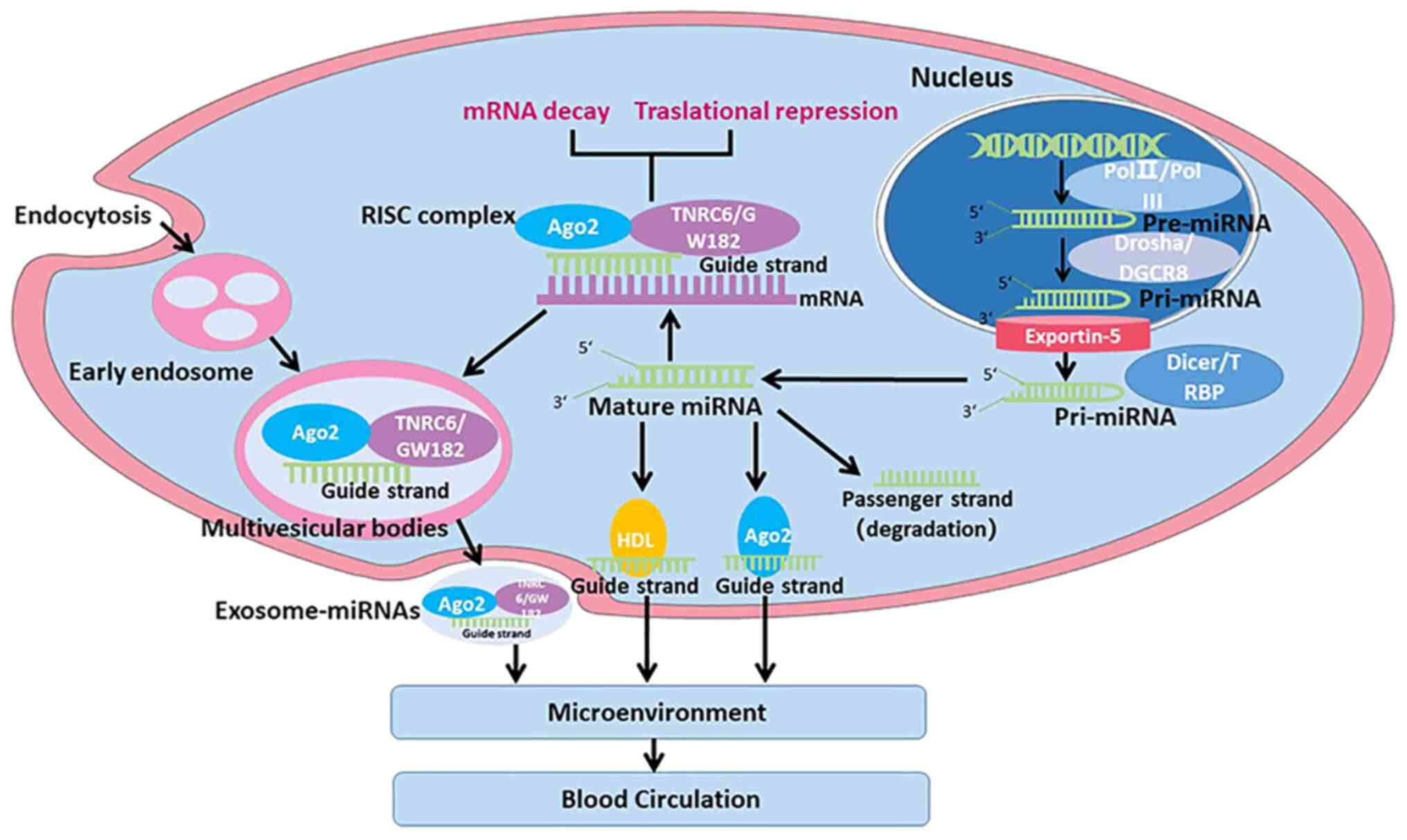
Cachexia is a complex syndrome featuring loss of
weight that results from reduced skeletal muscle mass (1). This syndrome usually appears in the
late stages of severe illnesses, including cancer, kidney disease,
human immunodeficiency virus, congestive heart failure and chronic
obstructive pulmonary disease (2,3).
Patients with cachexia are insensitive to treatment, have a low
quality of life and have a high mortality rate (4).
Cancer cachexia affects 50% of patients with cancer
and causes ~40% of cancer-associated mortalities (5). The incidence of cancer cachexia
changes with the stage and type of cancer (6). According to a previous cohort study
on patients with advanced tumors, those with pancreatic cancer are
at the greatest risk of developing cancer cachexia (~70%), followed
by colorectal, gastroesophageal, and head and neck cancer (~45%)
(7), while patients with breast
and prostate cancer are at the lowest risk of developing cachexia
(20-30%) (7). In addition, cancer
cachexia may result in inefficient chemotherapy, increased
treatment interruptions or decreased survival rates (8).
The diagnostic standard of cachexia is loss of
weight >5% or >2% among patients who have a body mass index
(BMI) less than 20 kg/m2 (9). In addition, neuroendocrine changes
occur in patients with cancer cachexia, leading to early satiety
and food aversion (10). The
Warburg effect is the catabolism of glucose to lactate to obtain
adenosine triphosphate (11).
Lactate is converted to glucose in the liver at a cost of energy.
When glucose is released into the bloodstream, cancer cells may use
it again for glycolysis. The Cori cycle is a fruitless
glucose-lactate shuttle that increases energy expenditure and
hepatic gluconeogenesis (12). As
a result, catabolic metabolism in fat and skeletal muscle provides
additional glucose precursors for gluconeogenesis. In cachexia, the
Warburg effect in myocytes contributes to muscle mass reduction
(13). Reduced food absorption and
excessive metabolism eventually lead to a negative energy balance
and mass loss, particularly skeletal muscle mass loss (5). Decreased skeletal muscle mass and
muscle function are found to negatively influence the life quality
among patients with cancer cachexia and have recently been widely
referred to as ‘sarcopenia’ (14,15).
Cancer cachexia may subsequently progress to refractory cachexia,
and interventions at such stage are unlikely to be successful.
Currently, there are limited options for the
treatment of cancer cachexia. There are two therapeutic concepts:
i) non-pharmacological options, which are focused on nutrition and
exercise interventions (3,16); and ii) chemotherapy, including the
usage of hormone therapy (e.g. gonadotropins), myostatin inhibitors
and anti-inflammatory drugs (17).
However, the effectiveness of these treatments remains unclear, as
clinical outcomes and long-term efficacy reports are insufficient
(18). Therefore, novel early
diagnostic biomarkers and therapeutic targets for cancer cachexia
are needed (19).
Several microRNAs (miRNAs or miRs), such as
let-7d-3p and miR-345-5p, were found to be markedly dysregulated
among patients with cachexia (6,20).
Furthermore, several miRNAs have been found to have a regulatory
effect on inflammatory pathways, and on the degradation and
synthesis of proteins in skeletal muscle, which makes miRNAs
potential novel therapeutic candidates in cancer cachexia therapy
(21,22). The present review summarizes miRNAs
differentially expressed in specimens derived from patients with
cancer cachexia, including muscle, adipose tissue and blood. In
addition, the present review proposes that miRNAs may be considered
as potential diagnostic markers or therapeutic targets for cancer
cachexia.
miRNAs are short RNAs that can regulate the
expression of ~60% of protein-encoding genes of human mRNAs
(23). miRNAs were firstly
identified in 1993, and additional types of miRNAs have been
identified and studied since then (24). The miRBase database contains
published miRNA sequences, and the up-to-date version of this
database contains >2,570 mature miRNAs from humans (25). The majority of miRNAs can be
transcribed by RNA polymerase (pol) II or pol III in the nucleus to
produce primary precursor miRNAs (pri-miRNAs) (60-100 nt) (Fig. 1) (26). The Drosha/DiGeorge critical region
8 ribonuclease complex divides pri-miRNAs to generate precursor
pre-miRNAs, which are later exported to the cytoplasm via the
exportin-5 complex (27). The
Dicer/TAR-RNA binding protein complex subsequently divides
pre-miRNAs to produce mature double-stranded miRNAs (28). To become functional,
double-stranded miRNAs are then disassembled to produce passenger
and guide strands. The passenger strand is degraded, while the
guide strand is loaded onto the RNA-induced silencing complex
(29,30). The primary function of miRNAs is to
inhibit the translation of target mRNAs.
miRNA expression profiling shows that changes in
miRNA expression are associated with various illnesses, including
primary muscle diseases, dexamethasone-induced atrophy, diabetes
and wasting diseases (such as cancer cachexia) (31,32).
In addition, various aspects of metabolic changes and inflammatory
responses are also regulated by miRNAs (33-35).
Hypermetabolism and systemic inflammation are typical symptoms of
cancer cachexia (36). Therefore,
miRNAs possibly impact cancer cachexia pathogenesis.
Cancer cells may produce inflammatory cytokines and
cause local and systemic inflammation in the host (37,38).
Previous studies have demonstrated that the tumor itself may be
capable of secreting exosomes containing miRNAs (39-42),
which can increase the synthesis of circulating inflammatory
factors (39). The levels of
circulating inflammatory cytokines, including tumor necrosis
factor-α (TNF-α), interferon-γ (IFN-γ), interleukin 1 (IL-1) and
IL-6, can be also altered in patients with cachexia (43,44).
miRNAs can be transported via exosomes, which can be secreted into
the serum, cerebrospinal fluid, urine and saliva (45). Exosomes from adipose tissue in the
tumor microenvironment may also promote the development of systemic
inflammation (46,47).
miR-182-5p, miR-183-5p, miR-21-5p, the miR-200
family, miR-7-5p, miR-125b-5p, miR-96-5p, miR-139-5p, miR-99a-5p,
miR-497-5p and miR-486-5p have been found to be altered in breast
cancer (BC) (48). A total of 26
differentially expressed miRNAs were found to interact with
frequently deregulated genes known to be involved in colorectal
cancer pathways (49). The
majority of these miRNAs could predict the prognosis of patients
with colorectal cancer in stages II and III (49). It has been demonstrated that miRNAs
can be used for the early detection of oral cancer (50). A total of 9 differentially
expressed miRNAs (miR-486-1, miR-486-2, miR-153, miR-210, miR-9-1,
miR-9-2, miR-9-3, miR-577 and miR-4732) have been identified, which
could be used as lung adenocarcinoma diagnostic biomarkers
(51).
In addition, miRNAs may have a prognostic value for
patients treated with a combination of interventions, including
diet and physical activity (48).
Differentially expressed extracellular vesicle (EV) miRNAs
resulting from the Mediterranean diet may be engaged in pathways
associated with cardiometabolic risk factors in overweight BC
survivors (52). In addition,
environmental factors such as pesticides may modify miRNA
expression and the DNA methylation status (53). Alteration of miRNA expression
profiles upon exposure to naturally occurring asbestiform fibers is
a diagnostic indicator of mesothelial neoplastic transformation
(54). In patients with colon
cancer, vascular endothelial growth factor (VEGF) may be an
independent predictor of weight loss (55). VEGF promotes the proliferation,
migration and tube formation of endothelial cells (ECs), and has
become a primary target of anti-angiogenic therapy (56-59).
Furthermore, VEGF is linked to systemic inflammation and
malnutrition, supporting the possible involvement of VEGF in cancer
cachexia pathogenesis (55). VEGF
is required for tumor angiogenesis, and inhibition of VEGF inhibits
angiogenesis and tumor growth (57,60-62).
miRNAs promote angiogenesis by facilitating the proliferation and
migration of ECs (63). The
hypoxia inducible factor-1α/VEGF signaling pathways regulated by
miR-210, miR-21 and miR-126 play a role in colon cancer initiation
(64). Overexpression of miR-638
could inhibit angiogenesis and tumor growth in hepatocellular
carcinoma by suppressing VEGF signaling (65). miRNAs produced from tumor cells,
such as miR-23a, miR-494 and miR-210, were reported to be packaged
into EVs and transported to recipient ECs (66). These miRNAs promote angiogenesis by
facilitating the proliferation and migration of ECs (63).
Patients with cancer cachexia can lose ≤75% of their
skeletal muscle mass, which may lead to poor prognosis and higher
mortality associated with cancer (67). Muscle protein degradation in cancer
cachexia is mediated mainly by the ubiquitin proteasome system,
induced by activation of E3 ligands (68). The Fork head box O (FoxO) signaling
pathway is involved in this process by inducing the transcription
of E3 ubiquitin ligases, of which there are three members in
skeletal muscle: FoxO3, FoxO1 and FoxO4(68). Inhibition of FoxO transcriptional
activity attenuates muscle fiber atrophy during cachexia (69). miRNA-486 reduces FoxO1 protein
expression and enhances FoxO1 phosphorylation to inhibit E3
ubiquitin ligase (70). miR-21
associates with and activates Toll-like receptor 7, which induces
apoptosis in muscle cells via the c-Jun N-terminal kinase pathway,
leading to atrophy (18).
Dysregulated expression of miRNAs (such as
myomiRNAs, a subset of miRNAs with high expression in skeletal
muscle) is associated with muscle atrophy, which is a hallmark of
cancer cachexia (71-74).
The expression profile of miRNAs in rectus abdominis muscle samples
was evaluated among patients with cancer who exhibited or not a
cachexia syndrome (6). In that
study, 8 miRNAs were upregulated among patients with cancer
cachexia, including let-7d-3p, miR-423-5p, miR-345-5p, miR-532-5p,
miR-3184-3p, miR-1296-5p, miR-423-3p and miR-199a-3p (6). Pathway analysis indicated that the
target miRNAs were enriched in the adipogenesis, myogenesis,
inflammation and innate immune response pathways (6). In another study, the expression
levels of 754 miRNAs in broad fascia biopsies of 8 healthy
individuals and 8 patients with non-small cell lung cancer who
exhibited cachexia were investigated (75). The expression of 28 miRNAs was
significantly changed, with 23 miRNAs being downregulated and 5
upregulated (75). In addition,
the genes of TNF, transforming growth factor-β, IL-6 and insulin
are among the 158 putative target genes identified using miRTarBase
(75). A total of 9 miRNAs were
found to be differentially expressed in muscles of a cancer
cachexia mouse model (20).
miRNA-mRNA co-sequencing revealed activation of the atrophy-related
transcription factors STAT3, NF-κB and FoxO, thus exposing
transcriptional and post-transcriptional regulatory networks
involved in muscle wasting (76).
The hallmarks of cancer cachexia are muscle loss,
browning of white adipose tissue (WAT) and lipolysis (77,78).
Increased levels of circulating inflammatory cytokines can also
induce lipolysis and proteolysis in adipose tissue and muscle,
respectively, as well as downregulate protein synthesis, which
causes a reduction in skeletal muscle mass and adipose tissue in
patients with cancer cachexia (21). WAT can promote the circulation of
inflammatory cytokines as well as regulate inflammatory processes
in immune cells and tissues by secreting miRNA-containing exosomes
(79-81).
miR-483-5p, miR-744, miR-23a and miR-99b were found to be
downregulated in the abdomen subcutaneous adipose tissue of
patients with gastrointestinal cancer and cachexia in contrast to
those of patients without cachexia syndrome, while the expression
of miR-378 was upregulated (82).
miRNAs in blood may serve as non-invasive biomarkers of cancer
malignancy, and miRNAs can remain highly stable in blood.
Exosomes are the most common type of EVs, which are
small membrane-bound vesicles between 30 and 150 nm in diameter
(89). The presence of miRNA-rich
circulating exosomes may promote the development and maintenance of
systemic chronic inflammation in patients with cancer cachexia
(21,89). Furthermore, a previous study
reported the upregulation of miR-155 in exosomes of BC cells (4T1),
which can target peroxisome proliferator-activated receptor-γ in
adipocytes, and promote adipocyte metabolism and browning
differentiation (90). In
conclusion, tumor-derived exosomal miRNAs may induce cancer
cachexia, and therefore exosomal miRNAs are considered potential
early diagnostic markers of cancer cachexia (90-94).
Dysregulation of specific miRNAs, such as let-7d-3p,
miR-345-5p, miR-532-5p, miR-378, miR-92a-3p, miR-21, is involved in
the development of cachexia. Cachexia may induce the differential
expression of miRNAs but it has not been validated. Dysregulated
expression of miRNAs was observed in muscle tissue, adipose tissue
and blood specimens from patients with cancer cachexia in contrast
to the findings in patients who did not exhibit cancer cachexia or
in healthy controls (Table I)
(6,75,82,87,88,95-97).
However, miRNAs directly obtained from adipose or muscle tissue
biopsies are not applicable as diagnostic markers of cancer
cachexia (84). Thus, the
diagnostic value of miRNAs for cancer cachexia should be restricted
to circulating miRNAs. miRNAs with high stability in body fluids
can be potentially used as non-invasive markers (98,99).
miRNAs from plasma/serum have been reported as biomarkers for the
early diagnosis of different types of tumor, including gastric
cancer (100), BC (101) and pancreatic cancer (102). Therefore, it can be proposed that
circulating miRNAs in the blood can be used as biomarkers to
differentiate patients at risk of developing cancer cachexia. For
example, circulating miRNAs such as miR-21 may serve as markers for
diagnosing cancer cachexia among patients likely to develop
colorectal cancer (88). However,
the application of using circulating miRNAs in patients with cancer
as biomarkers for diagnosis needs to be validated in future
clinical trials.
Multiple characteristics of miRNAs make them
potential targets for new treatments of cancer cachexia. Firstly,
miRNAs regulate the translation of mRNAs belonging to multiple
genes and signaling pathways that are dysregulated in cancer
cachexia, such as TNF, IFN signaling, STAT and NF-κB transcription
factors and associated target genes (15,103-105).
Secondly, miRNAs have been used to promote muscle development and
maintain muscle homeostasis (106). The expression of multiple miRNAs
has been found to be dysregulated in muscle wasting of cachexia
(107). Thirdly, treatment of
cancer cachexia with miRNAs can induce reversible and specific
changes in gene regulation without affecting the DNA (108). miRNAs can be used as knockdown
complementary mRNA targets (103). In knockdown therapy,
complement-specific miRNA drugs compete with their mRNA targets for
translation. Fourthly, EVs can prevent miRNAs from being degraded
in transfer and expedite their uptake via target cells (109,110). Finally, miRNAs can be efficiently
stabilized or concentrated using novel processing methods (103,111). However, no miRNA drugs have been
clinically used to date, although there are several ongoing
clinical trials on phases 1 and 2(112). For example, a phase I clinical
study that applied miR-16 mimics for the treatment of non-small
cell lung cancer or mesothelioma was accomplished, and may be
followed up by a phase II study (113). miRNAs have also been adopted for
targeting serum amyloid 1 and 2, which are lipoproteins usually
generated in response to inflammatory cytokines, and were shown to
successfully relieve muscle atrophy in a pre-clinical mouse model
(114). miRNA mimics already used
in clinical studies for cancer therapy, such as miR-16, can be
investigated in animal models of cancer cachexia to evaluate
whether they can improve weight loss and alleviate cancer cachexia
symptoms. The implications of miRNAs in the pathogenesis of cancer
cachexia make them attractive therapeutic targets. In addition,
miRNA-based therapies for cancer cachexia target specific pathways
that have the potential to restore homeostasis in chronically
dysfunctional networks and enable positive muscle responses to
exercise and diet.
Not applicable.
Funding: This study is supported by Shandong Provincial
Administration of Traditional Chinese Medicine (grant no. 2017 218)
and Department of Science and Technology of Shandong Province
(2019GSF108234).
Not applicable.
ZL conceived and designed the review. XL, LD and QL
wrote the first draft of the manuscript in light of the literature
data. Data authentication is not applicable. All authors
contributed to the article and approved the submitted version for
publication.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Evans WJ, Morley JE, Argilés J, Bales C,
Baracos V, Guttridge D, Jatoi A, Kalantar-Zadeh K, Lochs H,
Mantovani G, et al: Cachexia: A new definition. Clin Nutr.
27:793–799. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Holecek M: Muscle wasting in animal models
of severe illness. Int J Exp Pathol. 93:157–171. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Argilés JM, Busquets S, Stemmler B and
López-Soriano FJ: Cancer cachexia: Understanding the molecular
basis. Nat Rev Cancer. 14:754–762. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Nixon DW, Heymsfield SB, Cohen AE, Kutner
MH, Ansley J, Lawson DH and Rudman D: Protein-calorie
undernutrition in hospitalized cancer patients. Am J Med.
68:683–690. 1980.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fearon KC, Glass DJ and Guttridge DC:
Cancer cachexia: Mediators, signaling, and metabolic pathways. Cell
Metab. 16:153–166. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Narasimhan A, Ghosh S, Stretch C, Greiner
R, Bathe OF, Baracos V and Damaraju S: Small RNAome profiling from
human skeletal muscle: Novel miRNAs and their targets associated
with cancer cachexia. J Cachexia Sarcopenia Muscle. 8:405–416.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Anker MS, Holcomb R, Muscaritoli M, von
Haehling S, Haverkamp W, Jatoi A, Morley JE, Strasser F, Landmesser
U, Coats AJS and Anker SD: Orphan disease status of cancer cachexia
in the USA and in the European Union: A systematic review. J
Cachexia Sarcopenia Muscle. 10:22–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Caillet P, Liuu E, Raynaud Simon A,
Bonnefoy M, Guerin O, Berrut G, Lesourd B, Jeandel C, Ferry M,
Rolland Y and Paillaud E: Association between cachexia,
chemotherapy and outcomes in older cancer patients: A systematic
review. Clin Nutr. 36:1473–1482. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fearon K, Strasser F, Anker SD, Bosaeus I,
Bruera E, Fainsinger RL, Jatoi A, Loprinzi C, MacDonald N,
Mantovani G, et al: Definition and classification of cancer
cachexia: An international consensus. Lancet Oncol. 12:489–495.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Thoresen L, Frykholm G, Lydersen S,
Ulveland H, Baracos V, Prado CM, Birdsell L and Falkmer U:
Nutritional status, cachexia and survival in patients with advanced
colorectal carcinoma. Different assessment criteria for nutritional
status provide unequal results. Clin Nutr. 32:65–72.
2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Phypers B and Pierce JT: Lactate
physiology in health and disease. CEACCP. 6:128–132. 2001.
|
|
13
|
Der-Torossian H, Gourin CG and Couch ME:
Translational implications of novel findings in cancer cachexia:
The use of metabolomics and the potential of cardiac malfunction.
Curr Opin Support Palliat Care. 6:446–450. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Muscaritoli M, Anker SD, Argilés J, Aversa
Z, Bauer JM, Biolo G, Boirie Y, Bosaeus I, Cederholm T, Costelli P,
et al: Consensus definition of sarcopenia, cachexia and
pre-cachexia: Joint document elaborated by Special Interest Groups
(SIG) ‘cachexia-anorexia in chronic wasting diseases’ and
‘nutrition in geriatrics’. Clin Nutr. 29:154–159. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Freire PP, Fernandez GJ, Cury SS, de
Moraes D, Oliveira JS, de Oliveira G, Dal-Pai-Silva M, Dos Reis PP
and Carvalho RF: The pathway to cancer cachexia: MicroRNA-Regulated
networks in muscle wasting based on integrative meta-analysis. Int
J Mol Sci. 20(1962)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Schmidt SF, Rohm M, Herzig S and Berriel
Diaz M: Cancer cachexia: More than skeletal muscle wasting. Trends
Cancer. 4:849–860. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Argilés JM, Anguera A and Stemmler B: A
new look at an old drug for the treatment of cancer cachexia:
Megestrol acetate. Clin Nutr. 32:319–324. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
He WA, Calore F, Londhe P, Canella A,
Guttridge DC and Croce CM: Microvesicles containing miRNAs promote
muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci
USA. 111:4525–4529. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang YW, Ma X, Zhang YA, Wang MJ, Yatabe
Y, Lam S, Girard L, Chen JY and Gazdar AF: ITPKA gene body
methylation regulates gene expression and serves as an early
diagnostic marker in lung and other cancers. J Thorac Oncol.
11:1469–1481. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lee DE, Brown JL, Rosa-Caldwell ME,
Blackwell TA, Perry RA Jr, Brown LA, Khatri B, Seo D, Bottje WG,
Washington TA, et al: Cancer cachexia-induced muscle atrophy:
Evidence for alterations in microRNAs important for muscle size.
Physiol Genomics. 49:253–260. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Camargo RG, Quintas Teixeira Ribeiro H,
Geraldo MV, Matos-Neto E, Neves RX, Carnevali LC Jr, Donatto FF,
Alcântara PS, Ottoch JP and Seelaender M: Cancer cachexia and
MicroRNAs. Mediators Inflamm. 2015(367561)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li X, Wang S, Zhu R, Li H, Han Q and Zhao
RC: Lung tumor exosomes induce a pro-inflammatory phenotype in
mesenchymal stem cells via NFκB-TLR signaling pathway. J Hematol
Oncol. 9(42)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Stegeman S, Amankwah E, Klein K, O'Mara
TA, Kim D, Lin HY, Permuth-Wey J, Sellers TA, Srinivasan S, Eeles
R, et al: A Large-scale analysis of genetic variants within
putative miRNA binding sites in prostate cancer. Cancer Discov.
5:368–379. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Denli AM, Tops BB, Plasterk RH, Ketting RF
and Hannon GJ: Processing of primary microRNAs by the
Microprocessor complex. Nature. 432:231–235. 2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wilson RC, Tambe A, Kidwell MA, Noland CL,
Schneider CP and Doudna JA: Dicer-TRBP complex formation ensures
accurate mammalian microRNA biogenesis. Mol Cell. 57:397–407.
2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gregory RI, Chendrimada TP, Cooch N and
Shiekhattar R: Human RISC couples microRNA biogenesis and
posttranscriptional gene silencing. Cell. 123:631–640.
2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Thomson DW, Bracken CP and Goodall GJ:
Experimental strategies for microRNA target identification. Nucleic
Acids Res. 39:6845–6853. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Eisenberg I, Eran A, Nishino I, Moggio M,
Lamperti C, Amato AA, Lidov HG, Kang PB, North KN,
Mitrani-Rosenbaum S, et al: Distinctive patterns of microRNA
expression in primary muscular disorders. Proc Natl Acad Sci USA.
104:17016–17021. 2007.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Soares RJ, Cagnin S, Chemello F,
Silvestrin M, Musaro A, De Pitta C, Lanfranchi G and Sandri M:
Involvement of microRNAs in the regulation of muscle wasting during
catabolic conditions. J Biol Chem. 289:21909–21925. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou X, Hu S, Zhang Y, Du G and Li Y: The
mechanism by which noncoding RNAs regulate muscle wasting in cancer
cachexia. Precision Clin Med. 4:136–147. 2021.
|
|
35
|
Marceca GP, Nigita G, Calore F and Croce
CM: MicroRNAs in skeletal muscle and hints on their potential role
in muscle wasting during cancer cachexia. Front Oncol.
10(607196)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kim DH: Nutritional issues in patients
with cancer. Intest Res. 17:455–462. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhu X, Burfeind KG, Michaelis KA, Braun
TP, Olson B, Pelz KR, Morgan TK and Marks DL: MyD88 signalling is
critical in the development of pancreatic cancer cachexia. J
Cachexia Sarcopenia Muscle. 10:378–390. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Du L, Dong F, Guo L, Hou Y, Yi F, Liu J
and Xu D: Interleukin-1β increases permeability and upregulates the
expression of vascular endothelial-cadherin in human renal
glomerular endothelial cells. Mol Med Rep. 11:3708–3714.
2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lobb RJ, Lima LG and Möller A: Exosomes:
Key mediators of metastasis and pre-metastatic niche formation.
Semin Cell Dev Biol. 67:3–10. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Tomasetti M, Lee W, Santarelli L and
Neuzil J: Exosome-derived microRNAs in cancer metabolism: Possible
implications in cancer diagnostics and therapy. Exp Mol Med.
49(e285)2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Cordonnier M, Chanteloup G, Isambert N,
Seigneuric R, Fumoleau P, Garrido C and Gobbo J: Exosomes in cancer
theranostic: Diamonds in the rough. Cell Adh Migr. 11:151–163.
2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Song W, Yan D, Wei T, Liu Q, Zhou X and
Liu J: Tumor-derived extracellular vesicles in angiogenesis. Biomed
Pharmacother. 102:1203–1208. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bilir C, Engin H, Can M, Temi YB and
Demirtas D: The prognostic role of inflammation and hormones in
patients with metastatic cancer with cachexia. Med Oncol.
32(56)2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Batista ML Jr, Olivan M, Alcantara PS,
Sandoval R, Peres SB, Neves RX, Silverio R, Maximiano LF, Otoch JP
and Seelaender M: Adipose tissue-derived factors as potential
biomarkers in cachectic cancer patients. Cytokine. 61:532–539.
2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Nie M, Deng ZL, Liu J and Wang DZ:
Noncoding RNAs, emerging regulators of skeletal muscle development
and diseases. Biomed Res Int. 2015(676575)2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang Y, Yu M and Tian W: Physiological
and pathological impact of exosomes of adipose tissue. Cell Prolif.
49:3–13. 2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Lazar I, Clement E, Dauvillier S, Milhas
D, Ducoux-Petit M, LeGonidec S, Moro C, Soldan V, Dalle S, Balor S,
et al: Adipocyte exosomes promote melanoma aggressiveness through
fatty acid oxidation: A novel mechanism linking obesity and cancer.
Cancer Res. 76:4051–4057. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Falzone L, Grimaldi M, Celentano E,
Augustin LSA and Libra M: Identification of modulated MicroRNAs
associated with breast cancer, diet, and physical activity. Cancers
(Basel). 12(2555)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Fonseca A, Ramalhete SV, Mestre A, Pires
das Neves R, Marreiros A, Castelo-Branco P and Roberto VP:
Identification of colorectal cancer associated biomarkers: An
integrated analysis of miRNA expression. Aging (Albany NY).
13:21991–22029. 2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Falzone L, Lupo G, La Rosa GRM, Crimi S,
Anfuso CD, Salemi R, Rapisarda E, Libra M and Candido S:
Identification of novel MicroRNAs and their diagnostic and
prognostic significance in oral cancer. Cancers (Basel).
11(610)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ren ZP, Hou XB, Tian XD, Guo JT, Zhang LB,
Xue ZQ, Deng JQ, Zhang SW, Pan JY and Chu XY: Identification of
nine microRNAs as potential biomarkers for lung adenocarcinoma.
FEBS Open Bio. 9:315–327. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kwon YJ, Cho YE, Cho AR, Choi WJ, Yun S,
Park H, Kim HS, Cashion AK, Gill J, Lee H and Lee JW: The possible
influence of mediterranean diet on extracellular vesicle miRNA
expression in breast cancer survivors. Cancers (Basel).
12(1355)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Giambò F, Leone GM, Gattuso G, Rizzo R,
Cosentino A, Cinà D, Teodoro M, Costa C, Tsatsakis A, Fenga C and
Falzone L: Genetic and epigenetic alterations induced by pesticide
exposure: Integrated analysis of gene expression, microRNA
Expression, and DNA methylation datasets. Int J Environ Res Public
Health. 18(8697)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Filetti V, Falzone L, Rapisarda V,
Caltabiano R, Eleonora Graziano AC, Ledda C and Loreto C:
Modulation of microRNA expression levels after naturally occurring
asbestiform fibers exposure as a diagnostic biomarker of
mesothelial neoplastic transformation. Ecotoxicol Environ Saf.
198(110640)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Kemik O, Sumer A, Kemik AS, Hasirci I,
Purisa S, Dulger AC, Demiriz B and Tuzun S: The relationship among
acute-phase response proteins, cytokines and hormones in cachectic
patients with colon cancer. World J Surg Oncol.
8(85)2010.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Guo L, Dong F, Hou Y, Cai W, Zhou X, Huang
AL, Yang M, Allen TD and Liu J: Dihydroartemisinin inhibits
vascular endothelial growth factor-induced endothelial cell
migration by a p38 mitogen-activated protein kinase-independent
pathway. Exp Ther Med. 8:1707–1712. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wei T, Jia J, Wada Y, Kapron CM and Liu J:
Dose dependent effects of cadmium on tumor angiogenesis.
Oncotarget. 8:44944–44959. 2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Gao P, Wang LL, Liu J, Dong F, Song W,
Liao L, Wang B, Zhang W, Zhou X, Xie Q, et al: Dihydroartemisinin
inhibits endothelial cell tube formation by suppression of the
STAT3 signaling pathway. Life Sci. 242(117221)2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Liu J, Ren Y, Hou Y, Zhang C, Wang B, Li
X, Sun R and Liu J: Dihydroartemisinin induces endothelial cell
autophagy through suppression of the Akt/mTOR Pathway. J Cancer.
10:6057–6064. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Xie Q, Cheng Z, Chen X, Lobe CG and Liu J:
The role of Notch signalling in ovarian angiogenesis. J Ovarian
Res. 10(13)2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Kim KJ, Li B, Winer J, Armanini M, Gillett
N, Phillips HS and Ferrara N: Inhibition of vascular endothelial
growth factor-induced angiogenesis suppresses tumour growth in
vivo. Nature. 362:841–844. 1993.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Liu J, Li Y, Dong F, Li L, Masuda T, Allen
TD and Lobe CG: Trichostatin A suppresses lung adenocarcinoma
development in Grg1 overexpressing transgenic mice. Biochem Biophys
Res Commun. 463:1230–1236. 2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Muralidharan-Chari V, Clancy J, Plou C,
Romao M, Chavrier P, Raposo G and D'Souza-Schorey C: ARF6-regulated
shedding of tumor cell-derived plasma membrane microvesicles. Curr
Biol. 19:1875–1885. 2009.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Sabry D, El-Deek SEM, Maher M, El-Baz MAH,
El-Bader HM, Amer E, Hassan EA, Fathy W and El-Deek HEM: Role of
miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in
colorectal carcinoma: Impact of HIF-1α-VEGF signaling pathway. Mol
Cell Biochem. 454:177–189. 2019.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Cheng J, Chen Y, Zhao P, Liu X, Dong J, Li
J, Huang C, Wu R and Lv Y: Downregulation of miRNA-638 promotes
angiogenesis and growth of hepatocellular carcinoma by targeting
VEGF. Oncotarget. 7:30702–30711. 2016.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Yamada N, Tsujimura N, Kumazaki M,
Shinohara H, Taniguchi K, Nakagawa Y, Naoe T and Akao Y: Colorectal
cancer cell-derived microvesicles containing microRNA-1246 promote
angiogenesis by activating Smad 1/5/8 signaling elicited by PML
down-regulation in endothelial cells. Biochim Biophys Acta.
1839:1256–1272. 2014.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Tisdale MJ: Cancer cachexia. Curr Opin
Gastroenterol. 26:146–151. 2010.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Bilodeau PA, Coyne ES and Wing SS: The
ubiquitin proteasome system in atrophying skeletal muscle: Roles
and regulation. Am J Physiol Cell Physiol. 311:C392–C403.
2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Reed SA, Sandesara PB, Senf SM and Judge
AR: Inhibition of FoxO transcriptional activity prevents muscle
fiber atrophy during cachexia and induces hypertrophy. FASEB J.
26:987–1000. 2012.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Xu J, Li R, Workeneh B, Dong Y, Wang X and
Hu Z: Transcription factor FoxO1, the dominant mediator of muscle
wasting in chronic kidney disease, is inhibited by microRNA-486.
Kidney Int. 82:401–411. 2012.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Suzuki T and Springer J: MicroRNAs in
muscle wasting. J Cachexia Sarcopenia Muscle. 9:1209–1212.
2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Sutandyo N: The role of microRNA in cancer
cachexia and muscle wasting: A review article. Caspian J Intern
Med. 12:124–128. 2021.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Brzeszczyńska J, Brzeszczyński F, Hamilton
DF, McGregor R and Simpson AHRW: Role of microRNA in muscle
regeneration and diseases related to muscle dysfunction in atrophy,
cachexia, osteoporosis, and osteoarthritis. Bone Joint Res.
9:798–807. 2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Zhou L, Zhang T, Shao W, Lu R, Wang L, Liu
H, Jiang B, Li S, Zhuo H, Wang S, et al: Amiloride ameliorates
muscle wasting in cancer cachexia through inhibiting tumor-derived
exosome release. Skeletal muscle. 11(17)2021.PubMed/NCBI View Article : Google Scholar
|
|
75
|
van de Worp WRPH, Schols AMWJ, Schols
AMWJ, Dingemans AC, Op den Kamp CMH, Degens JHRJ, Kelders MCJM,
Coort S, Woodruff HC, Kratassiouk G, et al: Identification of
microRNAs in skeletal muscle associated with lung cancer cachexia.
J Cachexia Sarcopenia Muscle. 11:452–463. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Fernandez GJ, Ferreira JH, Vechetti IJ Jr,
de Moraes LN, Cury SS, Freire PP, Gutiérrez J, Ferretti R,
Dal-Pai-Silva M, Rogatto SR and Carvalho RF: MicroRNA-mRNA
Co-sequencing identifies transcriptional and post-transcriptional
regulatory networks underlying muscle wasting in cancer cachexia.
Front Genet. 11(541)2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Daas SI, Rizeq BR and Nasrallah GK:
Adipose tissue dysfunction in cancer cachexia. J Cell Physiol.
234:13–22. 2018.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Petruzzelli M, Schweiger M, Schreiber R,
Campos-Olivas R, Tsoli M, Allen J, Swarbrick M, Rose-John S, Rincon
M, Robertson G, et al: A switch from white to brown fat increases
energy expenditure in cancer-associated cachexia. Cell Metab.
20:433–447. 2014.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Neves RX, Rosa-Neto JC, Yamashita AS,
Matos-Neto EM, Riccardi DM, Lira FS, Batista ML Jr and Seelaender
M: White adipose tissue cells and the progression of cachexia:
Inflammatory pathways. J Cachexia Sarcopenia Muscle. 7:193–203.
2016.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Camargo RG, Riccardi DM, Ribeiro HQ,
Carnevali LC Jr, de Matos-Neto EM, Enjiu L, Neves RX, Lima JD,
Figuerêdo RG, de Alcântara PS, et al: NF-κBp65 and expression of
its pro-inflammatory target genes are upregulated in the
subcutaneous adipose tissue of cachectic cancer patients.
Nutrients. 7:4465–4479. 2015.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Aswad H, Forterre A, Wiklander OP, Vial G,
Danty-Berger E, Jalabert A, Lamazière A, Meugnier E, Pesenti S, Ott
C, et al: Exosomes participate in the alteration of muscle
homeostasis during lipid-induced insulin resistance in mice.
Diabetologia. 57:2155–2164. 2014.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Kulyté A, Lorente-Cebrián S, Gao H,
Mejhert N, Agustsson T, Arner P, Rydén M and Dahlman I: MicroRNA
profiling links miR-378 to enhanced adipocyte lipolysis in human
cancer cachexia. Am J Physiol Endocrinol Metab. 306:E267–E274.
2014.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Donzelli S, Farneti A, Marucci L, Ganci F,
Sacconi A, Strano S, Sanguineti G and Blandino G: Non-coding RNAs
as putative biomarkers of cancer-associated cachexia. Front Cell
Dev Biol. 8(257)2020.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Hamaguchi Y, Kaido T, Okumura S, Kobayashi
A, Hammad A, Tamai Y, Inagaki N and Uemoto S: Proposal for new
diagnostic criteria for low skeletal muscle mass based on computed
tomography imaging in Asian adults. Nutrition. 32:1200–1205.
2016.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Kaido T: Selection criteria and current
issues in liver transplantation for hepatocellular carcinoma. Liver
Cancer. 5:121–127. 2016.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Okugawa Y, Toiyama Y, Hur K, Yamamoto A,
Yin C, Ide S, Kitajima T, Fujikawa H, Yasuda H, Koike Y, et al:
Circulating miR-203 derived from metastatic tissues promotes
myopenia in colorectal cancer patients. J Cachexia Sarcopenia
Muscle. 10:536–548. 2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Okugawa Y, Yao L, Toiyama Y, Yamamoto A,
Shigemori T, Yin C, Omura Y, Ide S, Kitajima T, Shimura T, et al:
Prognostic impact of sarcopenia and its correlation with
circulating miR-21 in colorectal cancer patients. Oncol Rep.
39:1555–1564. 2018.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Wang H and Wang B: Extracellular vesicle
microRNAs mediate skeletal muscle myogenesis and disease. Biomed
Rep. 5:296–300. 2016.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Wu Q, Sun S, Li Z, Yang Q, Li B, Zhu S,
Wang L, Wu J, Yuan J, Yang C, et al: Tumour-originated exosomal
miR-155 triggers cancer-associated cachexia to promote tumour
progression. Mol Cancer. 17(155)2018.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Chitti SV, Fonseka P and Mathivanan S:
Emerging role of extracellular vesicles in mediating cancer
cachexia. Biochem Soc Trans. 46:1129–1136. 2018.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Du G, Zhang Y, Hu S, Zhou X and Li Y:
Non-coding RNAs in exosomes and adipocytes cause fat loss during
cancer cachexia. Noncoding RNA Res. 6:80–85. 2021.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Li L, Liu H, Tao W, Wen S, Fu X and Yu S:
Pharmacological inhibition of HMGB1 prevents muscle wasting. Front
Pharmacol. 12(731386)2021.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Wan Z, Chen X, Gao X, Dong Y, Zhao Y, Wei
M, Fan W, Yang G and Liu L: Chronic myeloid leukemia-derived
exosomes attenuate adipogenesis of adipose derived mesenchymal stem
cells via transporting miR-92a-3p. J Cell Physiol. 234:21274–21283.
2019.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Köberle V, Kronenberger B, Pleli T, Trojan
J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem
S, Piiper A and Waidmann O: Serum microRNA-1 and microRNA-122 are
prognostic markers in patients with hepatocellular carcinoma. Eur J
Cancer. 49:3442–3449. 2013.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Powrózek T, Mlak R, Brzozowska A, Mazurek
M, Gołębiowski P and Małecka-Massalska T: MiRNA-130a significantly
improves accuracy of SGA Nutritional assessment tool in prediction
of malnutrition and cachexia in radiotherapy-treated head and neck
cancer patients. Cancers (Basel). 10(294)2018.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Chen D, Goswami CP, Burnett RM, Anjanappa
M, Bhat-Nakshatri P, Muller W and Nakshatri H: Cancer affects
microRNA expression, release, and function in cardiac and skeletal
muscle. Cancer Res. 74:4270–4281. 2014.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Lin J, Li J, Huang B, Liu J, Chen X, Chen
XM, Xu YM, Huang LF and Wang XZ: Exosomes: Novel biomarkers for
clinical diagnosis. ScientificWorldJournal.
2015(657086)2015.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Belli R, Ferraro E, Molfino A, Carletti R,
Tambaro F, Costelli P and Muscaritoli M: Liquid biopsy for cancer
cachexia: Focus on muscle-derived microRNAs. Int J Mol Sci.
22(9007)2021.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo
X, Mao XH, Zou QM, Yu PW, Zuo QF, et al: Plasma microRNAs, miR-223,
miR-21 and miR-218, as novel potential biomarkers for gastric
cancer detection. PLoS One. 7(e41629)2012.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7(e29770)2012.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Wang J, Chen J, Chang P, LeBlanc A, Li D,
Abbruzzesse JL, Frazier ML, Killary AM and Sen S: MicroRNAs in
plasma of pancreatic ductal adenocarcinoma patients as novel
blood-based biomarkers of disease. Cancer Prev Res (Phila).
2:807–813. 2009.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Kottorou A, Dimitrakopoulos FI and Tsezou
A: Non-coding RNAs in cancer-associated cachexia: Clinical
implications and future perspectives. Transl Oncol.
14(101101)2021.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Yao P, Potdar AA, Arif A, Ray PS,
Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM and Fox PL:
Coding region polyadenylation generates a truncated tRNA synthetase
that counters translation repression. Cell. 149:88–100.
2012.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Gao P, Niu N, Wei T, Tozawa H, Chen X,
Zhang C, Zhang J, Wada Y, Kapron CM and Liu J: The roles of signal
transducer and activator of transcription factor 3 in tumor
angiogenesis. Oncotarget. 8:69139–69161. 2017.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Margolis LM and Rivas DA: Potential Role
of MicroRNA in the anabolic capacity of skeletal muscle with aging.
Exerc Sport Sci Rev. 46:86–91. 2018.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Hou B, Xu S, Xu Y, Gao Q, Zhang C, Liu L,
Yang H, Jiang X and Che Y: Grb2 binds to PTEN and regulates its
nuclear translocation to maintain the genomic stability in DNA
damage response. Cell Death Dis. 10(546)2019.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Carr RM, Enriquez-Hesles E, Olson RL,
Jatoi A, Doles J and Fernandez-Zapico ME: Epigenetics of
cancer-associated muscle catabolism. Epigenomics. 9:1259–1265.
2017.PubMed/NCBI View Article : Google Scholar
|
|
109
|
György B, Hung ME, Breakefield XO and
Leonard JN: Therapeutic applications of extracellular vesicles:
Clinical promise and open questions. Annu Rev Pharmacol Toxicol.
55:439–464. 2015.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Kalra H, Drummen GP and Mathivanan S:
Focus on extracellular vesicles: Introducing the next small big
thing. Int J Mol Sci. 17(170)2016.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Terasawa K, Shimizu K and Tsujimoto G:
Synthetic Pre-miRNA-Based shRNA as Potent RNAi Triggers. J Nucleic
Acids. 2011(131579)2011.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Bonneau E, Neveu B, Kostantin E, Tsongalis
GJ and De Guire V: How close are miRNAs from clinical practice? A
perspective on the diagnostic and therapeutic market. EJIFCC.
30:114–127. 2019.PubMed/NCBI
|
|
113
|
van Zandwijk N, Pavlakis N, Kao SC, Linton
A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey
DL, et al: Safety and activity of microRNA-loaded minicells in
patients with recurrent malignant pleural mesothelioma: A
first-in-man, phase 1, open-label, dose-escalation study. Lancet
Oncol. 18:1386–1396. 2017.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Ebner N, Anker SD and von Haehling S:
Recent developments in the field of cachexia, sarcopenia, and
muscle wasting: Highlights from the 12th cachexia conference. J
Cachexia Sarcopenia Muscle. 11:274–285. 2020.PubMed/NCBI View Article : Google Scholar
|