Introduction
Bone defects, which can be caused by trauma,
infection, tumors or congenital deformation, have been accepted as
difficult-to-treat conditions in medicine (1). To date, autologous bone grafting has
been regarded as the most common strategy for the treatment of bone
defects (2-4);
however, it is considered as a ‘wound-repairing-wound’ method with
disadvantages, such as donor site pain, infection, haemorrhage,
nerve damage and limited blood supply (5). The development of cell and molecular
biology as well as biomaterial science has enabled the introduction
of bone tissue engineering (BTE) as a promising strategy for bone
repair (6). During bone repair, the
regulation of seed cell osteogenic differentiation is important
(7). Mesenchymal stem cells (MSCs)
are frequently used for these purposes; therefore, further
investigation into the key genes and related signaling pathways of
MSCs is required to improve BTE.
Wnt signaling is one of the five main osteogenic
signaling pathways (Wnt, bone morphogenic protein (BMP), fibroblast
growth factor, Hedgehog and Notch) and includes the three following
branches (8): Wnt/β-catenin
(9),
Wnt/peridinin-chlorophyll-protein complex (10) and Wnt/Ca2+ (11) signaling pathways. Among the
different pathways, the Wnt/β-catenin (canonical Wnt) signaling
pathway is crucial for the maintenance of bone mass (12). Increasing evidence has indicated
that the canonical Wnt signaling pathway can repress MSC
chondrogenic and adipogenic differentiation (13-15),
and improve MSC osteogenic differentiation (15-17).
The canonical Wnt signaling pathway can be initiated by the binding
of Wnt ligands to the receptor complex, which is comprised of
Frizzled (FZD) and LDL receptor related protein (Lrp)5/Lrp6,
resulting in the accumulation of β-catenin in the cytoplasm and its
translocation to the nucleus to interact with transcription factor
1 (Tcf1)/lymphoid enhancer binding factor 1 (Lef1) (12). Lrp5 is a transmembrane receptor
involved in the aforementioned signaling pathway (18). Several recent studies have indicated
that inactivation of the canonical Wnt signaling pathway in Lrp5
knockout mice leads to symptoms of bone loss (19,20).
By contrast, Lrp5 mutations in mice can result in high bone mass
(21).
Casein kinase-2 interaction protein-1 (Ckip-1)
mediates interactions with various proteins to participate in
different signaling pathways (22).
Ckip-1 was initially identified as a negative regulator of the BMP
signaling pathway via the activation of Smad Specific E3 Ubiquitin
Protein Ligase 1 and the degradation of Smad1/5(23). However, a limited number of studies
have focused on other bone formation signaling pathways. The
present study investigated whether Ckip-1 interacted with the
canonical Wnt signaling pathway in MSCs, and the role of Lrp5 that
may be involved in it.
Materials and methods
Cell culture and osteogenic
induction
C3H10T1/2 cells (donated by the Tissue Engineering
Center of the Fourth Military Medical University of China) were
cultured at 37˚C with 5% CO2 in low glucose DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 0.1% streptomycin/penicillin.
The culture medium was changed every 2 days and the cells were
passaged at ~85% confluence. At ~85% confluence, osteogenic
differentiation was induced by culturing cells in osteogenic medium
(DMEM supplemented with 10% FBS, 10 nM dexamethasone, 10 mM
β-glycerophosphate and 50 µg/ml ascorbic acid; all purchased from
Sigma-Aldrich; Merck KGaA) for 7 or 14 days at 37˚C with 5%
CO2.
Cell transfection
C3H10T1/2 cells were seeded (1x105
cells/well) into 24-well plates and cultured overnight prior to
transfection. Lentivirus-packed Ckip-1 (Ckip-1 overexpression:
Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin; Ckip-1 knockdown:
hU6-MCS-Ubiquitin-EGFP-IRES-puromycin; 1x108 TU/ml, 20
µl; Genechem, Inc.) and Lrp5 knockdown shRNA plasmids
(GV248-hU6-MCS-Ubiquitin-EGFP-IRES-puromycin; 1x108
TU/ml, 20 µl; Genechem, Inc.) were transfected into C310T1/2 cells
(1x105 cells/well) using polybrene (50 µg/ml, 20 µl;
Genechem, Inc.) for 10 h at 37˚C. The Ckip-1 knockdown shRNA
sequences used were as follows: Ckip-1-shRNA1,
5'-CCTGAGTGACTATGAGAAG-3'; Ckip-1-shRNA2,
5'-AGTGCGAAGAGCTCCGGAAA-3'; Lrp5-shRNA1, 5'-GACCTAAAGCGAAUCGAAA-3';
Lrp5-shRNA2, 5'-CGACCTGATGGGACUCAAA-3'; negative control shRNA of
shCkip-1, 5'-TTCTCCGAACGTGTCACGT-3'; and negative control shRNA of
shLrp5, 5'-TTCTCCGAACGTGTCACGT-3'. The empty plasmid served as the
control of Ckip-1 overexpression group. Following culture for 72 h,
fluorescence microscopy and reverse transcription-quantitative PCR
(RT-qPCR) were used to verify the results of the transfection.
Immunofluorescence
Cells were seeded into 24-well plates at a density
of 70%. Cells were washed three times with PBS and fixed with 4%
paraformaldehyde at 37˚C for 30 min. Cells were permeabilized for
15 min with 0.2% Triton X-100 and sealed with 2% BSA (Sangon
Biotech Co., Ltd.) for 30 min at room temperature. Subsequently,
cells were incubated with an anti-Ckip-1 primary antibody (1:200;
cat. no. D122120; Sangon Biotech Co., Ltd.) overnight at 4˚C.
Following washing with PBS, cells were incubated with a goat
anti-rabbit fluorescein-conjugated secondary antibody (1:1,000;
cat. no. ab150079; Abcam) for 2 h in the dark. The nuclei were
labeled with DAPI (5 µg/ml; Sigma-Aldrich; Merck KGaA) for 10 min
at room temperature. Stained cells were visualized using a confocal
scanning microscope (magnification, x40; Nikon Corporation). To
observe cell localization, double immunofluorescence staining was
performed using rabbit anti-Ckip-1 (1:200; cat. no. D122120; Sangon
Biotech Co., Ltd.) and goat anti-Lrp5 (1:1,000; cat. no. ab36121;
Abcam) antibodies. Similar to the aforementioned protocol,
following fixation, permeabilization and sealing, cells were
incubated with anti-Ckip-1 and anti-Lrp5 primary antibodies
overnight at 4˚C. Subsequently, cells were incubated with a goat
red fluorescein-conjugated secondary antibody (1:1,000; cat. no.
ab150079; Abcam) for 2 h in the dark at room temperature. Following
gentle washing with PBS, cells were incubated with a donkey green
fluorescein-conjugated secondary antibody (1:1,000; cat. no.
ab150129; Abcam) for 2 h in the dark at room temperature. Following
washing with PBS, stained cells were visualized using a confocal
laser scanning microscope (magnification, x40; Nikon
Corporation).
Proliferation assay
Cell proliferation was analyzed by performing an MTT
assay (Sigma-Aldrich; Merck KGaA). Following transfection,
C3H10T1/2 cells were seeded (2x103 cells/well) in
96-well plates and cultured for 24 h. MTT (10 µl) was added to each
well for 4 h at 37˚C. Subsequently, the medium was removed and DMSO
was added to each well to dissolve the purple formazan. The
absorbance was measured at a wavelength of 570 nm using a Bio-Rad
680 microplate reader (Bio-Rad Laboratories, Inc.).
Alkaline phosphatase (ALP)
staining
Cells were seeded (5x105 cells/well) in
6-well plates. Following culture for 14 days, cells were washed
with PBS and fixed in 4% paraformaldehyde at 4˚C for 30 min. ALP
staining was performed using the BCIP/NBT ALP color development kit
(LeaGene Biotech Co., Ltd.) according to the manufacturer's
protocol. Stained cells were observed using a M205RA stereoscopic
light microscope (Leica Microsystems GmbH) at a magnification of
x200, and quantified using Image Pro Plus software (version 7.1;
Media Cybernetics, Inc.).
RT-qPCR
Cells were seeded (5x105 cells/well) into
6-well plates and cultured in DMEM. Subsequently, the medium was
removed and cells were washed three times with PBS. Total RNA was
extracted using RNAisoPlus (Takara Bio, Inc.) and reverse
transcribed into cDNA using Prime Script RT Master Mix (Takara Bio,
Inc.) in a 20-µl volume. Subsequently, qPCR was performed using a
SYBR PCR Master Mix kit (Takara Bio, Inc.) with 10 µM specific
primers in a 25 µl total reaction volume with the following
thermocycling conditions: Initial denaturation step at 95˚C for 1
min; followed by 35 cycles at 95˚C for 30 sec, 58˚C for 30 sec; and
a final extension step at 72˚C for 30 sec. All signals were
normalized to GAPDH, and the 2-ΔΔCq method was used for
quantification (24). The mRNA
expression levels of Ckip-1, RUNX family transcription factor 2
(Runx2), Osterix (Osx), type I collagen (Col1), bone sialoprotein
(Bsp), osteocalcin (Ocn), Lrp5, Lef1 and Tcf1 were assessed via
qPCR. The sequences of the primers used are presented in Table I. mRNA expression levels were
normalized to the internal reference gene GAPDH.
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5'→3') |
|---|
| GAPDH | F:
AGGTCGGTGTGAACGGATTTG |
| | R:
GTAGACCATGTAGTTGAGGTCA |
| Ckip-1 | F:
AACCGCTATGTGGTGCTGAA |
| | R:
CAGGGTGAACTTGCTGTGATTT |
| Runx2 | F:
GACTGTGGTTACCGTCATGGC |
| | R:
ACTTGGTTTTTCATAACAGCGGA |
| Osx | F:
CCCAGCCACCTTTACCTACA |
| | R:
TATGGAGTGCTGCTGGTCTG |
| Col-1 | F:
GAGGCATAAAGGGTCATCGTGG |
| | R:
CATTAGGCGCAGGAAGGTCAG |
| Bsp | F:
GAGCCAGGACTGCCGAAAGGAA |
| | R:
CCGTTGTCTCCTCCGCTGCTGC |
| Ocn | F:
CAGCTTGGTGCACACCTAGC |
| | R:
AGGGTTAAGCTCACACTGCTCC |
| Lrp5 | F:
CTGCCAGGATCGCTCTGATG |
| | R:
ACACTGTTGCTTGATGAGGACACAC |
| Lef1 | F:
GCCACCGATGAGATGATCCC |
| | R:
TTGATGTCGGCTAAGTCGCC |
| Tcf1 | F:
TGAATCACCACCCGGAATGG |
| | R:
CTGGGCCAACTTCACATCCC |
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (Sangon Biotech Co., Ltd.) and quantified using a BCA
protein kit (Sangon Biotech Co., Ltd.). Equal amounts of protein
(20 µg/lane) were separated via 10% SDS-PAGE and transferred onto
PVDF membranes, which were blocked in TBST (0.1% Tween-20) with 5%
non-fat milk for 1 h at room temperature. Subsequently, the
membranes were incubated with the following primary antibodies
overnight at 4˚C: anti-Ckip-1 (1:500; cat. no. D122120; Sangon
Biotech Co., Ltd.), anti-β-actin (1:3,000; cat. no. 4970; Cell
Signaling Technology, Inc.), anti-Lrp5 (1:1,000; cat. no. ab38311;
Abcam), anti-β-catenin (1:1,000; cat. no. 8480; Cell Signaling
Technology, Inc.) and anti-α-tubulin (1:2,000; cat. no. 2125; Cell
Signaling Technology, Inc.). Following primary incubation, the
membranes were incubated with a fluorescein-conjugated secondary
antibody (1:3,000; cat. no. ab150079; Abcam) for 2 h at room
temperature. Protein bands were visualized using ECL reagent
(Sangon Biotech Co., Ltd.) and Odyssey V3.0 image scanning (LI-COR
Biosciences).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 19.0; SPSS, Inc.). Data are presented as the mean
± SD from at least three independent experiments. Comparisons among
groups were analyzed using one-way ANOVA followed by Tukey's post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
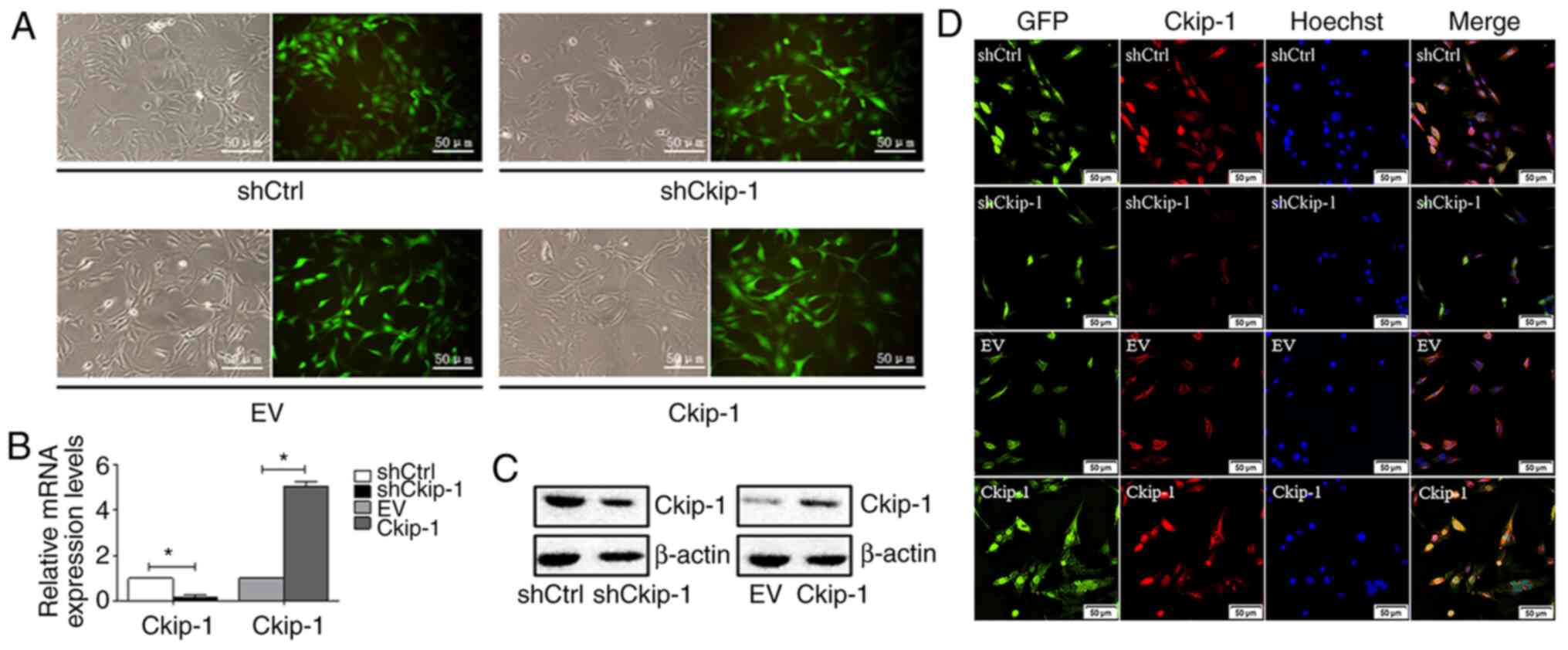
Ckip-1 knockdown and overexpression in
C3H10T1/2 cells
Following transfection of C3H10T1/2 cells with
different lentiviral expression vectors and subsequent puromycin
selection, transfection efficiency was determined. The majority of
cells displayed GFP expression and transfection efficiency was
estimated to be ≥90% (Fig. 1A). In
addition, the expression levels of Ckip-1 in different groups were
measured via RT-qPCR and western blotting. Ckip-1 mRNA (Fig. 1B) and protein (Fig. 1C) expression levels were markedly
decreased in the shCkip-1 group compared with the shCtrl group,
whereas the opposite results were observed in the Ckip-1 group
compared with the EV group, demonstrating successful and effective
Ckip-1 knockdown and overexpression, respectively. To further
detect the expression of Ckip-1 in C3H10T1/2 cells,
immunofluorescence analysis was conducted. The majority of cells
with green fluorescence in the shCtrl and EV groups displayed
merged signals with red fluorescence that were representative of
Ckip-1 detection (Fig. 1D).
Decreased expression levels of Ckip-1 were observed in the shCkip-1
group compared with the shCtrl group, whereas increased expression
levels of Ckip-1 were observed in the Ckip-1 group compared with
the EV group.
Effects of Ckip-1 on C3H10T1/2 cell
proliferation
To determine the effects of Ckip-1 knockdown and
overexpression on C3H10T1/2 cell proliferation, an MTT assay was
performed. On days 1, 3, 5 and 7, cell proliferation in the
shCkip-1 group was significantly increased compared with the shCtrl
group. By contrast, on day 5 and 7, cell proliferation in the
Ckip-1 group was significantly decreased compared with the EV group
(Fig. 2A).
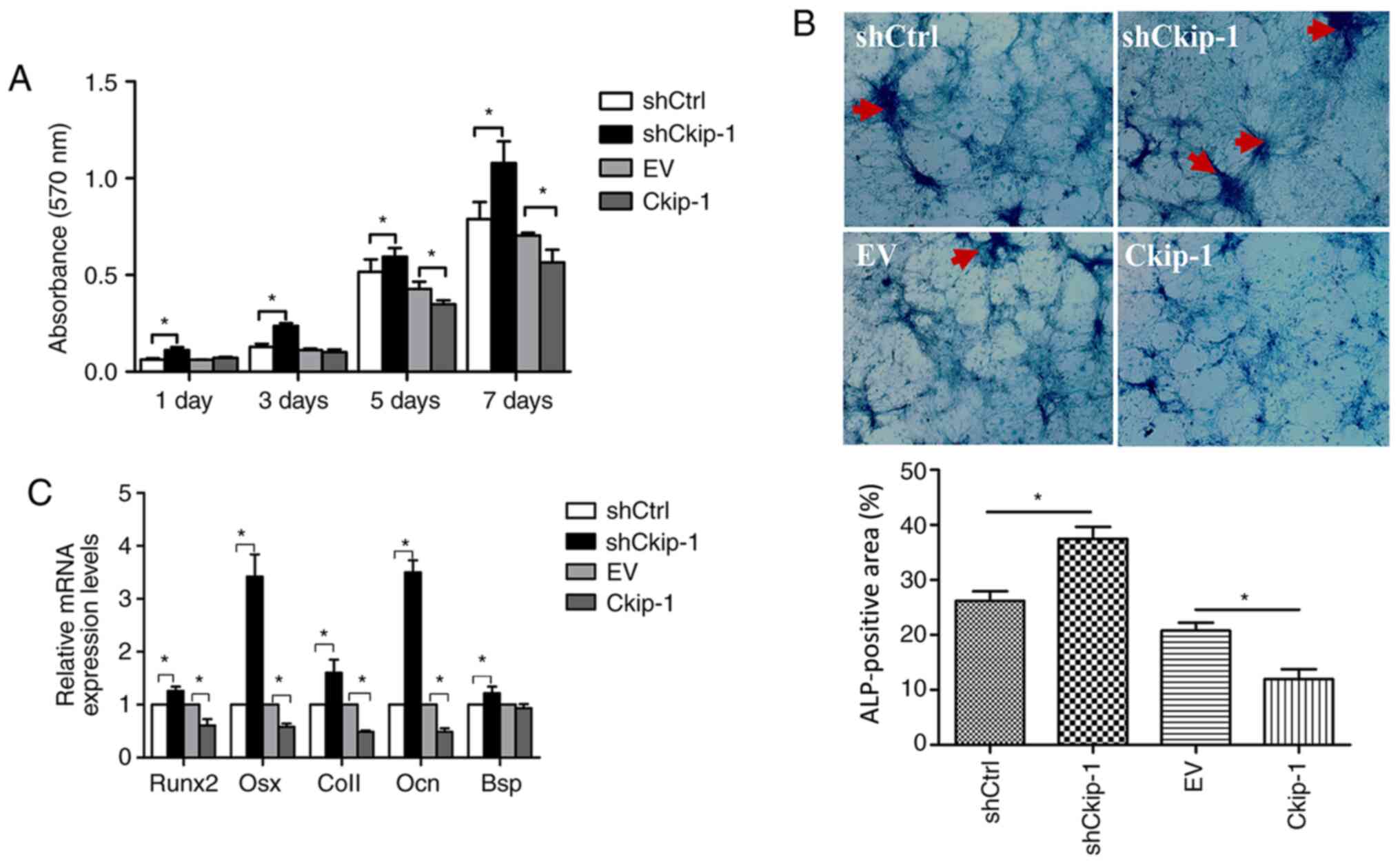 | Figure 2C3H10T1/2 cell proliferation and
osteogenic differentiation. (A) Cell proliferation was assessed by
performing an MTT assay on day 1, 3, 5 and 7. (B) Following 14-day
osteoinduction, ALP activity was assessed via ALP staining
(magnification, x200). Red arrows indicate the high positive black
precipitate. (C) mRNA expression levels of five osteogenic makers
(Runx2, Osx, Col1, Ocn and Bsp). *P<0.05. ALP,
alkaline phosphatase; Runx2, RUNX family transcription factor 2;
Osx, Osterix; Col1, type I collagen; Ocn, osteocalcin; Bsp, bone
sialoprotein; sh, short hairpin RNA; Ctrl, control; EV, empty
vector. |
Effects of Ckip-1 on C3H10T1/2 cell
osteogenic differentiation
An ALP activity assay and RT-qPCR were performed to
evaluate C3H10T1/2 cell osteogenic differentiation. Following
14-day osteoinduction, the shCkip-1 group displayed significantly
higher positive staining (dark purple) compared with the shCtrl
group (Fig. 2B). By contrast, the
Ckip-1 group displayed significantly decreased ALP staining
compared with the EV group. Moreover, gene expression analysis of
osteogenic markers (Runx2, Osx, Col1, Ocn and Bsp) in the shCkip-1
group demonstrated a significant increase in expression levels
compared with the shCtrl group (Fig.
2C). By contrast, with the exception of Bsp, the expression
levels of the markers in the Ckip-1 group were significantly
decreased compared with the EV group.
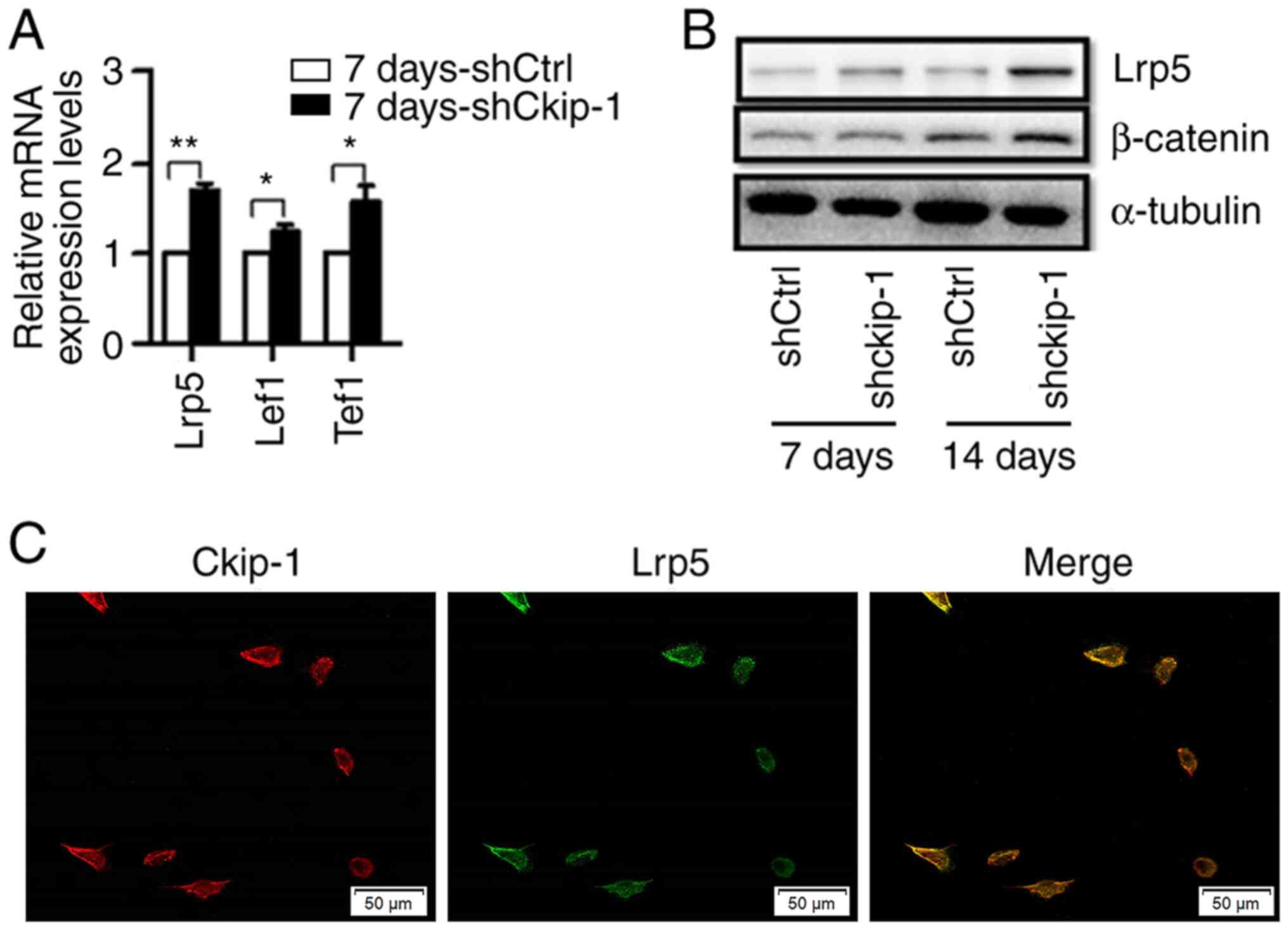
Ckip-1 knockdown and knockout increase
the expression levels of Lrp5 in C3H10T1/2 cells
The effects of Ckip-1 knockdown on the expression
levels of Lrp5, Tcf1 and Lef1 in C3H10T1/2 cells in the 7
d-shCkip-1 group were subsequently examined via RT-qPCR. The
results indicated that the expression levels of Lrp5, Tcf1 and Lef1
were significantly increased in the 7 d-shCkip-1 group compared
with the 7 d-shCtrl group (Fig.
3A). The western blotting results indicated that the expression
levels of Lrp5 were increased in the 7 d-shCkip-1 group compared
with the 7 d-shCtrl group, whereas β-catenin expression levels were
not altered. However, the expression levels of Lrp5 and β-catenin
were markedly increased in the 14 d-shCkip-1 group compared with
the shCtrl group (Fig. 3B). To
examine the cellular localization of Ckip-1 and Lrp5,
immunofluorescence staining was performed (Fig. 3C). Colocalization of Ckip-1 (red)
and Lrp5 (green) was observed in C3H10T1/2 cells under normal
conditions. Moreover, the results suggested that Ckip-1 highly
accumulated in the vicinity of the membrane of C3H10T1/2 cells,
which may imply an underlying membrane-associated role of
Ckip-1.
Decreased cell proliferation in the
Lrp5 knockdown group can be rescued by shCkip-1
Simultaneous Lrp5 and Ckip-1 knockdown was achieved
by lentiviral transfection and confirmed via RT-qPCR. In accordance
with the aforementioned results, the expression level of Lrp5 was
significantly increased in the shCkip-1 and shLrp5 + shCkip-1
groups compared with shCtrl group (Fig.
4A). C3H10T1/2 cell proliferation and osteogenic
differentiation were evaluated in the different groups. At 3 and 5
days, cell proliferation in the shLrp5 group was significantly
reduced compared with the shCtrl group (Fig. 4B). At 1, 3 and 5 days in the shLrp5
+ shCkip-1 group, cell proliferation was significantly higher
compared with the shLrp5 group.
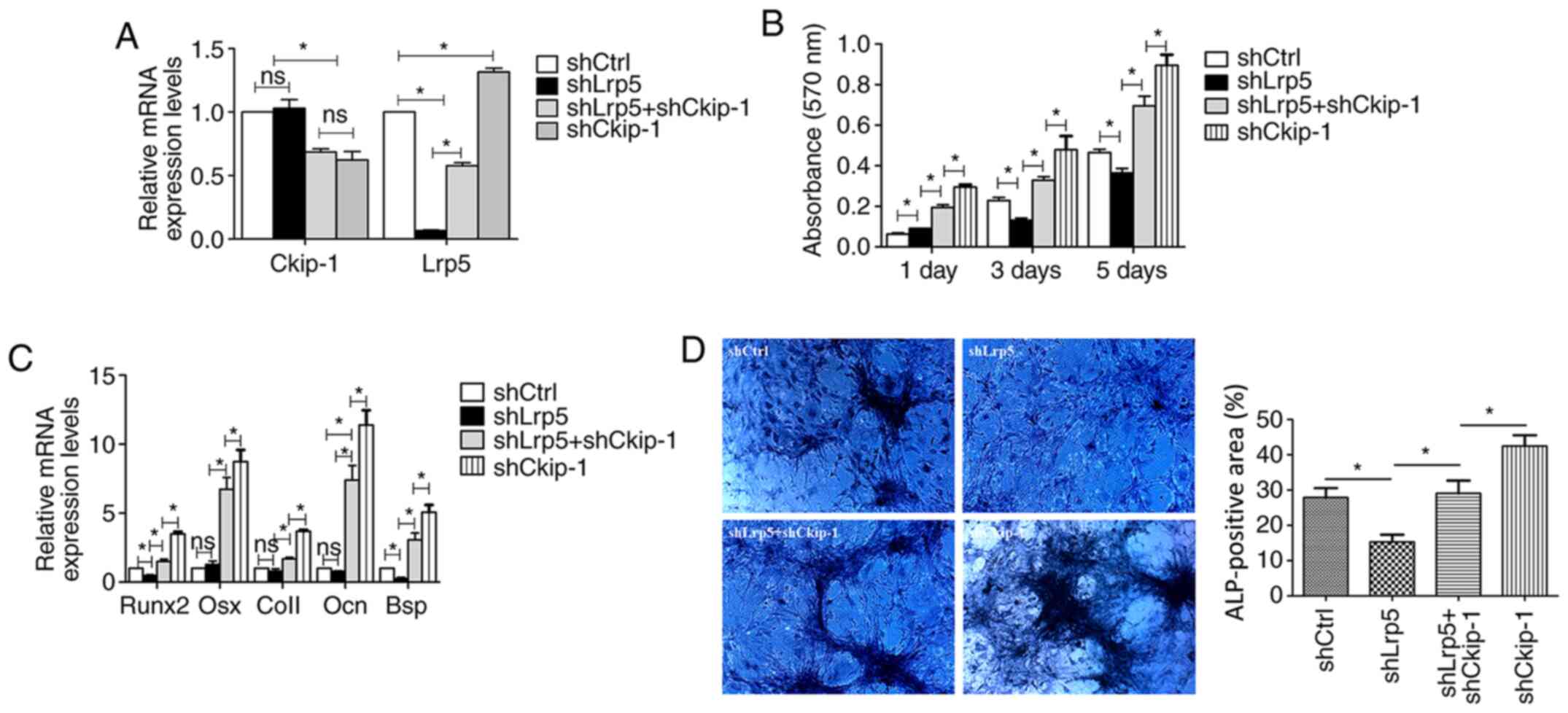 | Figure 4Rescue effect of shCkip-1 on
shLrp5-transfected C3H10T1/2 cell proliferation and osteogenic
differentiation. (A) Transfection efficiency of shLrp5 and
shCkip-1. (B) Cell proliferation was assessed by performing an MTT
assay on day 1, 3 and 5. (C) mRNA expression levels of five
osteogenic makers (Runx2, Osx, Col1, Ocn, Bsp). (D) ALP activity
was assessed via ALP staining (magnification, x200).
*P<0.05. sh, short hairpin RNA; Ckip-1, casein
kinase-2 interaction protein-1; Lrp5, LDL receptor related protein
5; Runx2, RUNX family transcription factor 2; Osx, Osterix; Col1,
type I collagen; Ocn, osteocalcin; Bsp, bone sialoprotein; ALP,
alkaline phosphatase; Ctrl, control. |
Involvement of Lrp5 in Ckip-1
knockdown-induced C3H10T1/2 cell osteogenic differentiation
The relative gene expression levels of osteogenic
markers in the shLrp5, shCkip-1, shLrp5 + shCkip-1 and shCtrl
groups were evaluated after 7-day osteoinduction. The majority of
the osteogenic markers were expressed at lower levels in the shLrp5
group compared with the shCtrl group, with significantly decreased
Runx2 and Bsp expression levels in the shLrp5 group compared with
the shCtrl group (Fig. 4C).
However, in the shLrp5 + shCkip-1 group, osteogenic-related marker
expression levels were significantly increased compared with the
shLrp5 group. Moreover, the expression levels of the osteogenic
markers in the shCkip-1 group were significantly higher compared
with the shLrp5 + shCkip-1 group. The ALP activity detection
results further indicated the rescue response of shCkip-1 in
C3H10T1/2 cell osteogenic differentiation following Lrp5 knockdown
(Fig. 4D).
Discussion
The stimulation of MSC proliferation and osteogenic
differentiation is a promising approach for the treatment of bone
defects (25,26). Ckip-1 can negatively regulate bone
formation, and the canonical Wnt signaling pathway is one of the
five osteogenesis signaling pathways (8,27). In
the present study, the role of Ckip-1 in the regulation of
osteogenic differentiation via the canonical Wnt signaling pathway
was assessed. Moreover, to the best of our knowledge, with the
exception of the BMP signaling pathway (22), the association between the
osteogenic signaling pathways and Ckip-1 has not been previous
reported. In the present study, the common C3H10T1/2 MSC-like
pluripotent cell line was used, which was obtained from mouse
embryos and established by Reznikoff et al in 1973(28). The effects of Ckip-1 on C3H10T1/2
cell proliferation and osteogenic differentiation were evaluated in
the present study.
The results of the MTT assay indicated that
C3H10T1/2 cell proliferation was enhanced and reduced in the
shCkip-1 and Ckip-1 groups compared with the shCtrl and EV groups,
respectively, indicating that Ckip-1 negatively regulated C3H10T1/2
cell proliferation. ALP activity is upregulated during osteogenesis
and is regarded as an early bone marker (29). The ALP staining and RT-qPCR assays
suggested that Ckip-1 also negatively regulated C3H10T1/2 cell
osteogenic differentiation.
Subsequently, the association between Ckip-1 and the
canonical Wnt signaling pathway was investigated. Following
immunofluorescence staining, the expression and localization of
Ckip-1 were examined in the cell membrane, cytoplasm and nucleus
under normal conditions. High accumulation of Ckip-1 was observed
at the cell membrane, which indicated colocalization of Ckip-1 with
the Lrp5 receptor. Previous studies have suggested that Ckip-1 is
localized near the cell membrane and distributed throughout the
cytoplasm or in the nucleus, suggesting that its function is
determined by its cellular location (30-33).
Therefore, it was hypothesized that membrane-associated Ckip-1
localization may account for the phenotype of the C3H10T1/2 cell
line or for the expression of the membrane receptors.
Ckip-1 knockdown significantly increased the
expression levels of the key molecules of the canonical Wnt
signaling pathway (Lrp5, Lef1 and Tcf1) in C3H10T1/2 cells compared
with the shCtrl group. Lrp5 knockdown was established in C3H10T1/2
cells, since the cell line exhibited high Lrp5 expression compared
with the shCtrl group (Fig. 3A and
B). Lrp5 is a transmembrane
receptor that can bind to Wnt ligands and FZD proteins to initiate
and activate the canonical Wnt signaling pathway (12). Related studies have demonstrated
that disruption of Lrp5 can result in a low bone mass phenotype in
postnatal mice, whereas Lrp5 knockdown can lead to a high bone mass
phenotype (34,35). The shCkip-1 group displayed
increased expression levels of Lrp5 compared with the shCtrl group,
as determined via RT-qPCR and western blotting. In addition,
β-catenin is an important downstream molecule of the canonical Wnt
signaling pathway (36,37). Previous studies have indicated that
deletion of β-catenin can negatively affect osteoblast
differentiation and bone formation (36,37).
The western blotting results indicated that β-catenin expression
levels were significantly increased on day 14 of osteoinduction in
the shCkip-1 group compared with the shCtrl group, which was
consistent with previous studies.
In the present study, the effect of Lrp5 knockdown
on C3H10T1/2 cell proliferation and osteogenic differentiation was
assessed. Moreover, the potential of Ckip-1 knockdown to reverse
Lrp5 knockdown-mediated effects was also examined. Following
lentiviral transfection, MTT and RT-qPCR assays were performed to
evaluate cell proliferation and osteogenic marker expression,
respectively. Cell proliferation was significantly decreased in the
shLrp5 group on day 3 compared with the shCtrl group, indicating
that Lrp5 may exert a potential positive effect on C3H10T1/2 cell
proliferation. The shLrp5 + shCkip-1 group displayed higher cell
proliferation compared with the shLrp5 group, which implied a
rescue role of shCkip-1. With regard to osteogenic differentiation,
the expression levels of the osteogenic-associated genes of the
shLrp5 + shCkip-1 group were significantly increased compared with
the shCtrl and shLrp5 groups. The shLrp5 and shCtrl groups
displayed no significant differences. However, the expression
levels of almost all the genes investigated were slightly decreased
in the shLrp5 group compared with the shCtrl group, which was
consistent with previous findings demonstrating that Lrp5 may serve
a positive regulatory role on bone formation (34,35).
Furthermore, it was hypothesized that the transmembrane receptor
Lrp5 may serve as a potential target of Ckip-1 based on the
membrane-associated colocalization of Ckip-1 and Lrp5 in C3H10T1/2
cells. However, further studies are required to verify the
hypothesis, potentially by using a Wnt inhibitor.
In conclusion, the present study evaluated the
effects of Ckip-1 on C3H10T1/2 cell proliferation and osteogenic
differentiation, as well as the potential underlying mechanism. The
results demonstrated that Ckip-1 negatively regulated C3H10T1/2 MSC
cell proliferation and osteogenic differentiation via the canonical
Wnt-signaling receptor Lrp5, which may provide a promising target
for the improvement of BTE.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81670803).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YS and LK conceived the study. XH and JL designed
the experiments. JL provided software. YG validated, formally
analyzed and investigated the data. YH provided resources and
performed experiments. XH wrote and reviewed the final manuscript.
YS provided supervision. LK provided project administration and
acquired funding. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li L, Lu H, Zhao Y, Luo J, Luo J, Yang L,
Liu W and He Q: Functionalized cell-free scaffolds for bone defect
repair inspired by self-healing of bone fractures: A review and new
perspectives. Mater Sci Eng C Mater Biol Appl. 98:1241–1251.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Liu M and Lv Y: Reconstructing bone with
natural bone graft: A review of in vivo studies in bone defect
animal model. Nanomaterials (Basel). 8(E999)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wei X, Liu B, Liu G, Yang F, Cao F, Dou X,
Yu W, Wang B, Zheng G, Cheng L, et al: Mesenchymal stem cell-loaded
porous tantalum integrated with biomimetic 3D collagen-based
scaffold to repair large osteochondral defects in goats. Stem Cell
Res Ther. 10(72)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chen X, Fan H, Deng X, Wu L, Yi T, Gu L,
Zhou C, Fan Y and Zhang X: Scaffold structural microenvironmental
cues to guide tissue regeneration in bone tissue applications.
Nanomaterials (Basel). 8(E960)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
White KA and Olabisi RM: Spatiotemporal
control strategies for bone formation through tissue engineering
and regenerative medicine approaches. Adv Healthc Mater.
8(e1801044)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
El-Rashidy AA, Roether JA, Harhaus L,
Kneser U and Boccaccini AR: Regenerating bone with bioactive glass
scaffolds: A review of in vivo studies in bone defect models. Acta
Biomater. 62:1–28. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Leyendecker Junior A, Gomes Pinheiro CC,
Lazzaretti Fernandes T and Franco Bueno D: The use of human dental
pulp stem cells for in vivo bone tissue engineering: A systematic
review. J Tissue Eng. 9(2041731417752766)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wehner D and Weidinger G: Signaling
networks organizing regenerative growth of the zebrafish fin.
Trends Genet. 31:336–343. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ren L, Chen H, Song J, Chen X, Lin C,
Zhang X, Hou N, Pan J, Zhou Z, Wang L, et al: MiR-454-3p-mediated
Wnt/β-catenin signaling antagonists suppression promotes breast
cancer metastasis. Theranostics. 9:449–465. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yuan K, Shamskhou EA, Orcholski ME, Nathan
A, Reddy S, Honda H, Mani V, Zeng Y, Ozen MO, Wang L, et al: Loss
of endothelial derived WNT5A is associated with reduced pericyte
recruitment and small vessel loss in pulmonary arterial
hypertension. Circulation. 139:1710–1724. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gong B, Shen W, Xiao W, Meng Y, Meng A and
Jia S: The Sec14-like phosphatidylinositol transfer proteins
Sec14l3/SEC14L2 act as GTPase proteins to mediate
Wnt/Ca2+ signaling. Elife. 6(e26362)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Baron R and Kneissel M: WNT signaling in
bone homeostasis and disease: From human mutations to treatments.
Nat Med. 19:179–192. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Taipaleenmäki H, Abdallah BM, Aldahmash A,
Säämänen AM and Kassem M: Wnt signalling mediates the cross-talk
between bone marrow derived pre-adipocytic and pre-osteoblastic
cell populations. Exp Cell Res. 317:745–756. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Day TF, Guo X, Garrett-Beal L and Yang Y:
Wnt/beta-catenin signaling in mesenchymal progenitors controls
osteoblast and chondrocyte differentiation during vertebrate
skeletogenesis. Dev Cell. 8:739–750. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
D'Alimonte I, Lannutti A, Pipino C, Di
Tomo P, Pierdomenico L, Cianci E, Antonucci I, Marchisio M, Romano
M, Stuppia L, et al: Wnt signaling behaves as a ‘master regulator’
in the osteogenic and adipogenic commitment of human amniotic fluid
mesenchymal stem cells. Stem Cell Rev Rep. 9:642–654.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zuo R, Liu M, Wang Y, Li J, Wang W, Wu J,
Sun C, Li B, Wang Z, Lan W, et al: BM-MSC-derived exosomes
alleviate radiation-induced bone loss by restoring the function of
recipient BM-MSCs and activating Wnt/β-catenin signaling. Stem Cell
Res Ther. 10(30)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fan J, An X, Yang Y, Xu H, Fan L, Deng L,
Li T, Weng X, Zhang J and Zhao RC: MiR-1292 targets FZD4 to
regulate senescence and osteogenic differentiation of stem cells in
TE/SJ/mesenchymal tissue system via the Wnt/β-catenin pathway.
Aging Dis. 9:1103–1121. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Williams BO: LRP5: From bedside to bench
to bone. Bone. 102:26–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li T, Li H, Wang Y, Li T, Fan J, Xiao K,
Zhao RC and Weng X: microRNA-23a inhibits osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
by targeting LRP5. Int J Biochem Cell Biol. 72:55–62.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Borrell-Pagès M, Romero JC and Badimon L:
LRP5 deficiency down-regulates Wnt signalling and promotes aortic
lipid infiltration in hypercholesterolaemic mice. J Cell Mol Med.
19:770–777. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Semenov MV and He X: LRP5 mutations linked
to high bone mass diseases cause reduced LRP5 binding and
inhibition by SOST. J Biol Chem. 281:38276–38284. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Nie J, Liu L, He F, Fu X, Han W and Zhang
L: CKIP-1: A scaffold protein and potential therapeutic target
integrating multiple signaling pathways and physiological
functions. Ageing Res Rev. 12:276–281. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lu K, Yin X, Weng T, Xi S, Li L, Xing G,
Cheng X, Yang X, Zhang L and He F: Targeting WW domains linker of
HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat
Cell Biol. 10:994–1002. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chen BY, Wang X, Chen LW and Luo ZJ:
Molecular targeting regulation of proliferation and differentiation
of the bone marrow-derived mesenchymal stem cells or mesenchymal
stromal cells. Curr Drug Targets. 13:561–571. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lee DJ, Kwon J, Current L, Yoon K, Zalal
R, Hu X, Xue P and Ko CC: Osteogenic potential of mesenchymal stem
cells from rat mandible to regenerate critical sized calvarial
defect. J Tissue Eng. 10(2041731419830427)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Moore ER and Jacobs CR: The primary cilium
as a signaling nexus for growth plate function and subsequent
skeletal development. J Orthop Res. 36:533–545. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Reznikoff CA, Brankow DW and Heidelberger
C: Establishment and characterization of a cloned line of C3H mouse
embryo cells sensitive to postconfluence inhibition of division.
Cancer Res. 33:3231–3238. 1973.PubMed/NCBI
|
|
29
|
Hashimi SM: Exogenous noggin binds the
BMP-2 receptor and induces alkaline phosphatase activity in
osteoblasts. J Cell Biochem. 120:13237–13242. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li D, Zhu H, Liang C, Li W, Xing G, Ma L,
Ding L, Zhang Y, He F and Zhang L: CKIP-1 suppresses the
adipogenesis of mesenchymal stem cells by enhancing
HDAC1-associated repression of C/EBPα. J Mol Cell Biol. 6:368–379.
2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Safi A, Vandromme M, Caussanel S, Valdacci
L, Baas D, Vidal M, Brun G, Schaeffer L and Goillot E: Role for the
pleckstrin homology domain-containing protein CKIP-1 in
phosphatidylinositol 3-kinase-regulated muscle differentiation. Mol
Cell Biol. 24:1245–1255. 2004.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang L, Xing G, Tie Y, Tang Y, Tian C, Li
L, Sun L, Wei H, Zhu Y and He F: Role for the pleckstrin homology
domain-containing protein CKIP-1 in AP-1 regulation and apoptosis.
EMBO J. 24:766–778. 2005.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Bosc DG, Graham KC, Saulnier RB, Zhang C,
Prober D, Gietz RD and Litchfield DW: Identification and
characterization of CKIP-1, a novel pleckstrin homology
domain-containing protein that interacts with protein kinase CK2. J
Biol Chem. 275:14295–14306. 2000.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kato M, Patel MS, Levasseur R, Lobov I,
Chang BHJ, Glass DA II, Hartmann C, Li L, Hwang TH, Brayton CF, et
al: Cbfa1-independent decrease in osteoblast proliferation,
osteopenia, and persistent embryonic eye vascularization in mice
deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 157:303–314.
2002.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cui Y, Niziolek PJ, MacDonald BT, Zylstra
CR, Alenina N, Robinson DR, Zhong Z, Matthes S, Jacobsen CM, Conlon
RA, et al: Lrp5 functions in bone to regulate bone mass. Nat Med.
17:684–691. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM and
Long F: Sequential roles of Hedgehog and Wnt signaling in
osteoblast development. Development. 132:49–60. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hill TP, Später D, Taketo MM, Birchmeier W
and Hartmann C: Canonical Wnt/beta-catenin signaling prevents
osteoblasts from differentiating into chondrocytes. Dev Cell.
8:727–738. 2005.PubMed/NCBI View Article : Google Scholar
|