications. Crit Rev
Eukaryot Gene Expr. 19:109–124. 2009.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Scheller EL and Rosen CJ: What's the
matter with MAT? Marrow adipose tissue, metabolism, and skeletal
health. Ann N Y Acad Sci. 1311:14–30. 2014.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Garcia-Gomez MC and Vilahur G:
Osteoporosis and vascular calcification: A shared scenario. Clin
Investig Arterioscler. 32:33–42. 2020.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Chen X, Hua W, Huang X, Chen Y, Zhang J
and Li G: Regulatory role of RNA N6-methyladenosine
modification in bone biology and osteoporosis. Front Endocrinol
(Lausanne). 10(911)2019.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang
Y, Li J, Sheng R, Deng P, Wang Y, et al: Mettl3-mediated
m6A RNA methylation regulates the fate of bone marrow
mesenchymal stem cells and osteoporosis. Nat Commun.
9(4772)2018.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Yu J, Shen L, Liu Y, Ming H, Zhu X, Chu M
and Lin J: The m6A methyltransferase METTL3 cooperates with
demethylase ALKBH5 to regulate osteogenic differentiation through
NF-κB signaling. Mol Cell Biochem. 463:203–210. 2020.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Tian C, Huang Y, Li Q, Feng Z and Xu Q:
Mettl3 regulates osteogenic differentiation and alternative
splicing of vegfa in bone marrow mesenchymal stem cells. Int J Mol
Sci. 20(551)2019.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Busilacchi A, Gigante A, Mattioli-Belmonte
M, Manzotti S and Muzzarelli RA: Chitosan stabilizes platelet
growth factors and modulates stem cell differentiation toward
tissue regeneration. Carbohydr Polym. 98:665–676. 2013.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Hu K and Olsen BR: Osteoblast-derived VEGF
regulates osteoblast differentiation and bone formation during bone
repair. J Clin Invest. 126:509–526. 2016.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Yan G, Yuan Y, He M, Gong R, Lei H, Zhou
H, Wang W, Du W, Ma T, Liu S, et al: m6A methylation of
precursor-miR-320/RUNX2 controls osteogenic potential of bone
marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids.
19:421–436. 2020.PubMed/NCBI View Article : Google Scholar
|
|
149
|
Li D, Cai L, Meng R, Feng Z and Xu Q:
METTL3 modulates osteoclast differentiation and function by
controlling RNA stability and nuclear export. Int J Mol Sci.
21(1660)2020.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Gerken T, Girard CA, Tung YC, Webby CJ,
Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill
LA, et al: The obesity-associated FTO gene encodes a
2-oxoglutarate-dependent nucleic acid demethylase. Science.
318:1469–1472. 2007.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Eyre DR: Bone biomarkers as tools in
osteoporosis management. Spine (Phila Pa 1976). 22 (24
Suppl):17S–24S. 1997.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Takada I, Kouzmenko AP and Kato S: Wnt and
PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev
Rheumatol. 5:442–447. 2009.PubMed/NCBI View Article : Google Scholar
|
|
153
|
Shen GS, Zhou HB, Zhang H, Chen B, Liu ZP,
Yuan Y, Zhou XZ and Xu YJ: The GDF11-FTO-PPARγ axis controls the
shift of osteoporotic MSC fate to adipocyte and inhibits bone
formation during osteoporosis. Biochim Biophys Acta Mol Basis Dis.
1864:3644–3654. 2018.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Zhang Q, Riddle RC, Yang Q, Rosen CR,
Guttridge DC, Dirckx N, Faugere MC, Farber CR and Clemens TL: The
RNA demethylase FTO is required for maintenance of bone mass and
functions to protect osteoblasts from genotoxic damage. Proc Natl
Acad Sci USA. 116:17980–17989. 2019.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Guo Y, Liu H, Yang TL, Li SM, Li SK, Tian
Q, Liu YJ and Deng HW: The fat mass and obesity associated gene,
FTO, is also associated with osteoporosis phenotypes. PLoS One.
6(e27312)2011.PubMed/NCBI View Article : Google Scholar
|
|
156
|
Li Y, Yang F, Gao M, Gong R, Jin M, Liu T,
Sun Y, Fu Y, Huang Q, Zhang W, et al: miR-149-3p regulates the
switch between adipogenic and osteogenic differentiation of BMSCs
by targeting FTO. Mol Ther Nucleic Acids. 17:590–600.
2019.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Mannerstrom B, Kornilov R, Abu-Shahba AG,
Chowdhury IM, Sinha S, Seppänen-Kaijansinkko R and Kaur S:
Epigenetic alterations in mesenchymal stem cells by
osteosarcoma-derived extracellular vesicles. Epigenetics.
14:352–364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Lorenzo J: Cytokines and bone:
Osteoimmunology. Handb Exp Pharmacol. 262:177–230. 2020.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Zhang Y, Gu X, Li D, Cai L and Xu Q:
METTL3 regulates osteoblast differentiation and inflammatory
response via Smad signaling and MAPK Signaling. Int J Mol Sci.
21(199)2019.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Yu R, Li Q, Feng Z, Cai L and Xu Q: m6A
reader YTHDF2 regulates LPS-induced inflammatory response. Int J
Mol Sci. 20(1323)2019.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Wang T and He C: TNF-α and IL-6: The link
between immune and bone system. Curr Drug Targets. 20:213–227.
2020.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Mathew AJ and Ravindran V: Infections and
arthritis. Best Pract Res Clin Rheumatol. 28:935–959.
2014.PubMed/NCBI View Article : Google Scholar
|
|
163
|
Harth M and Nielson WR: Pain and affective
distress in arthritis: Relationship to immunity and inflammation.
Expert Rev Clin Immunol. 15:541–552. 2019.PubMed/NCBI View Article : Google Scholar
|
|
164
|
Parkinson L, Waters DL and Franck L:
Systematic review of the impact of osteoarthritis on health
outcomes for comorbid disease in older people. Osteoarthritis
Cartilage. 25:1751–1770. 2017.PubMed/NCBI View Article : Google Scholar
|
|
165
|
Glyn-Jones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015.PubMed/NCBI View Article : Google Scholar
|
|
166
|
Abramoff B and Caldera FE: Osteoarthritis:
Pathology, diagnosis, and treatment options. Med Clin North Am.
104:293–311. 2020.PubMed/NCBI View Article : Google Scholar
|
|
167
|
Liossis SN and Tsokos GC: Cellular
immunity in osteoarthritis: Novel concepts for an old disease. Clin
Diagn Lab Immunol. 5:427–429. 1998.PubMed/NCBI View Article : Google Scholar
|
|
168
|
Sakata M, Tsuruha JI, Masuko-Hongo K,
Nakamura H, Matsui T, Sudo A, Nishioka K and Kato T: Autoantibodies
to osteopontin in patients with osteoarthritis and rheumatoid
arthritis. J Rheumatol. 28:1492–1495. 2001.PubMed/NCBI
|
|
169
|
Walker J, Gordon T, Lester S, Downie-Doyle
S, McEvoy D, Pile K, Waterman S and Rischmueller M: Increased
severity of lower urinary tract symptoms and daytime somnolence in
primary Sjogren's syndrome. J Rheumatol. 30:2406–2412.
2003.PubMed/NCBI
|
|
170
|
Kato T, Xiang Y, Nakamura H and Nishioka
K: Neoantigens in osteoarthritic cartilage. Curr Opin Rheumatol.
16:604–608. 2004.PubMed/NCBI View Article : Google Scholar
|
|
171
|
Zhao W, Wang T, Luo Q, Chen Y, Leung VY,
Wen C, Shah MF, Pan H, Chiu K, Cao X and Lu WW: Cartilage
degeneration and excessive subchondral bone formation in
spontaneous osteoarthritis involves altered TGF-β signaling. J
Orthop Res. 34:763–770. 2016.PubMed/NCBI View Article : Google Scholar
|
|
172
|
Liu Q, Li M, Jiang L, Jiang R and Fu B:
METTL3 promotes experimental osteoarthritis development by
regulating inflammatory response and apoptosis in chondrocyte.
Biochem Biophys Res Commun. 516:22–27. 2019.PubMed/NCBI View Article : Google Scholar
|
|
173
|
Goldring SR and Goldring MB: The role of
cytokines in cartilage matrix degeneration in osteoarthritis. Clin
Orthop Relat Res. (427 Suppl):S27–S36. 2004.PubMed/NCBI View Article : Google Scholar
|
|
174
|
Yang F, Zhou S, Wang C, Huang Y, Li H,
Wang Y, Zhu Z, Tang J and Yan M: Epigenetic modifications of
interleukin-6 in synovial fibroblasts from osteoarthritis patients.
Sci Rep. 7(43592)2017.PubMed/NCBI View Article : Google Scholar
|
|
175
|
Guo Q, Wang Y, Xu D, Nossent J, Pavlos NJ
and Xu J: Rheumatoid arthritis: Pathological mechanisms and modern
pharmacologic therapies. Bone Res. 6(15)2018.PubMed/NCBI View Article : Google Scholar
|
|
176
|
Li HB, Tong J, Zhu S, Batista PJ, Duffy
EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al:
m6A mRNA methylation controls T cell homeostasis by
targeting the IL-7/STAT5/SOCS pathways. Nature. 548:338–342.
2017.PubMed/NCBI View Article : Google Scholar
|
|
177
|
Wang J, Yan S, Lu H, Wang S and Xu D:
METTL3 attenuates LPS-induced inflammatory response in macrophages
via NF-κB signaling pathway. Mediators Inflamm.
2019(3120391)2019.PubMed/NCBI View Article : Google Scholar
|
|
178
|
Mo XB, Zhang YH and Lei SF: Genome-wide
identification of N6-methyladenosine (m6A)
SNPs associated with rheumatoid arthritis. Front Genet.
9(299)2018.PubMed/NCBI View Article : Google Scholar
|
|
179
|
Zheng Y, Nie P, Peng D, He Z, Liu M, Xie
Y, Miao Y, Zuo Z and Ren J: m6AVar: A database of functional
variants involved in m6A modification. Nucleic Acids Res.
46:D139–D145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
180
|
Smolen JS, Aletaha D, Barton A, Burmester
GR, Emery P, Firestein GS, Kavanaugh A, McInnes IB, Solomon DH,
Strand V and Yamamoto K: Rheumatoid arthritis. Nat Rev Dis Primers.
4(18001)2018.PubMed/NCBI View Article : Google Scholar
|
|
181
|
Kondo Y, Yokosawa M, Kaneko S, Furuyama K,
Segawa S, Tsuboi H, Matsumoto I and Sumida T: Review:
Transcriptional regulation of CD4+ T cell differentiation in
experimentally induced arthritis and rheumatoid arthritis.
Arthritis Rheumatol. 70:653–661. 2018.PubMed/NCBI View Article : Google Scholar
|
|
182
|
Noack M and Miossec P: Th17 and regulatory
T cell balance in autoimmune and inflammatory diseases. Autoimmun
Rev. 13:668–677. 2014.PubMed/NCBI View Article : Google Scholar
|
|
183
|
Hunt L, Hensor EM, Nam J, Burska AN,
Parmar R, Emery P and Ponchel F: T cell subsets: An immunological
biomarker to predict progression to clinical arthritis in
ACPA-positive individuals. Ann Rheum Dis. 75:1884–1889.
2016.PubMed/NCBI View Article : Google Scholar
|
|
184
|
Kumar BV, Connors TJ and Farber DL: Human
T cell development, localization, and function throughout life.
Immunity. 48:202–213. 2018.PubMed/NCBI View Article : Google Scholar
|
|
185
|
Abada A and Elazar Z: Getting ready for
building: Signaling and autophagosome biogenesis. Embo Rep.
15:839–852. 2014.PubMed/NCBI View Article : Google Scholar
|
|
186
|
Wang DW, Wu LW, Cao Y, Yang L, Liu W, E
XQ, Ji G and Bi ZG: A novel mechanism of mTORC1-mediated
serine/glycine metabolism in osteosarcoma development. Cell Signal.
29:107–114. 2017.PubMed/NCBI View Article : Google Scholar
|
|
187
|
Miao W, Chen J, Jia L, Ma J and Song D:
The m6A methyltransferase METTL3 promotes osteosarcoma progression
by regulating the m6A level of LEF1. Biochem Biophys Res Commun.
516:719–725. 2019.PubMed/NCBI View Article : Google Scholar
|
|
188
|
Nguyen DX, Chiang AC, Zhang XH, Kim JY,
Kris MG, Ladanyi M, Gerald WL and Massagué J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009.PubMed/NCBI View Article : Google Scholar
|
|
189
|
Jia P, Wei G, Zhou C, Gao Q, Wu Y, Sun X
and Li X: Upregulation of miR-212 inhibits migration and
tumorigenicity and inactivates Wnt/β-catenin signaling in human
hepatocellular carcinoma. Technol Cancer Res Treat.
17(1533034618765221)2018.PubMed/NCBI View Article : Google Scholar
|
|
190
|
Wu L, Zhao JC, Kim J, Jin HJ, Wang CY and
Yu J: ERG is a critical regulator of Wnt/LEF1 signaling in prostate
cancer. Cancer Res. 73:6068–6079. 2013.PubMed/NCBI View Article : Google Scholar
|
|
191
|
Ling Z, Chen L and Zhao J: m6A-dependent
up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth,
migration, and colony formation in osteosarcoma. Biosci Rep.
40(BSR20200282)2020.PubMed/NCBI View Article : Google Scholar
|
|
192
|
Coker H, Wei G and Brockdorff N: m6A
modification of non-coding RNA and the control of mammalian gene
expression. Biochim Biophys Acta Gene Regul Mech. 1862:310–318.
2019.PubMed/NCBI View Article : Google Scholar
|
|
193
|
Li J, Rao B, Yang J, Liu L, Huang M, Liu
X, Cui G, Li C, Han Q, Yang H, et al: Dysregulated m6A-related
regulators are associated with tumor metastasis and poor prognosis
in osteosarcoma. Front Oncol. 10(769)2020.PubMed/NCBI View Article : Google Scholar
|
|
194
|
Fan D, Xia Y, Lu C, Ye Q, Xi X, Wang Q,
Wang Z, Wang C and Xiao C: Regulatory role of the RNA
N6-methyladenosine modification in immunoregulatory
cells and immune-related bone homeostasis associated with
rheumatoid arthritis. Front Cell Dev Biol. 8(627893)2020.PubMed/NCBI View Article : Google Scholar
|
|
195
|
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y,
Cheng C, Li L, Pi J, Si Y, et al: The m6A reader YTHDF1 promotes
ovarian cancer progression via augmenting EIF3C translation.
Nucleic Acids Res. 48:3816–3831. 2020.PubMed/NCBI View Article : Google Scholar
|
|
196
|
Ma Z and Ji J: N6-methyladenosine (m6A)
RNA modification in cancer stem cells. Stem Cells: Sep 27, 2020
(Epub Ahead of Print).
|
|
197
|
Patil DP, Pickering BF and Jaffrey SR:
Reading m6A in the transcriptome: m6A-binding
proteins. Trends Cell Biol. 28:113–127. 2018.PubMed/NCBI View Article : Google Scholar
|
|
198
|
Dai DJ, Wang HY, Zhu LY, Jin HC and Wang
X: N6-methyladenosine links RNA metabolism to cancer
progression. Cell Death Dis. 9(124)2018.PubMed/NCBI View Article : Google Scholar
|
|
199
|
Wang HF, Kuang MJ, Han SJ, Wang AB, Qiu J,
Wang F, Tan BY and Wang DC: BMP2 modified by the m6A
demethylation enzyme ALKBH5 in the ossification of the ligamentum
flavum through the AKT signaling pathway. Calcified Tissue Int.
106:486–493. 2020.PubMed/NCBI View Article : Google Scholar
|
|
200
|
Zhang W, He L, Liu Z, Ren X, Qi L, Wan L,
Wang W, Tu C and Li Z: Multifaceted functions and novel insight
into the regulatory role of RNA N6-methyladenosine
modification in musculoskeletal disorders. Front Cell Dev Biol.
8(870)2020.PubMed/NCBI View Article : Google Scholar
|