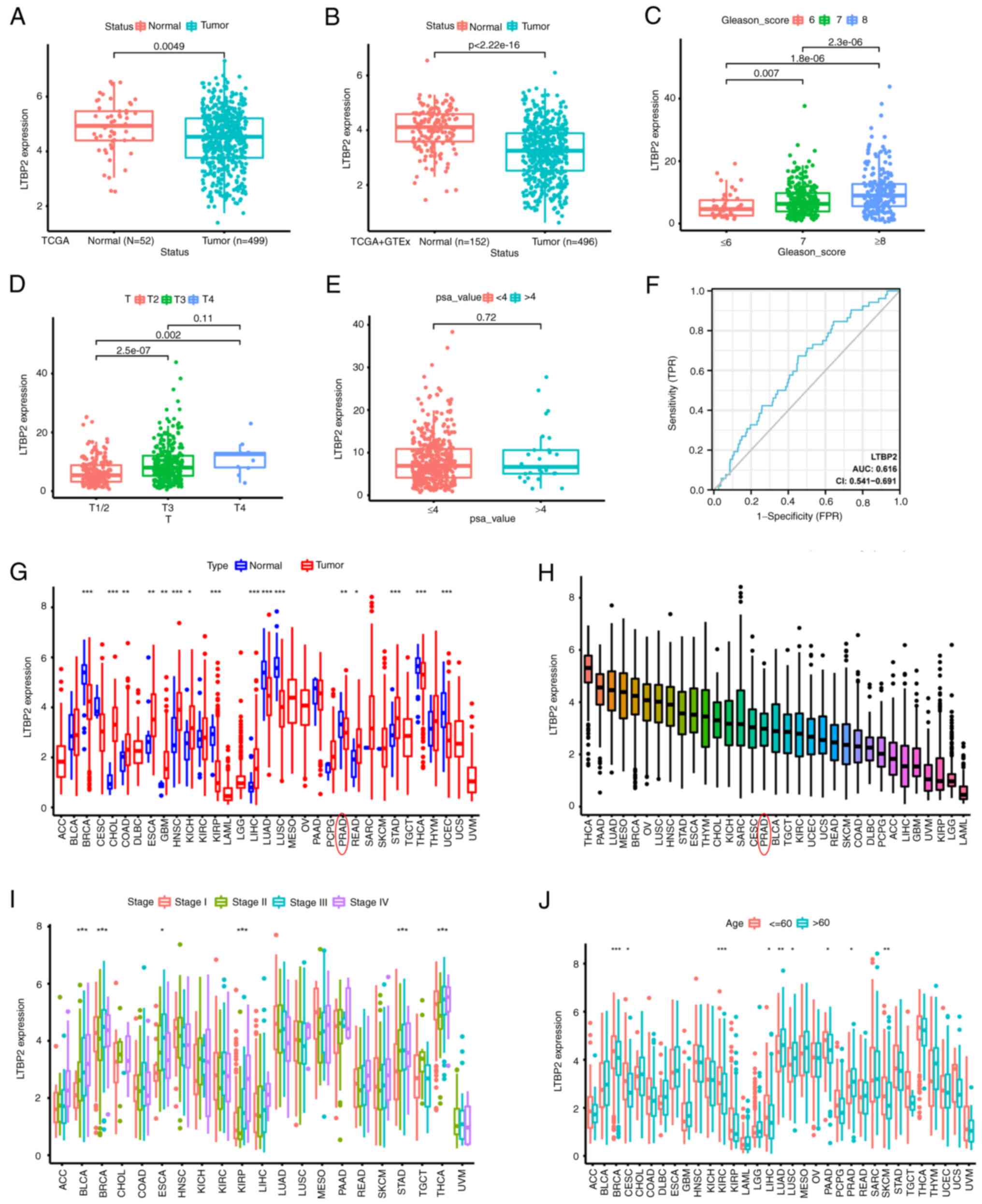
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nevedomskaya E, Baumgart SJ and Haendler
B: Recent advances in prostate cancer treatment and drug discovery.
Int J Mol Sci. 19(1359)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sundi D, Tosoian JJ, Nyame YA, Alam R,
Achim M, Reichard CA, Li J, Wilkins L, Schwen Z, Han M, et al:
Outcomes of very high-risk prostate cancer after radical
prostatectomy: Validation study from 3 centers. Cancer.
125:391–397. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cornford P, van den Bergh RCN, Briers E,
Van den Broeck T, Cumberbatch MG, Santis MD, Fanti S, Fossati N,
Gandaglia G, Gillessen S, et al: EAU-EANM-ESTRO-ESUR-SIOG
guidelines on prostate cancer. Part II-2020 update: Treatment of
relapsing and metastatic prostate cancer. Eur Urol. 79:263–282.
2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Van den Broeck T, van den Bergh RCN,
Briers E, Cornford P, Cumberbatch M, Tilki D, De Santis M, Fanti S,
Fossati N, Gillessen S, et al: Biochemical recurrence in prostate
cancer: The european association of urology prostate cancer
guidelines panel recommendations. Eur Urol Focus. 6:231–234.
2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Barry MJ and Simmons LH: Prevention of
prostate cancer morbidity and mortality: Primary prevention and
early detection. Med Clin North Am. 101:787–806. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim SH, Park WS, Park BR, Joo J, Joung JY,
Seo HK, Chung J and Lee KH: Psca, cox-2, and ki-67 are independent,
predictive markers of biochemical recurrence in clinically
localized prostate cancer: A retrospective study. Asian J Androl.
19:458–462. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gu P, Chen X, Xie R, Han J, Xie W, Wang B,
Dong W, Chen C, Yang M, Jiang J, et al: LncRNA HOXD-AS1 regulates
proliferation and chemo-resistance of castration-resistant prostate
cancer via recruiting wdr5. Mol Ther. 25:1959–1973. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gu P, Chen X, Xie R, Xie W, Huang L, Dong
W, Han J, Liu X, Shen J, Huang J and Lin T: A novel AR
translational regulator lncrna lbcs inhibits castration resistance
of prostate cancer. Mol Cancer. 18(109)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J,
Li J, Li F and Tan HB: Immune cells within the tumor
microenvironment: Biological functions and roles in cancer
immunotherapy. Cancer Lett. 470:126–133. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pitt JM, Marabelle A, Eggermont A, Soria
JC, Kroemer G and Zitvogel L: Targeting the tumor microenvironment:
Removing obstruction to anticancer immune responses and
immunotherapy. Ann Oncol. 27:1482–1492. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Riley RS, June CH, Langer R and Mitchell
MJ: Delivery technologies for cancer immunotherapy. Nat Rev Drug
Discov. 18:175–196. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tomita Y, Ikeda T, Sakata S, Saruwatari K,
Sato R, Iyama S, Jodai T, Akaike K, Ishizuka S, Saeki S and
Sakagami T: Association of probiotic clostridium butyricum therapy
with survival and response to immune checkpoint blockade in
patients with lung cancer. Cancer Immunol Res. 8:1236–1242.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bagchi S, Yuan R and Engleman EG: Immune
checkpoint inhibitors for the treatment of cancer: Clinical impact
and mechanisms of response and resistance. Annu Rev Pathol.
16:223–249. 2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Rui X, Shao S, Wang L and Leng J:
Identification of recurrence marker associated with immune
infiltration in prostate cancer with radical resection and build
prognostic nomogram. BMC Cancer. 19(1179)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hou Q, Bing ZT, Hu C, Li MY, Yang KH, Mo
Z, Xie XW, Liao JL, Lu Y, Horie S and Lou MW: Rankprod combined
with genetic algorithm optimized artificial neural network
establishes a diagnostic and prognostic prediction model that
revealed c1QTNF3 as a biomarker for prostate cancer. EBioMedicine.
32:234–244. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Mortensen MM, Høyer S, Lynnerup AS,
Ørntoft TF, Sørensen KD, Borre M and Dyrskjøt L: Expression
profiling of prostate cancer tissue delineates genes associated
with recurrence after prostatectomy. Sci Rep.
5(16018)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ross-Adams H, Lamb AD, Dunning MJ, Halim
S, Lindberg J, Massie CM, Egevad LA, Russell R, Ramos-Montoya A,
Vowler SL, et al: Integration of copy number and transcriptomics
provides risk stratification in prostate cancer: A discovery and
validation cohort study. EBioMedicine. 2:1133–1144. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jain S, Lyons CA, Walker SM, McQuaid S,
Hynes SO, Mitchell DM, Pang B, Logan GE, McCavigan AM, Rourke DO,
et al: Validation of a metastatic assay using biopsies to improve
risk stratification in patients with prostate cancer treated with
radical radiation therapy. Ann Oncol. 29:215–222. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Leek JT, Johnson WE, Parker HS, Jaffe AE
and Storey JD: The sva package for removing batch effects and other
unwanted variation in high-throughput experiments. Bioinformatics.
28:882–883. 2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for rna-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Engebretsen S and Bohlin J: Statistical
predictions with glmnet. Clin Epigenetics. 11(123)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Klosa J, Simon N, Westermark PO, Liebscher
V and Wittenburg D: Seagull: Lasso, group lasso and sparse-group
lasso regularization for linear regression models via proximal
gradient descent. BMC Bioinformatics. 21(407)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sun S, Shen Y, Wang J, Li J, Cao J and
Zhang J: Identification and validation of autophagy-related genes
in chronic obstructive pulmonary disease. Int J Chron Obstruct
Pulmon Dis. 16:67–78. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sanz H, Valim C, Vegas E, Oller JM and
Reverter F: SVM-RFE: Selection and visualization of the most
relevant features through non-linear kernels. BMC Bioinformatics.
19(432)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou X and Tuck DP: MSVM-RFE: Extensions
of SVM-RFE for multiclass gene selection on DNA microarray data.
Bioinformatics. 23:1106–1114. 2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li F, Zhao C, Xia Z, Wang Y, Zhou X and Li
GZ: Computer-assisted lip diagnosis on traditional chinese medicine
using multi-class support vector machines. BMC Complement Altern
Med. 12(127)2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xu Q, Xu H, Deng R, Wang Z, Li N, Qi Z,
Zhao J and Huang W: Multi-omics analysis reveals prognostic value
of tumor mutation burden in hepatocellular carcinoma. Cancer Cell
Int. 21(342)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wu X, Sui Z, Zhang H, Wang Y and Yu Z:
Integrated analysis of lncRNA-mediated ceRNA network in lung
adenocarcinoma. Front Oncol. 10(554759)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361.
2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liu P, Jiang W, Zhao J and Zhang H:
Integrated analysis of genome-wide gene expression and DNA
methylation microarray of diffuse large B-cell lymphoma with TET
mutations. Mol Med Rep. 16:3777–3782. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gambardella A, Licata G and Sohrt A: Dose
adjustment of biologic treatments for moderate-to-severe plaque
psoriasis in the real world: A systematic review. Dermatol Ther
(Heidelb). 11:1141–1156. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun.
4(2612)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang S, Zhang E, Long J, Hu Z, Peng J,
Liu L, Tang F, Li L, Ouyang Y and Zeng Z: Immune infiltration in
renal cell carcinoma. Cancer Sci. 110:1564–1572. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Pan JH, Zhou H, Cooper L, Huang JL, Zhu
SB, Zhao XX, Ding H, Pan YL and Rong L: LAYN is a prognostic
biomarker and correlated with immune infiltrates in gastric and
colon cancers. Front Immunol. 10(6)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The string database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4(2)2003.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of cerna crosstalk and competition. Nature.
505:344–352. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
Starbase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale clip-seq data. Nucleic Acids
Res. 42:D92–D97. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shannon P, Markiel A, Ozier O, Baliga NC,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hugo W, Zaretsky JM, Sun L, Song C, Moreno
BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G,
et al: Genomic and transcriptomic features of response to anti-PD-1
therapy in metastatic melanoma. Cell. 165:35–44. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Mancinelli S, Turcato A, Kisslinger A,
Bongiovanni A, Zazzu V, Lanati A and Liguori GL: Design of
transfections: Implementation of design of experiments for cell
transfection fine tuning. Biotechnol Bioeng. 118:4488–4502.
2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Taylor SC, Nadeau K, Abbasi M, Lachance C,
Nguyen M and Fenrich J: The ultimate qPCR experiment: Producing
publication quality, reproducible data the first time. Trends
Biotechnol. 37:761–774. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yu J, Mao W, Sun S, Hu Q, Wang C, Xu Z,
Liu R, Chen S, Xu B and Chen M: Identification of an m6A-related
lncRNA signature for predicting the prognosis in patients with
kidney renal clear cell carcinoma. Front Oncol.
11(663263)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chen S, Wang L, Xu C, Chen H, Peng B, Xu
Y, Yao X, Li L and Zheng J: Knockdown of regγ inhibits
proliferation by inducing apoptosis and cell cycle arrest in
prostate cancer. Am J Transl Res. 9:3787–3795. 2017.PubMed/NCBI
|
|
49
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative pcr and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Mao W, Wang K, Xu B, Zhang H, Sun S, Hu Q,
Zhang L, Liu C, Chen S, Wu J, et al: CiRS-7 is a prognostic
biomarker and potential gene therapy target for renal cell
carcinoma. Mol Cancer. 20(142)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Pulendran B and Davis MM: The science and
medicine of human immunology. Science. 369(eaay4014)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wang J, Liang WJ, Min GT, Wang HP, Chen W
and Yao N: LTBP2 promotes the migration and invasion of gastric
cancer cells and predicts poor outcome of patients with gastric
cancer. Int J Oncol. 52:1886–1898. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ren Y, Lu H, Zhao D, Ou Y, Yu K, Gu J,
Wang L, Jiang S, Chen M, Wang J, et al: LTPB2 acts as a prognostic
factor and promotes progression of cervical adenocarcinoma. Am J
Transl Res. 7:1095–1105. 2015.PubMed/NCBI
|
|
54
|
Wang J, Jiang C, Li N, Wang F, Xu Y, Shen
Z, Yang L, Li Z and He C: The circEPSTI1/mir-942-5p/LTBP2 axis
regulates the progression of oscc in the background of osf via emt
and the PI3K/Akt/mTOR pathway. Cell Death Dis.
11(682)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Pang XF, Lin X, Du JJ and Zeng DY: LTBP2
knockdown by siRNA reverses myocardial oxidative stress injury,
fibrosis and remodelling during dilated cardiomyopathy. Acta
Physiol (Oxf). 228(e13377)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Enomoto Y, Matsushima S, Shibata K,
Aoshima Y, Yagi H, Meguro S, Kawasaki H, Kosugi I, Fujisawa T,
Enomoto N, et al: LTBP2 is secreted from lung myofibroblasts and is
a potential biomarker for idiopathic pulmonary fibrosis. Clin Sci
(Lond). 132:1565–1580. 2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Huang Y, Wang G, Zhao C, Geng R, Zhang S,
Wang W, Chen J, Liu H and Wang X: High expression of LTBP2
contributes to poor prognosis in colorectal cancer patients and
correlates with the mesenchymal colorectal cancer subtype. Dis
Markers. 2019(5231269)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Rauf B, Irum B, Khan SY, Kabir F, Naeem
MA, Riazuddin S, Ayyagari R and Riazuddin SA: Novel mutations in
LTBP2 identified in familial cases of primary congenital glaucoma.
Mol Vis. 26:14–25. 2020.PubMed/NCBI
|
|
59
|
Bilusic M, Madan RA and Gulley JL:
Immunotherapy of prostate cancer: Facts and hopes. Clin Cancer Res.
23:6764–6770. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Gamat M and McNeel DG: Androgen
deprivation and immunotherapy for the treatment of prostate cancer.
Endocr Relat Cancer. 24:T297–T310. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Zhou Q, Chen X, He H, Peng S, Zhang Y,
Zhang J, Cheng L, Liu S, Huang R, Xie R, et al: Wd repeat domain 5
promotes chemoresistance and programmed death-ligand 1 expression
in prostate cancer. Theranostics. 11:4809–4824. 2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Zhang J, Zhou Q, Xie K, Cheng L, Peng S,
Xie R, Liu L, Zhang Y, Dong W, Han J, et al: Targeting WD repeat
domain 5 enhances chemosensitivity and inhibits proliferation and
programmed death-ligand 1 expression in bladder cancer. J Exp Clin
Cancer Res. 40(203)2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Turtoi A, Musmeci D, Wang Y, Dumont B,
Somja J, Bevilacqua G, De Pauw E, Delvenne P and Castronovo V:
Identification of novel accessible proteins bearing diagnostic and
therapeutic potential in human pancreatic ductal adenocarcinoma. J
Proteome Res. 10:4302–4313. 2011.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Chen H, Ko JMY, Wong VCL, Hyytiainen M,
Keski-Oja J, Chua D, Nicholls JM, Cheung FMF, Lee AWM, Kwong DLW,
et al: LTBP-2 confers pleiotropic suppression and promotes dormancy
in a growth factor permissive microenvironment in nasopharyngeal
carcinoma. Cancer Lett. 325:89–98. 2012.PubMed/NCBI View Article : Google Scholar
|