Liver fibrosis (LF) results from impaired wound
healing caused by acute or chronic exposure to detrimental factors,
including alcohol, viral diseases, drugs, cholestasis, toxins and
metabolic disorders (1,2). Slight or transient fibrosis is
necessary for wound healing and maintenance of tissue architecture
integrity (3). Fibrosis is a
reversible process that may be halted by removing the harmful
stimulus (4,5). However, severe or advanced fibrosis is
characterized by abnormal connective tissue hyperplasia and
extracellular matrix (ECM) protein deposition (6), leading to liver structural destruction
and even organ failure (7). The
accumulation of ECM proteins results in the distortion of hepatic
architecture due to scar formation, along with the appearance of
regenerating hepatocyte nodules, which define cirrhosis (8). Cirrhosis is characterized by
hepatocellular dysfunction, portal hypertension, hepatocellular
carcinoma (HCC) and eventual liver failure (9,10).
Liver fibrogenesis is initiated by hepatic stellate
cells (HSCs). Under specific conditions, quiescent HSCs are
transformed into myofibroblasts (MFs) to generate ECM proteins,
tissue inhibitors of metalloproteinases (TIMPs) and matrix
metalloproteinases (MMPs) (11-14).
HSC activation involves a systemic and complex pathological process
involving multiple cytokines and multiple cellular signaling
pathways (15).
The healthy liver has a strong regenerative
potential due to the unlimited proliferative potential of
cholangiocytes and hepatocytes (HCs). Hepatic stem/progenitor cells
(HpSCs) are positioned within the canals of Hering (16). These cells are quiescent in the
healthy liver but may be activated in response to liver injury by
proliferating and differentiating towards cholangiocytes and HCs
(17). In injured tissue, activated
HSCs, macrophages and MFs produce a variety of signals through
signaling pathways including the Wnt and Notch pathways to drive
HpSC proliferation and differentiation (18). HpSCs are also able to activate
stellate cells through signaling pathways, including the Hedgehog
(Hh) pathway, resulting in the release of various types of matrix
components during liver regeneration (19).
Studies have confirmed that fibrosis is regulated
through the expression of various genes (20). As key regulators of multiple
biological processes, long non-coding RNAs (lncRNAs), commonly
defined as RNAs longer than 200 nucleotides without any
protein-coding capacity, have attracted much research interest
(21). Although the classification
system of lncRNAs is currently incomplete, they are generally
divided into two broad types according to their position relative
to protein-coding genes: i) Intergenic lncRNAs and ii) coding
gene-overlapping lncRNAs (22).
lncRNAs regulate gene expression and protein synthesis through
multiple mechanisms (23) and are
considered to include ~30,000 different transcripts, accounting for
a large portion of the non-coding transcriptome in humans (24). Unlike mRNAs, most lncRNAs are
expressed at low levels, cell type-specific, associated with a high
number of coding genes (24) and
are present at specific positions within the nucleus (25). lncRNAs are essential in the
regulation of cell migration, apoptosis, differentiation and
proliferation processes (26,27).
Various lncRNAs have been confirmed to be involved in multiple
diseases and certain lncRNAs have been identified as disease
biomarkers (28). In addition,
recent studies have revealed that lncRNAs are linked to the complex
pathophysiological changes observed in LF. Although numerous
lncRNAs have been identified and proposed as promising targets for
anti-fibrosis therapies, the underlying mechanisms of the functions
of lncRNAs in LF have remained elusive.
In the present review, the role of lncRNAs in
modulating cellular signaling pathways in LF was explored. The
potential utility of lncRNAs as non-invasive biomarkers and novel
therapeutic targets for LF was also proposed.
lncRNA-activated by TGF-β (lncRNA-ATB) is an
important regulator of the TGF-β/Smad signaling pathway. Studies
have reported that lncRNA-ATB, Smad2 and TGFβRII share a common
microRNA (miRNA) response element (MRE) for miRNA (miR)-425-5p.
lncRNA-ATB was indicated to induce the expression of Smad2 and
TGF-βRII by inhibiting the expression of endogenous miR-425-5p in
hepatitis C virus (HCV)-induced hepatic fibrosis in a study on
hepatic stellate LX-2 cells treated with hepatoblastoma HepG2 cells
carrying the HCV core protein. Consequently, lncRNA-ATB caused
hepatic fibrosis by enhancing collagen I synthesis and stimulating
HSCs through competitive binding to miR-425-5p (36).
LF-associated lncRNA1 (lnc-LFAR1) was indicated to
facilitate the interaction between Smad2/3 and TGFβRI and promote
Smad2/3 phosphorylation in carbon tetrachloride
(CCl4)/bile duct ligation (BDL)-induced LF. In addition,
lnc-LFAR1 is able to directly bind to Smad2/3. On this basis, the
TGFβ1/Smad2/3/lnc-LFAR1 signaling pathway creates an active
feedback loop that enhances Smad2/3 functions in hepatic fibrosis
(37).
HOXA distal transcript antisense RNA (HOTTIP) has
been implicated in liver fibrogenesis (38). Li et al (39) determined that HOTTIP was upregulated
in mice with hepatic fibrosis and that inhibition of HOTTIP by
adenoviral delivery of short hairpin RNA-HOTTIP markedly reduced LF
(39). miR-148a participates in the
initiation and progression of HCC in the presence of LF and HOTTIP
inhibits miR-148a expression (40).
miR-148a may regulate TGFβRI/TGFβRII, subsequently decreasing their
expression levels, in human and mouse HSCs. Collectively, these
results suggest that HOTTIP may promote LF by downregulating
miR-148a and upregulating TGFβRI and TGFβRII.
The lncRNA Gm5091 significantly negatively regulates
HSCs in mice with alcoholic hepatic fibrosis (AHF) (45). Zhou et al (45) reported that Gm5091 downregulates
cell migration, collagen I expression and HSC activation marker
expression, including Desmin and α-smooth muscle actin. In
addition, data based on a bioinformatic analysis revealed that the
Gm5091 sequence contains binding sites for miR-24, miR-23b and
miR-27b. Full-length Gm5091 decreases the expression levels of
miR-27b/23b/24(45). In addition,
miR-27, miR-24 and miR-23 are positive regulators of HSC
proliferation and differentiation, as they activate TGF-β and Smad4
in mice with LF (46,47). As a ceRNA, Gm5091 sponges
miR-27b/23b/24, which alleviates liver injury and the progression
of AHF in mice by inhibiting HSC activation.
Nuclear paraspeckle assembly transcript 1 (NEAT1) is
critical for the formation of paraspeckles (48) and hence to the initiation of tumors.
It is highly expressed in activated HSCs and liver tissue of mice
with CCl4-induced LF. In human fibrotic liver samples,
upregulated expression levels of NEAT1 are positively correlated
with fibrosis markers (49). In a
mouse model of CCl4-induced fibrosis, NEAT1
overexpression promoted HSC activation in vivo, and similar
results were obtained in vitro (50). A previous study suggested that
miR-122 inhibits HSC activation and the expression of
fibrosis-associated genes induced by TGF-β (51). Stimulated expression of Kruppel-like
factor 6 (KLF6), an immediate-early gene in LF, induces the
expression of TGFβRI and TGFβRII in activated HSCs (52). Furthermore, miR-122 targets NEAT1 as
well as KLF6(53). Collectively,
these results suggest that NEAT1 competitively binds to miR-122 and
regulates KLF6 expression in hepatic fibrosis, indicating that the
NEAT1/miR-122/KLF6 axis promotes HSC activation.
The lncRNA ENSMUST00000158992 (SCARNA10) is
upregulated in liver tissues of fibrotic mice as well as in the
serum and liver tissue of humans with advanced LF (54). Several studies have indicated that
SCARNA10 is a positive regulator of LF, as it induces HC apoptosis
and HSC activation (55,56). Mechanistically, SCARNA10 functions
as a mediator of LF by inhibiting the binding of polycomb
repressive complex 2 to the promoters of genes involved in the
TGF-β pathway, thereby promoting the transcription of these
genes.
The Hh signaling pathway is a morphogenic pathway
that has multiple roles in cell proliferation, apoptosis, migration
and differentiation. It was first reported in Drosophila by
Nüsslein-Volhard and Wieschaus (57) in 1980. The Hh pathway comprises
Glioblastoma (GLI) family transcription factors (GLI1, GLI2, GLI3),
Smoothened (SMO), sonic Hh and Patched 1 (PTCH1) (58) and is driven by PTCH receptors that
are activated by Hh ligands, which abolish the inhibitory effect of
PTCH1 on SMO. In turn, SMO transduces Hh signals to regulate gene
expression via GLI transcription factors (59,60).
Normal adult HCs generally do not produce Hh ligands but hepatic
synthesis of Hh ligands is increased in liver injury (61). In addition, several resident liver
cells, including HCs, HSCs, cholangiocytes, macrophages, natural
killer T cells and liver sinusoidal endothelial cells, are able to
produce Hh ligands (62,63). Stimuli that contribute to liver
regeneration/remodeling promote the expression of Hh ligands in the
liver. HSCs become highly activated by Hh ligands, which enhances
their fibrogenic and proliferative capabilities (64-66).
Activation of the Hh pathway is influenced by liver regeneration,
hepatic accumulation of inflammatory cells, liver fibrogenesis and
vascular remodelling (67). In
addition, the Hh pathway activity is positively associated with the
fibrosis stage (68). Data from
Sicklick et al (69)
indicate that inactivation of Hh signaling in HSCs inhibits HSC
activation in vitro. Approaches that may block the Hh
pathway may not only reduce HSC activation and fibrosis but also
prevent the accumulation of hepatic progenitor cells (70). Furthermore, EMT is a key event in
HSC activation and is regulated by the Hh pathway (71,72).
Plasmacytoma variant translocation 1 (PVT1) promotes
HSC activation through the Hh pathway and EMT process (73). PTCH1 is a negative modulator of Hh
signalling (58). PVT1 knockdown
increases the expression of PTCH1. Inhibition of PTCH1 during liver
fibrogenesis results in PTCH1 methylation, whereas silencing of
PTCH1 expression promotes activation of the Hh pathway in
CCl4-induced LF (74).
Furthermore, PVT1 inhibits miR-152 through a post-transcriptional
mechanism, while miR-152 promotes hypomethylation of PTCH1 by
suppressing its direct target DNA methyltransferase 1(75). Demethylation of PTCH1 by miR-152
modulates the effect of PVT1 inhibition on PTCH1 levels and
treatment with a miR-152 antagonist abolishes these changes.
Therefore, PVT1 inhibits PTCH1 through competitive binding with
miR-152 and promotes EMT in hepatic fibrosis.
The lncRNA-maternally expressed gene 3 (MEG3) was
indicated to be downregulated in hepatic fibrosis in vitro
and in vivo, and its overexpression alleviates fibrogenesis
(76,77). Previous studies have reported that
MEG3 suppresses hepatic fibrosis via p53(78). In particular, overexpression of MEG3
inhibits HSC activation by promoting the EMT process (79). In addition, deletion of the SMO
binding site in MEG3 fails to block the effects of MEG3 on the EMT
process and GLI3 in mice treated with CCl4 (76). miR-212 is significantly
downregulated in MEG3-overexpressing cells and is able to target
PTCH1. Furthermore, MEG3 induces Hh pathway activation by sponging
miR-212, promoting PTCH1 expression and decreasing SMO
expression.
Wnt signaling modulates cellular apoptosis,
proliferation and differentiation. Wnt proteins are 350-400 amino
acids long with a conserved cysteine-rich binding domain containing
23-24 cysteine residues (80). Of
note, two cell surface receptor families participate in the
reception and transduction of Wnt signals: The low-density
lipoprotein receptor-related protein (LRP) family and members of
the Frizzled (Fz) gene family (81). When Wnt binds to its receptor,
either Fz or a complex formed by Fz and LRP5/6, a signal is
transduced to the cytoplasmic phosphoprotein disheveled (82). Mammals have three types of Dsh
proteins (Dsh-3, Dsh-2 and Dsh-1) (83). Wnt signaling is divided into three
independent pathways according to the affected Dsh protein: The
canonical ‘Wnt/β-catenin’ pathway, the ‘Wnt/polarity’ pathway (also
called the ‘planar cell polarity’ pathway) and the
‘Wnt/Ca2+’ pathway (84-86).
In these three pathways, Dsh is a key transducer of the Wnt
signal.
Long intergenic non-coding RNA-p21 (lincRNA-p21)
inhibits Wnt/β-catenin signaling, which mediates the effects of
salvianolic acid B (Sal B) on HSC activation. The inhibitory
effects of Sal B on Wnt/β-catenin signaling are abolished by
lincRNA-p21 suppression. lincRNA-p21 regulated by miR-17-5p is an
inhibitor of miR-17-5p. Collectively, these results indicate that
lincRNA-p21 inhibits HSC activation through the
miR-17-5p/Wnt/β-catenin axis (107).
Small nuclear RNA host gene 7 (SNHG7) has been
reported to be an oncogene in various cancer types. Yu et al
(108) reported that the
expression levels of SNHG7 are upregulated in the liver tissue of
CCl4 mice and that silencing of SNHG7 inhibits HSCs.
Furthermore, SNHG7 is able to regulate miR-378a-3p. Downregulation
of miR-378a-3p reduces the effects of SNHG7 loss on HSC activation
(109). SNHG7 enhances
Wnt/β-catenin signaling, leading to LF, characterized by a decline
in the phosphorylated β-catenin level and enhancement of T cell
factor activity (110).
SNHG7-mediated activation of the Wnt/β-catenin pathway is regulated
by miR-378a-regulated disheveled segment polarity protein 2 (DVL2).
Therefore, miR-378a-3p is a regulator of DVL2. DVL2 inhibition
abolishes SNHG7-induced HSC activation. Collectively, these results
indicate that SNHG7 inhibits miR-378a-3p and moderates its effects
on DVL2, leading to enhanced Wnt/β-catenin signaling and thereby
promoting hepatic fibrosis.
The NF-κB pathway has a crucial role in innate and
adaptive immunity. NF-κB is a eukaryotic transcription factor that
exists in almost all cell types and is involved in various liver
pathologies (111). The survival
and activation of HSCs and hepatic MFs is modulated by NF-κB
(112). In 1986, NF-κB was
discovered to regulate immunoglobulin κ light chain expression in B
cells (113). NF-κB dimers are
sequestered in the cytoplasm by inhibitor of NF-κB proteins in most
resting cells (114). NF-κB
activation is associated with at least 2 signal transduction routes
named the canonical and non-canonical pathways (115).
In fibrotic livers and activated HSCs, the lncRNA
taurine upregulated gene 1 (TUG1) is highly expressed, unlike in
normal HCs. miR-29b has been confirmed as a TUG1-targeting miRNA
(127). Previous research has
demonstrated that miR-29b inhibits HSC activation and ameliorates
CCl4/BDL-induced LF, as well as human advanced hepatic
fibrosis (128). Treatment with a
miR-29b mimic eliminates the effects of TUG1 overexpression on
cellular physiological responses and inactivation of Janus
kinase/STAT and NF-κB signaling in LPS-pretreated H9c2 cells
(127). miR-29b downregulates
fibrogenic genes associated with the NF-kB and TGF-β pathways in
HSCs (129,130). Murine miR-29b suppresses the
expression of collagen in HSCs and is downregulated in activated
HSCs in a manner dependent on NF-kB and TGF-β signalling (131). Thus, TUG1 is a positive regulator
of profibrogenic gene expression in HSCs, as it downregulates
miR-29b in HSCs (132).
The Notch signaling pathway is an elementary and
highly conserved pathway associated with liver development,
physiology and pathophysiology (133,134). Early studies based on C.
elegans and Drosophila genetic models identified a gene
locus that was correlated with the phenotype of a mutant fly with a
wing indentation (18,135). This locus was indicated to
participate in cell fate processes during Drosophila
embryogenesis and was later named Notch. The Notch pathway consists
of receptors (Notch1-Notch4), ligands (δ-like 1, 3 and 4, as well
as Jagged 1 and 2), transcriptional complex components and
downstream genes, including hairy-enhancer of split (Hes)-related
with YRPW motif (Hey) and Hes (136,137). Notch signaling promotes LF by
regulating the inflammatory response and the function of
macrophages (138,139). Xie et al (140) reported that Notch-Hh crosstalk
influences the pathogenesis of cirrhosis by regulating MFs/HSCs
through EMT in vitro and in vivo. In a mouse model of
liver steatosis and LF, sustained Notch activation induces hepatic
fibrosis in the presence of high lipid levels and inhibition of
Notch signaling or a reduction in liver fat content may ameliorate
hepatic fibrosis (141,142).
In mouse models of liver fibrogenesis, the protein
and mRNA levels of Notch2, Notch3, Hes1 and Hey2 are increased in
lnc-LFAR1-overexpressing HSCs, while downregulation of lnc-LFAR1
reverses these effects (37). Hes
is considered the prototype Notch target gene and encodes a basic
helix-loop-helix inhibitory transcription factor involved in the
self-renewal of target cells by inhibiting differentiation
(143,144). Furthermore, lentivirus-mediated
knockdown of lnc-LFAR1 reduced the expression levels of Hey2,
Notch2 and Notch3. Hes1 inhibits CCl4/BDL-induced
expression of these genes; therefore, lnc-LFAR1 may activate HSCs
and subsequently accelerate LF through modulation of Notch
signaling.
Further signaling pathways are involved in LF.
Homeobox transcript antisense RNA (HOTAIR) expression is
upregulated in HSCs during LF. HOTAIR modulates PTEN levels and
contributes to the activation of the ERK and AKT pathways through
miR-29b (145). In addition, the
lncRNA growth arrest-specific transcript 5 inhibits LF by targeting
miR-23a through the PTEN/PI3K/Akt signaling pathway in a rat model
of CCl4-induced hepatic fibrosis (146). Future research is expected to
focus increasingly on the association of lncRNAs with LF.
Regeneration of damaged mature liver tissue is
driven by multiple signaling pathways, including the TGF-β/Smad,
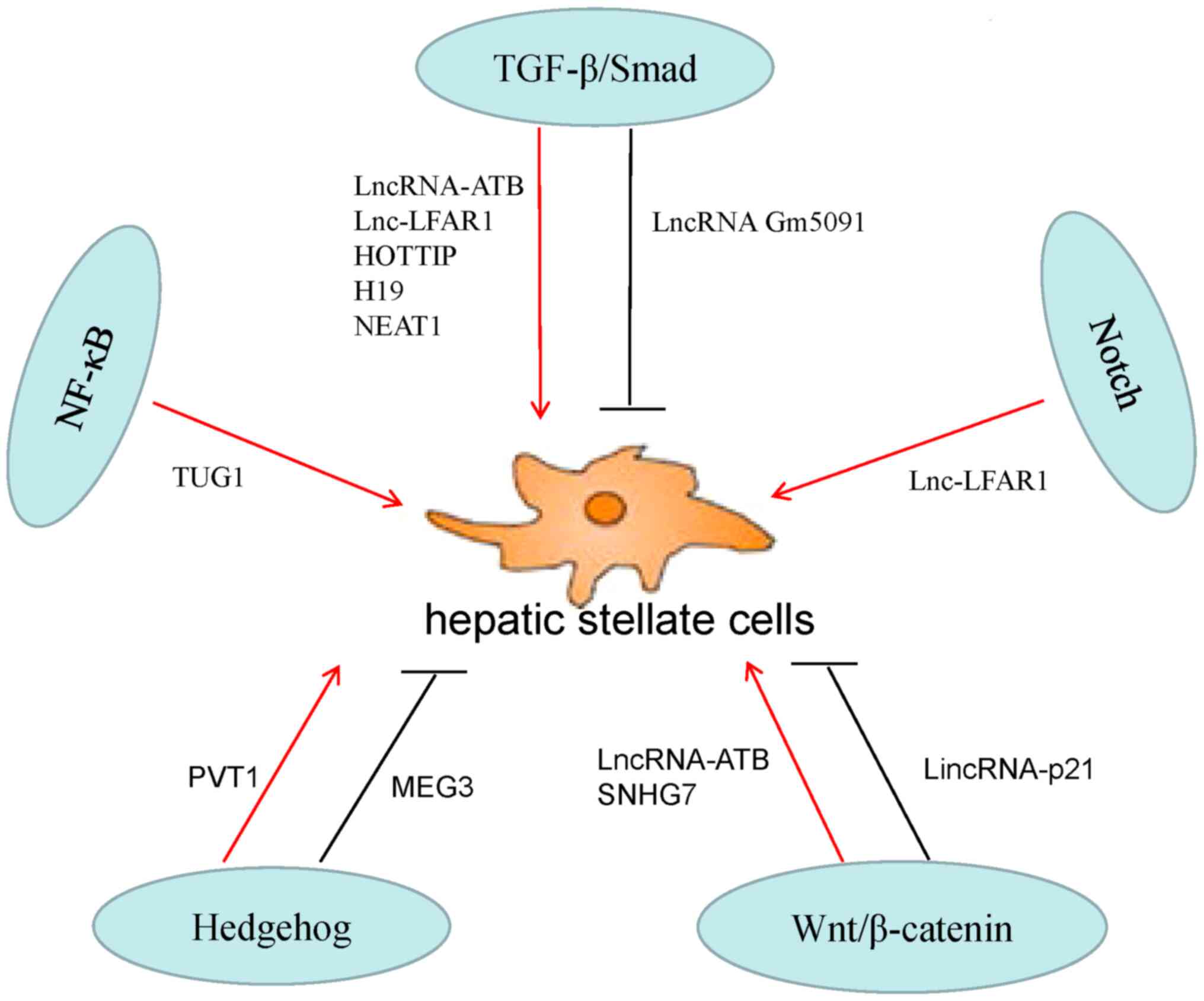
Hh, Wnt, NF-κB and Notch pathways. A summary is provided in
Table I and Fig. 1. The complex but delicate networks
interconnecting these molecular signals regulate cellular
proliferation, differentiation and apoptosis and thus the
pathological process of fibrosis. Due to the development of
high-throughput sequencing technologies, numerous lncRNAs have been
identified. These lncRNAs may act on oncogenes or tumor suppressors
and certain lncRNAs have been well characterized and proven to be
associated with LF. In the present review, these signals and
intracellular events were summarized that independently or
cooperatively drive HSC activation. The roles and possible
mechanisms of action of selected lncRNAs in LF were also reviewed.
The potential utility of lncRNAs as therapeutic agents and
biomarkers is promising, although the exact mechanisms of action
behind most lncRNAs remain elusive. Given the numerous potential
therapeutic anti-fibrosis strategies targeting factors that promote
fibrosis, combination therapies including these lncRNAs may produce
improved clinical outcomes. However, additional functions and
regulatory mechanisms of action of these lncRNAs require further
study prior to their use as clinically applicable biomolecules.
Not applicable.
Funding: This research was funded by the National Science and
Technology Major Project (no. 2017ZX10204401).
Not applicable.
ZW, SH and LT designed the article. ZW, SH, XZ and
SG wrote the first draft of the manuscript. QX, YG, JZ and BF
reviewed the literature. LT critically revised the manuscript. All
authors read and approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Friedman SL: Liver fibrosis-from bench to
bedside. J Hepatol. 38 (Suppl 1):S38–S53. 2003.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456.
2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cao L, Nicosia J, Larouche J, Zhang Y,
Bachman H, Brown AC, Holmgren L and Barker TH: Detection of an
integrin-binding mechanoswitch within fibronectin during tissue
formation and fibrosis. ACS Nano. 11:7110–7117. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kong D, Zhang F, Zhang Z, Lu Y and Zheng
S: Clearance of activated stellate cells for hepatic fibrosis
regression: Molecular basis and translational potential. Biomed
Pharmacother. 67:246–250. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Friedman SL: Fibrogenic cell reversion
underlies fibrosis regression in liver. Proc Natl Acad Sci USA.
109:9230–9231. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Schuppan D: Structure of the extracellular
matrix in normal and fibrotic liver: Collagens and glycoproteins.
Semin Liver Dis. 10:1–10. 1990.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Herrera J, Henke CA and Bitterman PB:
Extracellular matrix as a driver of progressive fibrosis. J Clin
Invest. 128:45–53. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Anthony PP, Ishak KG, Nayak NC, Poulsen
HE, Scheuer PJ and Sobin LH: The morphology of cirrhosis.
Recommendations on definition, nomenclature, and classification by
a working group sponsored by the World Health Organization. J Clin
Pathol. 31:395–414. 1978.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ginès P, Cárdenas A, Arroyo V and Rodés J:
Management of cirrhosis and ascites. N Engl J Med. 350:1646–1654.
2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhou WC, Zhang QB and Qiao L: Pathogenesis
of liver cirrhosis. World J Gastroenterol. 20:7312–7324.
2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Friedman SL, Roll FJ, Boyles J and Bissell
DM: Hepatic lipocytes: The principal collagen-producing cells of
normal rat liver. Proc Natl Acad Sci USA. 82:8681–8685.
1985.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Moreira RK: Hepatic stellate cells and
liver fibrosis. Arch Pathol Lab Med. 131:1728–1734. 2007.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yin C, Evason KJ, Asahina K and Stainier
DY: Hepatic stellate cells in liver development, regeneration, and
cancer. J Clin Invest. 123:1902–1910. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bataller R and Brenner DA: Hepatic
stellate cells as a target for the treatment of liver fibrosis.
Semin Liver Dis. 21:437–451. 2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tsuchida T and Friedman SL: Mechanisms of
hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol.
14:397–411. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lanzoni G, Cardinale V and Carpino G: The
hepatic, biliary, and pancreatic network of stem/progenitor cell
niches in humans: A new reference frame for disease and
regeneration. Hepatology. 64:277–286. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kitade M, Kaji K and Yoshiji H:
Relationship between hepatic progenitor cell-mediated liver
regeneration and non-parenchymal cells. Hepatol Res. 46:1187–1193.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Boulter L, Govaere O, Bird TG, Radulescu
S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, Van
Rooijen N, et al: Macrophage-derived Wnt opposes Notch signaling to
specify hepatic progenitor cell fate in chronic liver disease. Nat
Med. 18:572–579. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Carpino G, Renzi A, Franchitto A,
Cardinale V, Onori P, Reid L, Alvaro D and Gaudio E:
Stem/progenitor cell niches involved in hepatic and biliary
regeneration. Stem Cells Int. 2016(3658013)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Grimaldi V, De Pascale MR, Zullo A,
Soricelli A, Infante T, Mancini FP and Napoli C: Evidence of
epigenetic tags in cardiac fibrosis. J Cardiol. 69:401–408.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Khorkova O, Hsiao J and Wahlestedt C:
Basic biology and therapeutic implications of lncRNA. Adv Drug
Deliv Rev. 87:15–24. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
El Khodiry A, Afify M and El Tayebi HM:
Behind the curtain of non-coding RNAs; long non-coding RNAs
regulating hepatocarcinogenesis. World J Gastroenterol. 24:549–572.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jiang X, Lei R and Ning Q: Circulating
long noncoding RNAs as novel biomarkers of human diseases. Biomark
Med. 10:757–769. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hellerbrand C, Stefanovic B, Giordano F,
Burchardt ER and Brenner DA: The role of TGFbeta1 in initiating
hepatic stellate cell activation in vivo. J Hepatol. 30:77–87.
1999.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Inagaki Y and Okazaki I: Emerging insights
into Transforming growth factor beta Smad signal in hepatic
fibrogenesis. Gut. 56:284–292. 2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Dooley S and ten Dijke P: TGF-β in
progression of liver disease. Cell Tissue Res. 347:245–256.
2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fabregat I, Moreno-Càceres J, Sánchez A,
Dooley S, Dewidar B, Giannelli G and Ten Dijke P: IT-LIVER
Consortium. TGF-β signalling and liver disease. FEBS J.
283:2219–2232. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Breitkopf K, Godoy P, Ciuclan L, Singer MV
and Dooley S: TGF-beta/Smad signaling in the injured liver. Z
Gastroenterol. 44:57–66. 2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Border WA and Noble NA: Evidence that
TGF-beta should be a therapeutic target in diabetic nephropathy.
Kidney Int. 54:1390–1391. 1998.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Fu N, Niu X, Wang Y, Du H, Wang B, Du J,
Li Y, Wang R, Zhang Y, Zhao S, et al: Role of LncRNA-activated by
transforming growth factor beta in the progression of hepatitis C
virus-related liver fibrosis. Discov Med. 22:29–42. 2016.PubMed/NCBI
|
|
37
|
Zhang K, Han X, Zhang Z, Zheng L, Hu Z,
Yao Q, Cui H, Shu G, Si M, Li C, et al: The liver-enriched
lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch
pathways. Nat Commun. 8(144)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zheng J, Mao Y, Dong P, Huang Z and Yu F:
Long noncoding RNA HOTTIP mediates SRF expression through sponging
miR-150 in hepatic stellate cells. J Cell Mol Med. 23:1572–1580.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li Z, Wang J, Zeng Q, Hu C, Zhang J, Wang
H, Yan J, Li H and Yu Z: Long noncoding RNA HOTTIP promotes mouse
hepatic stellate cell activation via downregulating miR-148a. Cell
Physiol Biochem. 51:2814–2828. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Jung KH, Zhang J, Zhou C, Shen H, Gagea M,
Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK and Beretta L:
Differentiation therapy for hepatocellular carcinoma: Multifaceted
effects of miR-148a on tumor growth and phenotype and liver
fibrosis. Hepatology. 63:864–879. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Keniry A, Oxley D, Monnier P, Kyba M,
Dandolo L, Smits G and Reik W: The H19 lincRNA is a developmental
reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell
Biol. 14:659–665. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen X, Yamamoto M, Fujii K, Nagahama Y,
Ooshio T, Xin B, Okada Y, Furukawa H and Nishikawa Y: Differential
reactivation of fetal/neonatal genes in mouse liver tumors induced
in cirrhotic and non-cirrhotic conditions. Cancer Sci. 106:972–981.
2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang L, Zhou F, Drabsch Y, Gao R,
Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu
CX and ten Dijke P: USP4 is regulated by AKT phosphorylation and
directly deubiquitylates TGF-β type I receptor. Nat Cell Biol.
14:717–726. 2012.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhu J, Luo Z, Pan Y, Zheng W, Li W, Zhang
Z, Xiong P, Xu D, Du M, Wang B, et al: H19/miR-148a/USP4 axis
facilitates liver fibrosis by enhancing TGF-β signaling in both
hepatic stellate cells and hepatocytes. J Cell Physiol.
234:9698–9710. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhou B, Yuan W and Li X: LncRNA Gm5091
alleviates alcoholic hepatic fibrosis by sponging miR-27b/23b/24 in
mice. Cell Biol Int. 42:1330–1339. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Rogler CE, Matarlo JS, Kosmyna B, Fulop D
and Rogler LE: Knockdown of miR-23, miR-27, and miR-24 alters fetal
liver development and blocks fibrosis in mice. Gene Expr.
17:99–114. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhu D, Lyu L, Shen P, Wang J, Chen J, Sun
X, Chen L, Zhang L, Zhou Q and Duan Y: rSjP40 protein promotes
PPARγ expression in LX-2 cells through microRNA-27b. FASEB J.
32:4798–4803. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kong Y, Huang T, Zhang H, Zhang Q, Ren J,
Guo X, Fan H and Liu L: The lncRNA NEAT1/miR-29b/Atg9a axis
regulates IGFBPrP1-induced autophagy and activation of mouse
hepatic stellate cells. Life Sci. 237(116902)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yu F, Jiang Z, Chen B, Dong P and Zheng J:
NEAT1 accelerates the progression of liver fibrosis via regulation
of microRNA-122 and Kruppel-like factor 6. J Mol Med (Berl).
95:1191–1202. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zeng C, Wang YL, Xie C, Sang Y, Li TJ,
Zhang M, Wang R, Zhang Q, Zheng L and Zhuang SM: Identification of
a novel TGF-β-miR-122-fibronectin 1/serum response factor signaling
cascade and its implication in hepatic fibrogenesis. Oncotarget.
6:12224–12233. 2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kim Y, Ratziu V, Choi SG, Lalazar A,
Theiss G, Dang Q, Kim SJ and Friedman SL: Transcriptional
activation of transforming growth factor beta1 and its receptors by
the Kruppel-like factor Zf9/core promoter-binding protein and Sp1.
Potential mechanisms for autocrine fibrogenesis in response to
injury. J Biol Chem. 273:33750–33758. 1998.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ,
Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, et al: MicroRNA-122
plays a critical role in liver homeostasis and
hepatocarcinogenesis. J Clin Invest. 122:2884–2897. 2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhang K, Han Y, Hu Z, Zhang Z, Shao S, Yao
Q, Zheng L, Wang J, Han X, Zhang Y, et al: SCARNA10, a
nuclear-retained long non-coding RNA, promotes liver fibrosis and
serves as a potential biomarker. Theranostics. 9:3622–3638.
2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Tu X, Zhang H, Zhang J, Zhao S, Zheng X,
Zhang Z, Zhu J, Chen J, Dong L, Zang Y, et al: MicroRNA-101
suppresses liver fibrosis by targeting the TGFβ signalling pathway.
J Pathol. 234:46–59. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Rapicavoli NA, Qu K, Zhang J, Mikhail M,
Laberge RM and Chang HY: A mammalian pseudogene lncRNA at the
interface of inflammation and anti-inflammatory therapeutics.
Elife. 2(e00762)2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Nusslein-Volhard C and Wieschaus E:
Mutations affecting segment number and polarity in
Drosophila. Nature. 287:795–801. 1980.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Omenetti A, Choi S, Michelotti G and Diehl
AM: Hedgehog signaling in the liver. J Hepatol. 54:366–373.
2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Machado MV and Diehl AM: Hedgehog
signalling in liver pathophysiology. J Hepatol. 68:550–562.
2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Gorojankina T: Hedgehog signaling pathway:
A novel model and molecular mechanisms of signal transduction. Cell
Mol Life Sci. 73:1317–1332. 2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Sicklick JK, Li YX, Melhem A, Schmelzer E,
Zdanowicz M, Huang J, Caballero M, Fair JH, Ludlow JW, McClelland
RE, et al: Hedgehog signaling maintains resident hepatic
progenitors throughout life. Am J Physiol Gastrointest Liver
Physiol. 290:G859–G870. 2006.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Xie G, Choi SS, Syn WK, Michelotti GA,
Swiderska M, Karaca G, Chan IS, Chen Y and Diehl AM: Hedgehog
signalling regulates liver sinusoidal endothelial cell
capillarisation. Gut. 62:299–309. 2013.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Syn WK, Agboola KM, Swiderska M,
Michelotti GA, Liaskou E, Pang H, Xie G, Philips G, Chan IS, Karaca
GF, et al: NKT-associated hedgehog and osteopontin drive
fibrogenesis in non-alcoholic fatty liver disease. Gut.
61:1323–1329. 2012.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Yang L, Wang Y, Mao H, Fleig S, Omenetti
A, Brown KD, Sicklick JK, Li YX and Diehl AM: Sonic hedgehog is an
autocrine viability factor for myofibroblastic hepatic stellate
cells. J Hepatol. 48:98–106. 2008.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Gao L, Zhang Z, Zhang P, Yu M and Yang T:
Role of canonical Hedgehog signaling pathway in liver. Int J Biol
Sci. 14:1636–1644. 2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Chen Y, Choi SS, Michelotti GA, Chan IS,
Swiderska-Syn M, Karaca GF, Xie G, Moylan CA, Garibaldi F, Premont
R, et al: Hedgehog controls hepatic stellate cell fate by
regulating metabolism. Gastroenterology. 143:1319–1329.e11.
2012.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Briscoe J and Thérond PP: The mechanisms
of Hedgehog signalling and its roles in development and disease.
Nat Rev Mol Cell Biol. 14:416–429. 2013.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Syn WK, Jung Y, Omenetti A, Abdelmalek M,
Guy CD, Yang L, Wang J, Witek RP, Fearing CM, Pereira TA, et al:
Hedgehog-mediated epithelial-to-mesenchymal transition and
fibrogenic repair in nonalcoholic fatty liver disease.
Gastroenterology. 137:1478–1488.e8. 2009.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sicklick JK, Li YX, Choi SS, Qi Y, Chen W,
Bustamante M, Huang J, Zdanowicz M, Camp T, Torbenson MS, et al:
Role for hedgehog signaling in hepatic stellate cell activation and
viability. Lab Invest. 85:1368–1380. 2005.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Omenetti A, Yang L, Li YX, McCall SJ, Jung
Y, Sicklick JK, Huang J, Choi S, Suzuki A and Diehl AM:
Hedgehog-mediated mesenchymal-epithelial interactions modulate
hepatic response to bile duct ligation. Lab Invest. 87:499–514.
2007.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Choi SS, Syn WK, Karaca GF, Omenetti A,
Moylan CA, Witek RP, Agboola KM, Jung Y, Michelotti GA and Diehl
AM: Leptin promotes the myofibroblastic phenotype in hepatic
stellate cells by activating the hedgehog pathway. J Biol Chem.
285:36551–36560. 2010.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Choi SS, Omenetti A, Witek RP, Moylan CA,
Syn WK, Jung Y, Yang L, Sudan DL, Sicklick JK, Michelotti GA, et
al: Hedgehog pathway activation and epithelial-to-mesenchymal
transitions during myofibroblastic transformation of rat hepatic
cells in culture and cirrhosis. Am J Physiol Gastrointest Liver
Physiol. 297:G1093–G1106. 2009.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Zheng J, Yu F, Dong P, Wu L, Zhang Y, Hu Y
and Zheng L: Long non-coding RNA PVT1 activates hepatic stellate
cells through competitively binding microRNA-152. Oncotarget.
7:62886–62897. 2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Yang JJ, Tao H, Huang C, Shi KH, Ma TT,
Bian EB, Zhang L, Liu LP, Hu W, Lv XW and Li J: DNA methylation and
MeCP2 regulation of PTCH1 expression during rats hepatic fibrosis.
Cell Signal. 25:1202–1211. 2013.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Yu F, Lu Z, Chen B, Wu X, Dong P and Zheng
J: Salvianolic acid B-induced microRNA-152 inhibits liver fibrosis
by attenuating DNMT1-mediated Patched1 methylation. J Cell Mol Med.
19:2617–2632. 2015.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Yu F, Geng W, Dong P, Huang Z and Zheng J:
LncRNA-MEG3 inhibits activation of hepatic stellate cells through
SMO protein and miR-212. Cell Death Dis. 9(1014)2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Haertle L, Maierhofer A, Böck J, Lehnen H,
Böttcher Y, Blüuher M, Schorsch M, Potabattula R, El Hajj N,
Appenzeller S and Haaf T: Hypermethylation of the non-imprinted
maternal MEG3 and paternal MEST alleles is highly variable among
normal individuals. PLoS One. 12(e0184030)2017.PubMed/NCBI View Article : Google Scholar
|
|
78
|
He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu
BM, Xu FY, Zhang L, Lv XW and Li J: Inhibitory effects of long
noncoding RNA MEG3 on hepatic stellate cells activation and liver
fibrogenesis. Biochim Biophys Acta. 1842:2204–2215. 2014.PubMed/NCBI View Article : Google Scholar
|
|
79
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Bejsovec A: Wnt signaling: An
embarrassment of receptors. Curr Biol. 10:R919–R922.
2000.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Habas R and Dawid IB: Dishevelled and Wnt
signaling: Is the nucleus the final frontier? J Biol.
4(2)2005.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Miller JR, Hocking AM, Brown JD and Moon
RT: Mechanism and function of signal transduction by the
Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene. 18:7860–7872.
1999.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Kühl M, Sheldahl LC, Park M, Miller JR and
Moon RT: The Wnt/Ca2+ pathway: A new vertebrate Wnt
signaling pathway takes shape. Trends Genet. 16:279–283.
2000.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Veeman MT, Axelrod JD and Moon RT: A
second canon. Functions and mechanisms of beta-catenin-independent
Wnt signaling. Dev Cell. 5:367–377. 2003.PubMed/NCBI View Article : Google Scholar
|
|
86
|
van Amerongen R, Mikels A and Nusse R:
Alternative wnt signaling is initiated by distinct receptors. Sci
Signal. 1(re9)2008.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Monga SP: beta-catenin signaling and roles
in liver homeostasis, injury, and tumorigenesis. Gastroenterology.
148:1294–1310. 2015.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Rios-Esteves J and Resh MD: Stearoyl CoA
desaturase is required to produce active, lipid-modified Wnt
proteins. Cell Rep. 4:1072–1081. 2013.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Zhao C, Zhang M, Liu W, Wang C, Zhang Q
and Li W: β-catenin knockdown inhibits pituitary adenoma cell
proliferation and invasion via interfering with AKT and gelatinases
expression. Int J Oncol. 46:1643–1650. 2015.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Xu W and Kimelman D: Mechanistic insights
from structural studies of beta-catenin and its binding partners. J
Cell Sci. 120:3337–3344. 2007.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Zhu Y, Tan J, Xie H, Wang J, Meng X and
Wang R: HIF-1α regulates EMT via the Snail and beta-catenin
pathways in paraquat poisoning-induced early pulmonary fibrosis. J
Cell Mol Med. 20:688–697. 2016.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Thompson MD and Monga SP: WNT/beta-catenin
signaling in liver health and disease. Hepatology. 45:1298–1305.
2007.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Miller JR: The Wnts. Genome Biol.
3(REVIEWS3001)2002.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Miao CG, Yang YY, He X, Huang C, Huang Y,
Zhang L, Lv XW, Jin Y and Li J: Wnt signaling in liver fibrosis:
Progress, challenges and potential directions. Biochimie.
95:2326–2335. 2013.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Ge WS, Wang YJ, Wu JX, Fan JG, Chen YW and
Zhu L: β-catenin is overexpressed in hepatic fibrosis and blockage
of Wnt/β-catenin signaling inhibits hepatic stellate cell
activation. Mol Med Rep. 9:2145–2151. 2014.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Osawa Y, Oboki K, Imamura J, Kojika E,
Hayashi Y, Hishima T, Saibara T, Shibasaki F, Kohara M and Kimura
K: Inhibition of cyclic adenosine monophosphate (cAMP)-response
element-binding protein (CREB)-binding protein (CBP)/β-catenin
reduces liver fibrosis in mice. EBioMedicine. 2:1751–1758.
2015.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Kordes C, Sawitza I and Haussinger D:
Canonical Wnt signaling maintains the quiescent stage of hepatic
stellate cells. Biochem Biophys Res Commun. 367:116–123.
2008.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Yin X, Yi H, Wang L, Wu W, Wu X and Yu L:
RSPOs facilitated HSC activation and promoted hepatic fibrogenesis.
Oncotarget. 7:63767–63778. 2016.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Corbett L, Mann J and Mann DA:
Non-canonical Wnt predominates in activated rat hepatic stellate
cells, influencing HSC survival and paracrine stimulation of
kupffer cells. PLoS One. 10(e0142794)2015.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Chatani N, Kamada Y, Kizu T, Ogura S,
Furuta K, Egawa M, Hamano M, Ezaki H, Kiso S, Shimono A, et al:
Secreted frizzled-related protein 5 (Sfrp5) decreases hepatic
stellate cell activation and liver fibrosis. Liver Int.
35:2017–2026. 2015.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y
and Jiang X: LncRNA-ATB: An indispensable cancer-related long
noncoding RNA. Cell Prolif. 50(e12381)2017.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-beta promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Liu J, Ruan B, You N, Huang Q, Liu W, Dang
Z, Xu W, Zhou T, Ji R, Cao Y, et al: Downregulation of miR-200a
induces EMT phenotypes and CSC-like signatures through targeting
the β-catenin pathway in hepatic oval cells. PLoS One.
8(e79409)2013.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Su J, Zhang A, Shi Z, Ma F, Pu P, Wang T,
Zhang J, Kang C and Zhang Q: MicroRNA-200a suppresses the
Wnt/β-catenin signaling pathway by interacting with β-catenin. Int
J Oncol. 40:1162–1170. 2012.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Fu N, Zhao SX, Kong LB, Du JH, Ren WG, Han
F, Zhang QS, Li WC, Cui P, Wang RQ, et al:
LncRNA-ATB/microRNA-200a/β-catenin regulatory axis involved in the
progression of HCV-related hepatic fibrosis. Gene. 618:1–7.
2017.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Yu F, Zhou G, Huang K, Fan X, Li G, Chen
B, Dong P and Zheng J: Serum lincRNA-p21 as a potential biomarker
of liver fibrosis in chronic hepatitis B patients. J Viral Hepat.
24:580–588. 2017.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Yu F, Dong P, Mao Y, Zhao B, Huang Z and
Zheng J: Loss of lncRNA-SNHG7 Promotes the Suppression of Hepatic
Stellate Cell Activation via miR-378a-3p and DVL2. Mol Ther Nucleic
Acids. 17:235–244. 2019.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Chen W, Zhao W, Yang A, Xu A, Wang H, Cong
M, Liu T, Wang P and You H: Integrated analysis of microRNA and
gene expression profiles reveals a functional regulatory module
associated with liver fibrosis. Gene. 636:87–95. 2017.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Yao X, Liu C, Liu C, Xi W, Sun S and Gao
Z: lncRNA SNHG7 sponges miR-425 to promote proliferation,
migration, and invasion of hepatic carcinoma cells via
Wnt/β-catenin/EMT signalling pathway. Cell Biochem Funct.
37:525–533. 2019.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Chakraborty JB and Mann DA: NF-kappaB
signalling: Embracing complexity to achieve translation. J Hepatol.
52:285–291. 2010.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Ghosh S and Karin M: Missing pieces in the
NF-kappaB puzzle. Cell. 109 (Suppl):S81–S96. 2002.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Sen R and Baltimore D: Multiple nuclear
factors interact with the immunoglobulin enhancer sequences. Cell.
46:705–716. 1986.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Taniguchi K and Karin M: NF-κB,
inflammation, immunity and cancer: Coming of age. Nat Rev Immunol.
18:309–324. 2018.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Gilmore TD: Introduction to NF-kappaB:
Players, pathways, perspectives. Oncogene. 25:6680–6684.
2006.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Olefsky JM and Glass CK: Macrophages,
inflammation, and insulin resistance. Annu Rev Physiol. 72:219–246.
2010.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Luedde T and Schwabe RF: NF-κB in the
liver-linking injury, fibrosis and hepatocellular carcinoma. Nat
Rev Gastroenterol Hepatol. 8:108–118. 2011.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Duffield JS, Forbes SJ, Constandinou CM,
Clay S, Partolina M, Vuthoori S, Wu S, Lang R and Iredale JP:
Selective depletion of macrophages reveals distinct, opposing roles
during liver injury and repair. J Clin Invest. 115:56–65.
2005.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Pradere JP, Kluwe J, De Minicis S, Jiao
JJ, Gwak GY, Dapito DH, Jang MK, Guenther ND, Mederacke I, Friedman
R, et al: Hepatic macrophages but not dendritic cells contribute to
liver fibrosis by promoting the survival of activated hepatic
stellate cells in mice. Hepatology. 58:1461–1473. 2013.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Lv P, Luo HS, Zhou XP, Xiao YJ, Paul SC,
Si XM and Zhou YH: Reversal effect of thalidomide on established
hepatic cirrhosis in rats via inhibition of nuclear
factor-kappaB/inhibitor of nuclear factor-kappaB pathway. Arch Med
Res. 38:15–27. 2007.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Oakley F, Meso M, Iredale JP, Green K,
Marek CJ, Zhou X, May MJ, Millward-Sadler H, Wright MC and Mann DA:
Inhibition of inhibitor of kappaB kinases stimulates hepatic
stellate cell apoptosis and accelerated recovery from rat liver
fibrosis. Gastroenterology. 128:108–120. 2005.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Wright MC, Issa R, Smart DE, Trim N,
Murray GI, Primrose JN, Arthur MJ, Iredale JP and Mann DA:
Gliotoxin stimulates the apoptosis of human and rat hepatic
stellate cells and enhances the resolution of liver fibrosis in
rats. Gastroenterology. 121:685–698. 2001.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Seki E, De Minicis S, Osterreicher CH,
Kluwe J, Osawa Y, Brenner DA and Schwabe RF: TLR4 enhances TGF-beta
signaling and hepatic fibrosis. Nat Med. 13:1324–1332.
2007.PubMed/NCBI View
Article : Google Scholar
|
|
124
|
Sunami Y, Leithauser F, Gul S, Fiedler K,
Guldiken N, Espenlaub S, Holzmann KH, Hipp N, Sindrilaru A, Luedde
T, et al: Hepatic activation of IKK/NFκB signaling induces liver
fibrosis via macrophage-mediated chronic inflammation. Hepatology.
56:1117–1128. 2012.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Shen H, Sheng L, Chen Z, Jiang L, Su H,
Yin L, Omary MB and Rui L: Mouse hepatocyte overexpression of
NF-κB-inducing kinase (NIK) triggers fatal macrophage-dependent
liver injury and fibrosis. Hepatology. 60:2065–2076.
2014.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Son G, Iimuro Y, Seki E, Hirano T, Kaneda
Y and Fujimoto J: Selective inactivation of NF-kappaB in the liver
using NF-kappaB decoy suppresses CCl4-induced liver injury and
fibrosis. Am J Physiol Gastrointest Liver Physiol. 293:G631–G639.
2007.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Zhang H, Li H, Ge A, Guo E, Liu S and
Zhang L: Long non-coding RNA TUG1 inhibits apoptosis and
inflammatory response in LPS-treated H9c2 cells by down-regulation
of miR-29b. Biomed Pharmacother. 101:663–669. 2018.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Roderburg C, Urban GW, Bettermann K, Vucur
M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi
M, et al: Micro-RNA profiling reveals a role for miR-29 in human
and murine liver fibrosis. Hepatology. 53:209–218. 2011.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Sekiya Y, Ogawa T, Yoshizato K, Ikeda K
and Kawada N: Suppression of hepatic stellate cell activation by
microRNA-29b. Biochem Biophys Res Commun. 412:74–79.
2011.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Ogawa T, Iizuka M, Sekiya Y, Yoshizato K,
Ikeda K and Kawada N: Suppression of type I collagen production by
microRNA-29b in cultured human stellate cells. Biochem Biophys Res
Commun. 391:316–321. 2010.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Xing TJ, Jiang DF, Huang JX and Xu ZL:
Expression and clinical significance of miR-122 and miR-29 in
hepatitis B virus-related liver disease. Genet Mol Res.
13:7912–7918. 2014.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Han X, Hong Y and Zhang K: TUG1 is
involved in liver fibrosis and activation of HSCs by regulating
miR-29b. Biochem Biophys Res Commun. 503:1394–1400. 2018.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Geisler F and Strazzabosco M: Emerging
roles of Notch signaling in liver disease. Hepatology. 61:382–392.
2015.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Morell CM and Strazzabosco M: Notch
signaling and new therapeutic options in liver disease. J Hepatol.
60:885–890. 2014.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Siebel C and Lendahl U: Notch signaling in
development, tissue homeostasis, and disease. Physiol Rev.
97:1235–1294. 2017.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Wakabayashi N, Chartoumpekis DV and
Kensler TW: Crosstalk between Nrf2 and Notch signaling. Free Radic
Biol Med. 88:158–167. 2015.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Ni MM, Wang YR, Wu WW, Xia CC, Zhang YH,
Xu J, Xu T and Li J: Novel Insights on Notch signaling pathways in
liver fibrosis. Eur J Pharmacol. 826:66–74. 2018.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Kimball AS, Joshi AD, Boniakowski AE,
Schaller M, Chung J, Allen R, Bermick J, Carson WF IV, Henke PK,
Maillard I, et al: Notch regulates macrophage-mediated inflammation
in diabetic wound healing. Front Immunol. 8(635)2017.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Wang T, Xiang Z, Wang Y, Li X, Fang C,
Song S, Li C, Yu H, Wang H, Yan L, et al: (-)-Epigallocatechin
gallate targets notch to attenuate the inflammatory response in the
immediate early stage in human macrophages. Front Immunol.
8(433)2017.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Xie G, Karaca G, Swiderska-Syn M,
Michelotti GA, Kruger L, Chen Y, Premont RT, Choi SS and Diehl AM:
Cross-talk between Notch and Hedgehog regulates hepatic stellate
cell fate in mice. Hepatology. 58:1801–1813. 2013.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Romeo S: Notch and nonalcoholic fatty
liver and fibrosis. N Engl J Med. 380:681–683. 2019.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Zhu C, Kim K, Wang X, Bartolome A, Salomao
M, Dongiovanni P, Meroni M, Graham MJ, Yates KP, Diehl AM, et al:
Hepatocyte Notch activation induces liver fibrosis in nonalcoholic
steatohepatitis. Sci Transl Med. 10(eaat0344)2018.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Iso T, Kedes L and Hamamori Y: HES and
HERP families: Multiple effectors of the Notch signaling pathway. J
Cell Physiol. 194:237–255. 2003.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Kageyama R, Ohtsuka T, Hatakeyama J and
Ohsawa R: Roles of bHLH genes in neural stem cell differentiation.
Exp Cell Res. 306:343–348. 2005.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Yu F, Chen B, Dong P and Zheng J: HOTAIR
epigenetically modulates PTEN expression via MicroRNA-29b: A novel
mechanism in regulation of liver fibrosis. Mol Ther. 25:205–217.
2017.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Dong Z, Li S, Wang X, Si L, Ma R, Bao L
and Bo A: lncRNA GAS5 restrains CCl4-induced hepatic fibrosis by
targeting miR-23a through the PTEN/PI3K/Akt signaling pathway. Am J
Physiol Gastrointest Liver Physiol. 316:G539–G550. 2019.PubMed/NCBI View Article : Google Scholar
|