Introduction
Pancreatic cancer is a common malignant tumor with
almost equivalent morbidity and mortality rates (~15/100,000), and
is third leading cause of cancer-related mortality (1). Gemcitabine (GEM) is regarded as the
first-line chemotherapy drug for pancreatic cancer treatment
(2). However, GEM-resistance in
pancreatic cancer significantly shortens progression-free survival
in patients treated with GEM (3,4).
Cancer stem cells (CSCs) are closely related to GEM-resistance in
pancreatic cancer (5-7).
CSCs are a subset of cancer cells capable of self-renewal and are
involved in the development, metastasis and relapse of pancreatic
cancer (8-10).
CSCs are present in GEM-resistant pancreatic tumor cells (11) and the process of
epithelial-mesenchymal transition (EMT) enhances the migration and
invasion of CSCs (6,11). The progression of pancreatic cancer
is facilitated by the phenomenon of GEM-resistance that is
originating from both CSCs and EMT (7). Therefore, exploring the relative
mechanisms of GEM resistance induced by CSCs and EMT is pivotal for
the treatment of GEM-resistant pancreatic cancer.
EMT is a necessary step for embryonic development
and is involved in multiple physiological and pathological
processes, such as injury repair, inflammation, fibrosis and
tumorigenesis (12). Cell
migration, stem-like characteristics and chemotherapy resistance
can be induced by EMT, which further contributes to immune,
senescent and apoptotic escape (13). An association between CSCs and EMT
has been widely reported. Mani et al (14) induced EMT progression in human
mammary epithelial cells using EMT transcription factors such as
the snail family transcriptional repressor 1 (Snail), the basic
helix-loop-helix transcription factor (Twist) or TGF-β1. Following
EMT activation, upregulation of stem cell biomarkers, formation of
mammary stem cell spheres and soft agar colonies and enhanced
tumorigenesis are observed and considered as an indication of CSCs
(15). In addition, in stem cells
extracted from rodent and human breast epithelium and tumor
tissues, a high expression level of the EMT phenotype is observed
(16). Therefore, EMT progression
and the associated formation of CSCs could be important targets for
the treatment of GEM-resistant pancreatic cancer.
Emodin (Emo; 1,3,8-trihydroxy-6-methylanthraquinone)
is an anthoquinone derivative extracted from the rhizome of
knotweed (Polygonum cuspidatum) and rhubarb (Rheum
officinale baill) and has multiple bioactivities, such as
anti-inflammatory, antibacterial, anti-oxidative and antitumor
activities (17,18). Emo induces cell apoptosis in
paclitaxel-resistant A2780 cells by reducing the expression of
apoptotic molecules such as survivin and X-linked inhibitor of
apoptosis (19). In addition, Emo
significantly inhibits EMT progression in colorectal cancer,
cervical cancer and head and neck squamous cells (20-22).
The present study aimed to investigate the inhibitory effect of Emo
on GEM-resistant pancreatic cancer, as well as the underlying
mechanism of action, by exploring the impact of Emo on the growth
of CSCs and EMT progression.
Methods and materials
Pancreatic cancer cell lines
Human pancreatic cancer cell line SW1990 was
obtained from Combioer Biosciences Co., Ltd. and cultured in DMEM
(Procell Life Science & Technology Co., Ltd.) supplemented with
10% fetal bovine serum (FBS; Procell Life Science & Technology
Co., Ltd.) at 37˚C with 5% CO2.
Establishment of gemcitabine-resistant
pancreatic cancer cell line (SW1990/GZ)
The human pancreatic cancer SW1990 cell line was
seeded in six-well plates at a density of 1x106
cells/well and cultured in DMEM for 48 h at 37˚C followed by
incubation for 7 days with different concentrations of GEM at 37˚C.
Following incubation with GEM, the cell viability was measured
using the CCK-8 assay and the GEM IC50 value was
calculated and further used to incubate SW1990 cells for 24 h. To
establish the SW1990/GZ cell line, SW1990 cells were incubated with
GEM at 2-fold IC50 for 24 h, followed by being replaced
with the drug-free medium. Subsequently, cells were incubated until
80% confluent, followed by incubation with GEM at 4-fold
IC50 for 24 h. The aforementioned procedure was repeated
and the incubation concentration of GEM was increased by 2-fold
each cycle until the final concentration reached ~1,000 µg/ml,
followed by incubation in blank medium for 3 months. Finally, a
CCK-8 assay was used to confirm GEM-resistance.
Transmission electron microscope
(TEM)
Cells were collected following centrifugation at 300
x g at room temperature for 10 min, washed twice with PBS buffer,
fixed overnight with 4.0% glutaraldehyde in PBS at 4˚C and then
embedded in epoxy resin. Subsequently, ultrathin sections
(thickness, 50-70 nm) were cut and collected on copper grids,
followed by counterstaining with 3% aqueous uranyl acetate for 1 h
at room temperature. Sections were then incubated with 2%
phosphotungstic acid for 1 h at room temperature and 1% Reynolds'
lead citrate for 20 min at room temperature. Finally, a TEM
(JEM-1400Flash; JEOL, Ltd.) was used for examination.
CCK-8 assay
Cell viability was measured using a CCK-8 assay kit
(Nanjing Jiancheng Bioengineering Research Institute Co., Ltd.).
Cells were seeded in 96-well plates at a density of
1x105 cells/well and incubated at 37˚C for 24 h.
Subsequently, 10 µl CCK-8 reagent was added to each well and
incubated for 3 h at 37˚C. The absorbance of each was at 450 nm was
measured using a microplate reader (PerkinElmer, Inc.) and the
IC50 value was calculated using GraphPad Prism 8
software (GraphPad Software, Inc.). Finally, the resistant index
(RI) was calculated according to the following formula:
RI=IC50(SW1990/GZ)/IC50(SW1990).
Sphere formation assay
Cells were centrifuged at 300 x g for 5 min at room
temperature, collected and resuspended in PBS Subsequently,
1x103 cells were transferred into each well of a
six-well plate and then cultured with serum-free DMEM-F12 medium
(Procell Life Science & Technology Co., Ltd.). Following
incubation at 37˚C for 7 days, a transparent tape with mesh was
overlaid to the bottom of the wells and an inverted microscope
(Ts2; Nikon Corporation) was used to count the stem cell
spheres.
Colony-formation assay
Cells were seeded in a 6-well plate at a
2x102 cells/well density and incubated for 10 days.
Subsequently, the cells were fixed and stained with 6% (w/v)
glutaraldehyde and 0.5% crystal violet (Sigma-Aldrich; Merck KGaA)
for 60 min at room temperature, followed by washing and air drying
at room temperature. Finally, images were captured using an
inverted microscope (Olympus, Corporation).
Analysis of apoptosis
Apoptosis of cells was evaluated using a flow
cytometry assay. In brief, the cells were seeded in 6-well plates
and incubated with 195 µl Annexin V-fluorescein isothiocyanate
(Wuhan Punosai Life Technology Co., Ltd.), followed by the addition
of 5 µl propidium iodide (BD Biosciences) and incubation for 10 min
at room temperature in the dark. Finally, the samples were loaded
onto a flow cytometer (BD Bioscience) for apoptosis analysis.
Transwell assay
A total of 1.5x105 cells were plated in
the upper chambers of Transwell plates using serum-free DMEM
(Corning, Inc.). Complete DMEM supplemented with 20% FBS was added
into the lower chamber. Following incubation at 37˚C for 24 h, the
migratory cells were stained with 0.1% crystal violet at room
temperature for 10 min. Finally, stained cells were counted in 3
random fields using an optical microscope (Ts2; Nikon
Corporation).
Transfection
Snail-overexpressing SW1990/GZ cells were
established by transfecting the cells implanted in 6-well plates at
a density of 1x106 cells/well with pcDNA3.1-Snail
(Genscript) using 2 µg Lipofectamine® 2000 (Thermo
Fisher Scientific, Inc.). pcDNA3.1-NC (Genscript) was used as the
negative control. After incubation for 48 h at 37˚C, transfection
efficacy was measured through western blot analysis.
Western blot assay
Total protein from cells was extracted using RIPA
Lysis Buffer (Sigma-Aldrich; Merck KGaA) and quantified using a BCA
kit (Sigma-Adrich; Merck KGaA). Total protein (~30 µg/lane) was
separated utilizing SDS-PAGE on a 12% gel and subsequently
transferred onto a PVDF membrane. The membrane was blocked with 5%
BSA (Beyotime Institute of Biotechnology) at room temperature for 2
h to remove non-specific binding proteins, followed by incubation
with primary antibodies against CD44 (1:800; cat. no. DF6392;
Affinity Biosciences, Ltd.), Aldehyde dehydrogenase 1 (ALDH1;
1:1,000; cat. no. DF6625; Affinity Biosciences, Ltd.), Nanog
(1:1,000; cat. no. AF5388; Affinity Biosciences, Ltd.), E-cadherin
(1:800; cat. no. AF0131; Affinity Biosciences, Ltd.), vimentin
(1:1,000; cat. no. AF7013; Affinity Biosciences, Ltd.), Snail
(1:1,000; cat. no. AF6032; Affinity Biosciences, Ltd.) and GAPDH
(1:5,000; cat. no. AF7021; Affinity Biosciences, Ltd.) at 4˚C for
12 h. The membrane was then washed and incubated with the
anti-rabbit IgG HRP-linked secondary antibody (1:3,000, cat. no.
7074, CST Biological Reagents Co., Ltd.) at room temperature for
1.5 h. Finally, the membrane was incubated with ECL solution
(Beyotime Institute of Biotechnology) and exposed to a Tanon
5200-multi (Tanon Science and Technology Co., Ltd.). The relative
expression levels of the target proteins were quantified using
ImageJ 1.8.0.172 software (National Institutes of Health) with
GAPDH as the loading control.
Xenograft experiments
A total of 18 5-6-week-old male SPF BABL/c nude mice
weighing 16-20 g were purchased from Shanghai Lingchang Biological
Technology Co., Ltd. Animals were housed in individually ventilated
cages (Suzhou Suhang Technology Equipment Co., Ltd.) at 20-24˚C,
40-60% humidity, minimum air change rate of 15/h, minimum pressure
gradient of 10 Pa, ammonia concentration ≤14 mg/m2,
noise ≤60 db, 12-h light/dark cycle and with ad libitum
access to food and water.
For the in vivo experiment, six groups were
used: GEM + Snail OE + EmoControl, Snail OE, GEM, GEM + Snail OE,
GEM + Emo, and GEM + Snail OE + Emo. For the general procedure of
xenograft model establishment, 1x107 cells were
subcutaneously injected into the axilla of each mouse. Animals in
the control group were xenografted with SW1990/GZ cells, followed
by intraperitoneal injection with an equal volume of normal saline.
In the Snail OE group, animals were xenografted with
Snail-overexpressing SW1990/GZ cells, followed by intraperitoneal
injection with an equal volume of normal saline. The animals in the
GEM and GEM + Snail OE groups, nude mice were injected with
SW1990/GZ cells and Snail-overexpressing SW1990/GZ cells,
respectively, followed by intraperitoneal injection with 60 mg/kg
GEM at a volume of 10 ml/kg. Animals in the GEM + Emo and GEM +
Snail OE + Emo groups were xenografted with SW1990/GZ cells and
Snail-overexpressing SW1990/GZ cells, respectively, followed by
intraperitoneal injection of 60 mg/kg GEM and subcutaneous
injection of 1 mg/kg Emo at a volume of 10 ml/kg (23). A total of three mice were used for
each group. All administrations were initiated when the tumor
volume reached 100 mm3. GEM was administered on days 1,
4, 7, 10, 14 and 16, whereas Emo was administered daily. Tumor
volume was measured and calculated using the following formula:
V=LxW2/2, where V represents volume in mm3, L
represents the greatest diameter in mm and W represents the lowest
diameter in mm. The animals were euthanized using the method of
CO2 euthanasia and then the tumors were explanted,
weighed and images captured.
H&E staining
The tumor tissues were collected, washed with
sterile water for 2 h and dehydrated with 70, 80 and 90% ethanol
series. Subsequently, the tissues were incubated with equal quality
ethanol and xylene for 15 min, and then incubated with xylene of
equal quality for another 15 min. This incubation procedure was
repeated until the tissues were cleared. Finally, the tissues were
embedded in paraffin, sectioned at a 5-mm thickness and stained
with H&E. Finally, tissues were observed in five
randomly-selected fields under an inverted microscope (Olympus
Corporation), with images captured.
Immunohistochemical assay
Tissue sections were deparaffinized by rinsing with
xylene for 10 min, followed by hydration using a 2-min incubation
with absolute ethyl alcohol, 95% ethyl alcohol, 85% ethyl alcohol
and 75% ethyl alcohol, successively. Sections were rinsed with PBS
and incubated with endogenous peroxidase blocking solution (P0100A;
Beyotime Institute of Biotechnology) at room temperature for 10
min, followed by incubation with 10% goat serum (Gibco; Thermo
Fisher Scientific, Inc.) blocking solution at room temperature for
2 h. Specimens were then incubated with primary antibodies against
E-cadherin (1:10; cat. no. ab231303; Abcam), vimentin (1:50; cat.
no. ab20346; Abcam), Snail (1:50; cat. no. AF6032; Affinity
Biosciences, Ltd.), Nanog (1:100; cat. no. 14295-1-AP; Proteintech
Group, Inc.; Wuhan Sanying Biotechnology), CD44 (1:100, 15675-1-AP,
Proteintech Group, Inc.; Wuhan Sanying Biotechnology), or ALDH1
(1:100, 15910-1-AP, Proteintech Group, Inc.; Wuhan Sanying
Biotechnology) at 4˚C for 24 h, followed by washing with PBS.
Following primary antibody incubation, the tissues were incubated
with the goat anti-rabbit IgG H&L (HRP) preadsorbed secondary
antibody (1:100; cat. no. ab97080; Abcam) was added and incubated
at 4˚C for 24 h, followed by rinsing and staining with
3,3'-diaminobenzidine dye (MilliporeSigma). Finally, images were
captured using an inverted light microscope (Eclipse E100; Nikon
Corporation).
Ethics statements
All animal experiments performed in this study were
authorized by the ethical committee of Zhejiang Cancer Hospital
(approval no. 2019-07-006) and conducted according to the
guidelines for care and use of laboratory animals and the
principles of laboratory animal care and protection.
Statistical analysis
Statistical analysis was performed using SPSS
(version 25.0; IBM Corp.). The Kruskal-Wallis test was used for
unequal distribution of tumor weights and single comparisons were
performed using the Mann-Whitney test. For analysis on other data,
one-way ANOVA followed by Tukey's post hoc test was applied.
P<0.05 was considered to indicate a statistically significant
difference.
Results
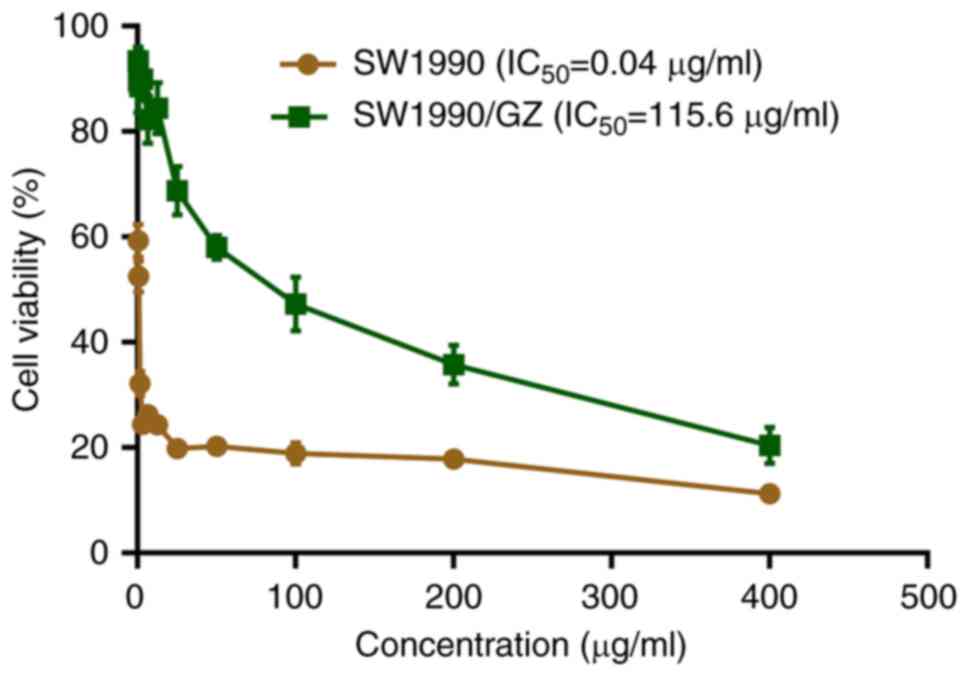
GEM-resistant SW1990 (SW1990/GZ) cell
line is successfully established
SW1990/GZ cell line was successfully established by
treating SW1990 cells with increasing concentrations of GEM as
confirmed using the CCK-8 assay. Compared with that of SW1990
cells, IC50 of GEM changed from 0.04 to 115.6 µg/ml,
with a ~3,000-fold increase (Fig.
1).
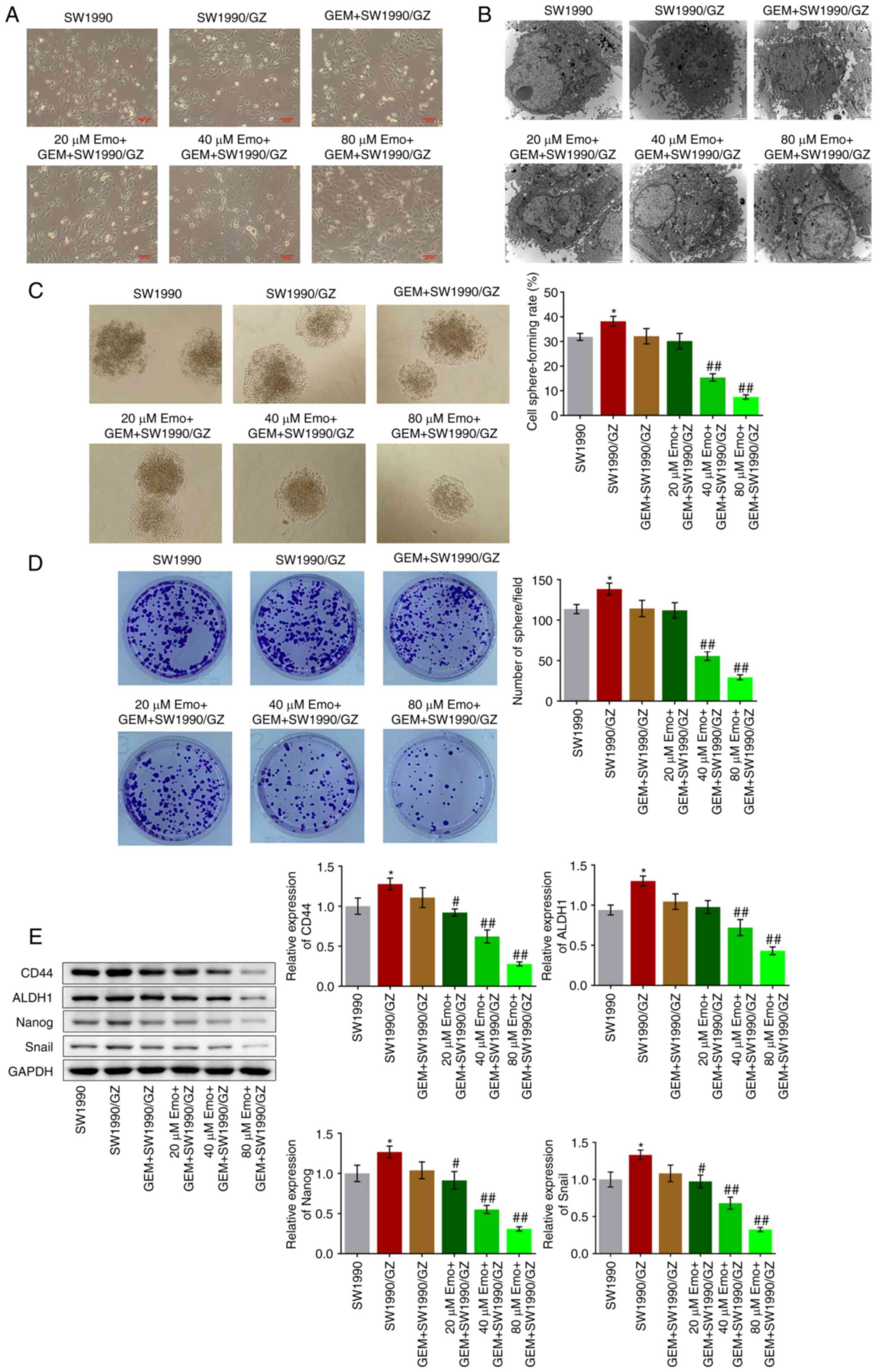
Emo significantly enhances the
inhibitory effect of GEM on the self-renewal of SW1990/GZ
cells
A total of 6 groups were used to explore the in
vitro antitumor effects of Emo: SW1990 (naïve SW1990 cells
treated with blank medium), SW1990/GZ (SW1990/GZ cells incubated
with blank medium), GEM + SW1990/GZ (SW1990/GZ cells incubated with
0.04 µg/ml GEM), 20 µM Emo + GEM + SW1990/GZ (SW1990/GZ cells
incubated with 0.04 µg/ml GEM and 20 µM Emo), 40 µM Emo + GEM +
SW1990/GZ (SW1990/GZ cells incubated with 0.04 µg/ml GEM and 40 µM
Emo) and 80 µM Emo + GEM + SW1990/GZ (SW1990/GZ cells incubated
with 0.04 µg/ml GEM and 80 µM Emo). The cellular morphology of each
group is shown in Fig. 2A.
Compared with the SW1990 cells (Fig.
2B), atypical nuclei, large nucleoli and organelle hypertrophy
were observed in the SW1990/GZ, GEM + SW1990/GZ and 20 µM Emo + GEM
+ SW1990/GZ groups, which were dramatically ameliorated by the
introduction of 40 and 80 µM Emo, indicating that the malignancy of
SW1990/GZ cells was significantly inhibited by GEM in the presence
of 40 and 80 µM Emo.
The self-renewal of SW1990/GZ cells was further
investigated by using sphere formation and colony-formation assays.
Compared with the SW1990 group, the cell sphere-forming rate
increased from 31.9 to 38.2% in the SW1990/GZ group, which further
declined to 32.1% after treatment with GEM, with no significant
difference (P<0.05 vs. SW1990; Fig.
2C). Compared with the GEM + SW1990/GZ group, the cell
sphere-forming rate was suppressed to 30.2, 15.4 and 7.5% by
co-treatment with 20, 40 and 80 µM Emo, respectively (P<0.01 vs.
GEM + SW1990/GZ; Fig. 2C). In
addition, the number of spheres was significantly greater in the
SW1990/GZ group than that in the SW1990 group (P<0.05 vs.
SW1990; Fig. 2D). After the
introduction of 40 µM and 80 µM Emo, the number of spheres
decreased significantly (P<0.01 vs. GEM + SW1990/GZ; Fig. 2D). The expression levels of stem
cell biomarkers were determined through western blot analysis.
Compared with SW1990 cells, the expression levels of CD44, ALDH1,
Nanog and Snail were significantly increased in SW1990/GZ cells
(P<0.05 vs. SW1990; Fig. 2E).
After the introduction of 20, 40 and 80 µM Emo, the expression
levels of CD44, ALDH1, Nanog and Snail were decreased (P<0.05
vs. and P<0.01 vs. GEM + SW1990/GZ; Fig. 2E).
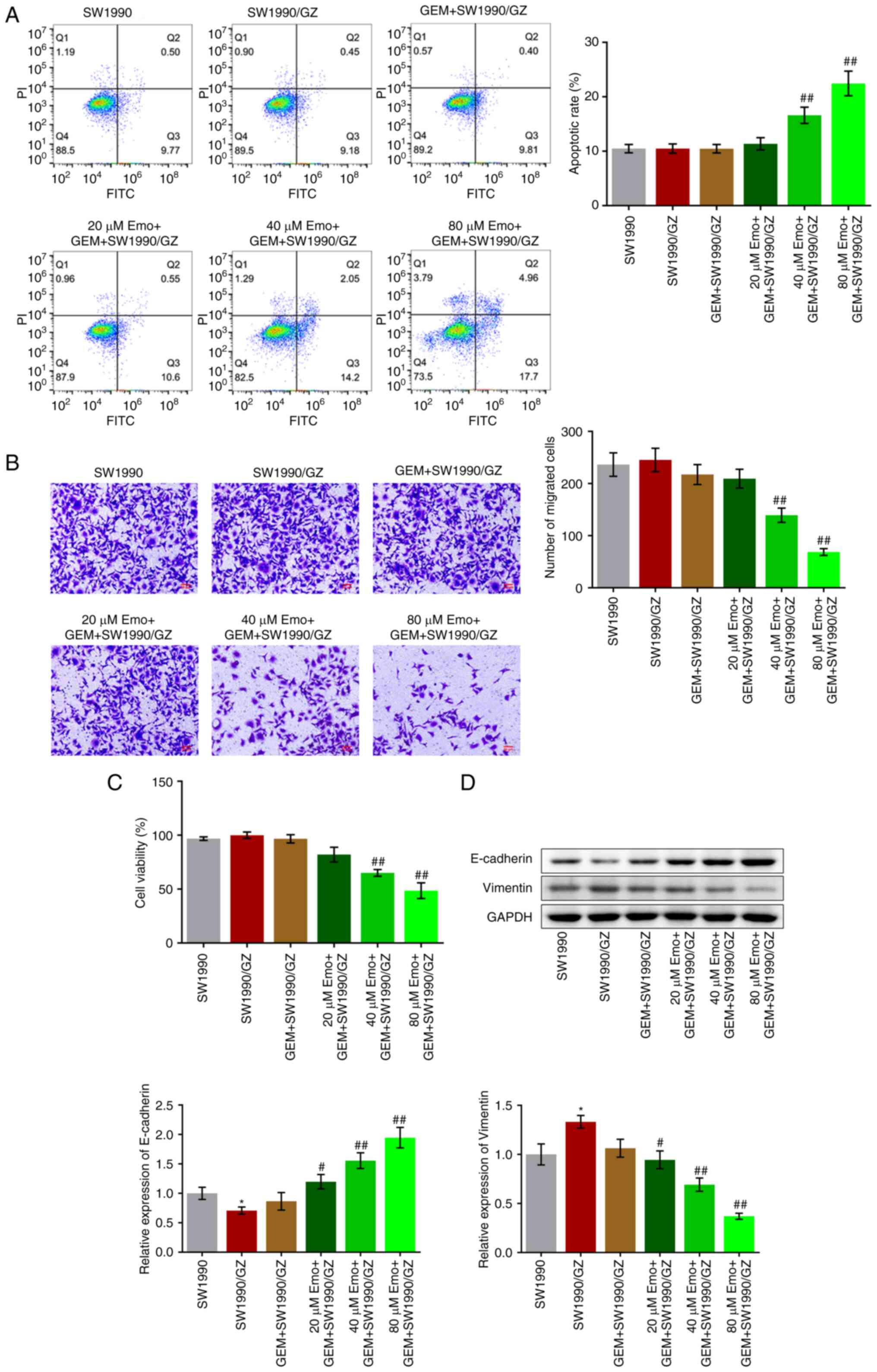
Inhibitory effect of GEM on the
proliferation, metastasis and EMT progression of SW1990/GZ cells is
enhanced by Emo
The apoptotic rate in the SW1990, SW1990/GZ and GEM
+ SW1990/GZ groups was 10.48, 10.49 and 10.46%, respectively
(Fig. 3A). Compared with the GEM +
SW1990/GZ group, the apoptotic rate increased to 11.37, 16.61 and
22.45% after co-treatment with 20, 40 and 80 µM Emo, respectively
(P<0.01 vs. GEM + SW1990/GZ). In addition, no significant
difference in the number of migrated cells was observed among the
SW1990, SW1990/GZ, GEM + SW1990/GZ and 20 µM Emo + GEM + SW1990/GZ
groups. Compared with the GEM + SW1990/GZ group, the number of
migrated cells decreased with the co-introduction of 40 and 80 µM
Emo (P<0.01 vs. GEM + SW1990/GZ). No significant differences in
cell viability were observed among the SW1990, SW1990/GZ, GEM +
SW1990/GZ and 20 µM Emo + GEM + SW1990/GZ groups (Fig. 3C). After treatment with 40 and 80
µM Emo, cell viability was significantly reduced (P<0.01 vs. GEM
+ SW1990/GZ). Finally, expression levels of the EMT biomarkers were
measured. Compared with the SW1990 group, E-cadherin was
significantly downregulated and vimentin was significantly
upregulated in the SW1990/GZ group (Fig. 3D). Compared with the GEM +
SW1990/GZ group, E-cadherin expression was upregulated, while
vimentin was downregulated by co-treatment with 20, 40 and 80 µM
Emo, respectively (P<0.05 vs. SW1990; P<0.05 vs. GEM +
SW1990/GZ; P<0.01 vs. GEM + SW1990/GZ).
Emo represses self-renewal and EMT
progression of SW1990/GZ cells
Compared with SW1990/GZ cells, significantly
decreased cell sphere-forming rate and number of spheres were
observed in 80 µM Emo-treated SW1990/GZ cells (Fig. S1A and B). Furthermore, compared with the
SW1990/GZ cells, significantly lower expression of CD44, ALDH1,
Nanog and Snail was observed with 80 µM Emo-treated SW1990/GZ cells
(P<0.05 vs. SW1990/GZ; Fig.
S1C). However, no significant changes were observed in the
apoptotic rate (Fig. S1D) and the
number of migrated cells (Fig.
S1E) after treatment with 80 µM Emo. Compared with SW1990/GZ
cells, upregulated E-cadherin and downregulated vimentin (Fig. S1F) were observed in 80 µM
Emo-treated SW1990/GZ cells (P<0.05 and P<0.01 vs.
SW1990/GZ).
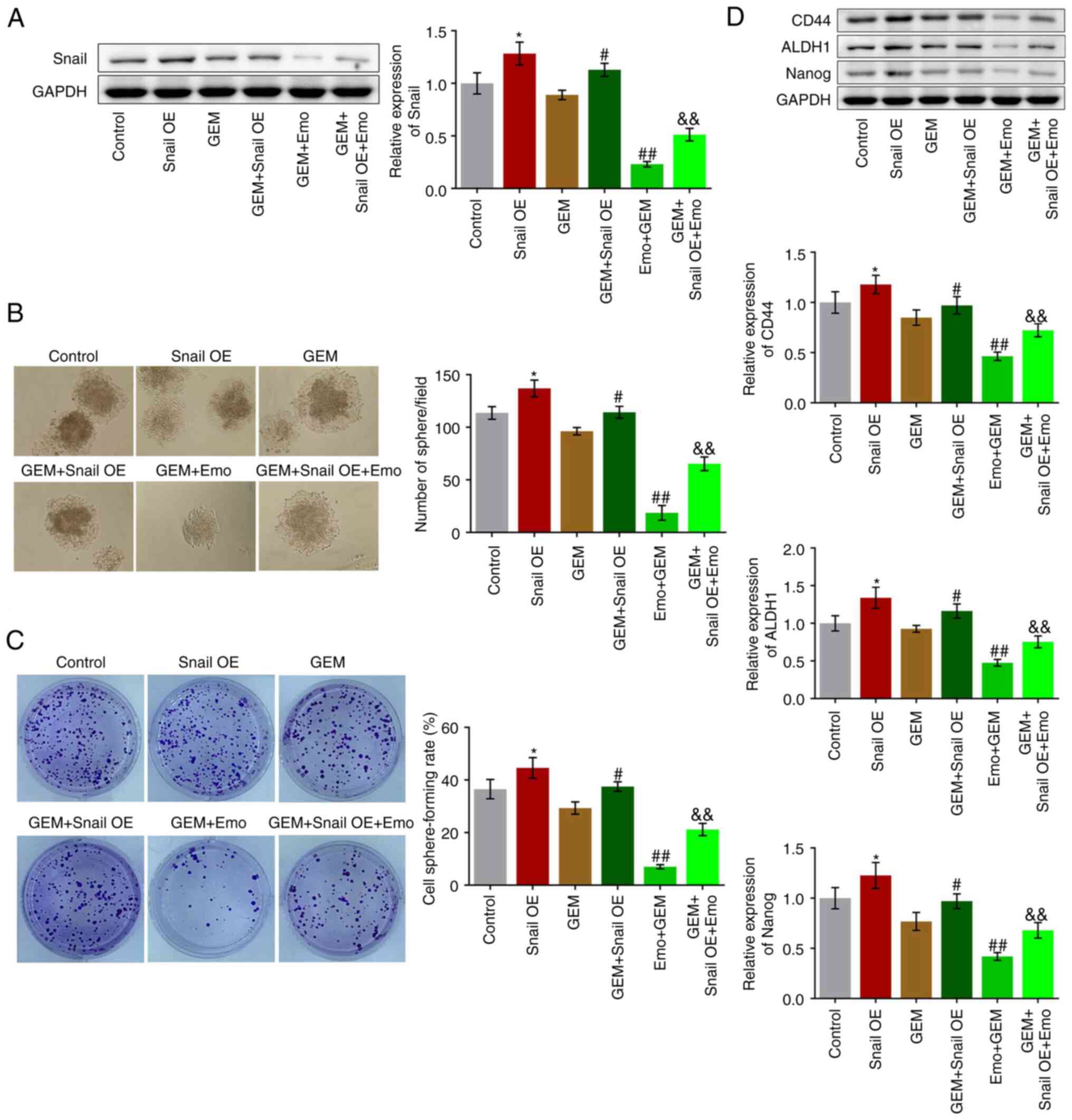
Snail overexpression abolishes the
effects of Emo on self-renewal of GEM-treated SW1990/GZ cells
SW1990/GZ cells overexpression Snail were
constructed to explore the potential mechanism underlying the
effects of Emo. Firstly, the successfully establishment of
Snail-overexpressed SW1990/GZ cells was verified by Western
blotting assay, which was shown in Fig. S2. Six groups were compared:
Control (SW1990/GZ cells cultured in blank medium), Snail OE
(Snail-overexpressed SW1990/GZ cells cultured in blank medium), GEM
(SW1990/GZ cells treated with 0.04 µg/ml GEM), GEM + Snail OE
(Snail-overexpressed SW1990/GZ cells treated with 0.04 µg/ml GEM),
GEM + Emo (SW1990/GZ cells treated with 0.04 µg/ml GEM and 80 µM
Emo) and GEM + Snail OE + Emo (Snail-overexpressed SW1990/GZ cells
treated with 0.04 µg/ml GEM and 80 µM Emo).
Compared with the control, Snail was significantly
upregulated in the Snail OE group, indicating the successful
establishment of Snail-overexpressing SW1990/GZ cells (P<0.05
vs. control; Fig. 4A). Compared
with the GEM group, Snail was significantly upregulated in the GEM
+ Snail OE group and downregulated in the Emo + GEM group, while
Snail was significantly upregulated in the GEM + Snail OE + Emo
group compared with the Emo + GEM group (P<0.05 and P<0.01
vs. GEM group; P<0.01 vs. Emo + GEM). Compared with the control,
the cell sphere-forming rate was significantly elevated from 36.5
to 44.6% in the Snail OE group (P<0.05; Fig. 4B). Compared with the GEM group, the
cell sphere-forming rate was increased from 29.3 to 37.5% in the
GEM + Snail OE group and decreased from 29.3 to 7.01% in the Emo +
GEM group, while was further increased to 21.20% in the Emo + Snail
OE + GEM group (P<0.05 vs. control; P<0.05 vs. GEM; P<0.01
vs. GEM; P<0.01 vs. Emo + GEM: Fig.
4B). In addition, compared with the control, the number of
spheres dramatically increased in the Snail OE group (P<0.05;
Fig. 4C). Compared with the GEM
group, the number of spheres was increased in the GEM + Snail OE
group and decreased in the Emo + GEM group, which was further
increased in the GEM + Snail OE + Emo group compared with the Emo +
GEM group (P<0.05 vs. GEM; P<0.01 vs. GEM; P<0.01 vs. Emo
+ GEM: Fig. 4C).
Finally, the expression levels of the stem cell
biomarkers were determined. Compared with the control, CD44, ALDH1
and Nanog were significantly upregulated in the Snail OE group
(P<0.05; Fig. 4D). Compared
with the GEM group, CD44, ALDH1 and Nanog were significantly
upregulated in the GEM + Snail OE group, downregulated in the Emo +
GEM group, while was further upregulated in the Emo + Snail OE +
GEM group compared with the Emo + GEM group (P<0.05 vs. GEM;
P<0.01 vs. GEM; P<0.01 vs. Emo + GEM; Fig. 4D).
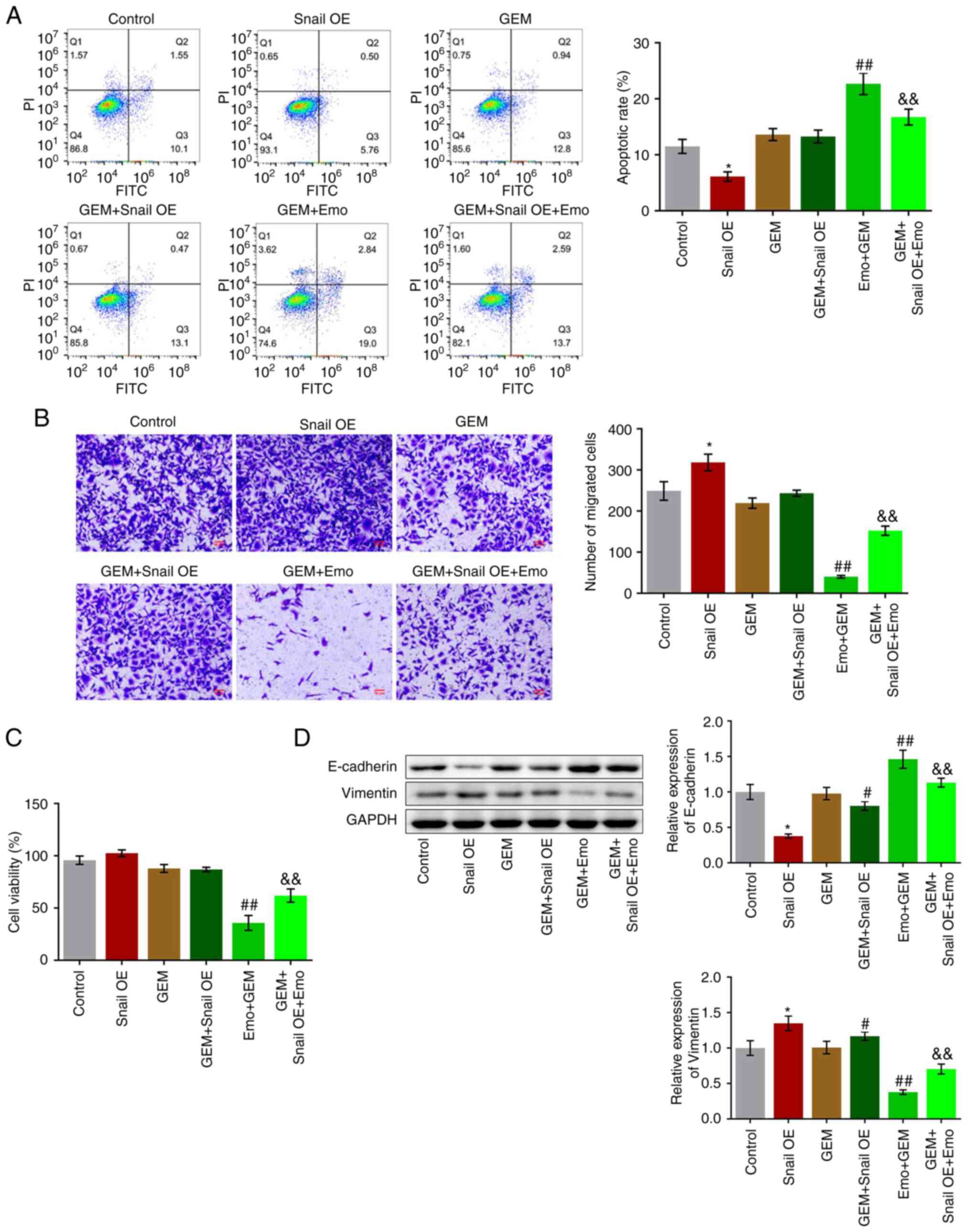
Snail overexpression abolishes the
effect of Emo on proliferation, metastasis and EMT progression in
GEM-treated SW1990/GZ cells
The apoptotic rate significantly decreased from
11.65 to 6.26% in the Snail OE group (P<0.05; Fig. 5A), while it was increased to 13.74%
in GEM group (Fig. 5A). Compared
with the GEM group, no significant difference was observed in the
apoptotic rate in the GEM + Snail OE group and the apoptotic rate
was significantly increased to 22.66% in the Emo + GEM group, while
it was further decreased to 16.29% in the Emo + Snail OE + GEM
group compared with the Emo + GEM group (P<0.01 vs. GEM;
P<0.01 vs. Emo + GEM). In addition, compared with the control,
the number of migrated cells was significantly increased in the
Snail OE group (P<0.05 vs. control; Fig. 5B). Compared with the GEM group, the
number of migrated cells was slightly increased in the GEM + Snail
OE group and decreased in the Emo + GEM group, while it was
significantly elevated in the GEM + Snail OE + Emo group compared
with the Emo + GEM group (P<0.01 vs. GEM; P<0.01 vs. Emo +
GEM). No significant difference in cell viability was observed
among the control, Snail OE, GEM and GEM + Snail OE groups.
Compared with the GEM group, cell viability was significantly
reduced in the Emo + GEM group, which while it was significantly
increased in the Emo + Snail OE + GEM group compared with the Emo +
GEM group (P<0.01 vs. GEM; P<0.01 vs. Emo + GEM; Fig. 5C). Finally, the expression levels
of the EMT biomarkers were evaluated. Compared with the control,
E-cadherin was significantly downregulated and vimentin was
significantly upregulated in the Snail OE group (P<0.05;
Fig. 5D). Compared with the GEM
group, the expression level of E-cadherin was significantly
decreased and that of vimentin was significantly increased in the
GEM + Snail OE group. Compared with the GEM group, the expression
levels of E-cadherin and vimentin were significantly increased and
decreased, respectively, while the expression levels of E-cadherin
and vimentin were significantly decreased and increased,
respectively, in the Emo + Snail OE + GEM group compared with the
Emo + GEM group (P<0.05 and P<0.01 vs. GEM; P<0.01 vs. Emo
+ GEM).
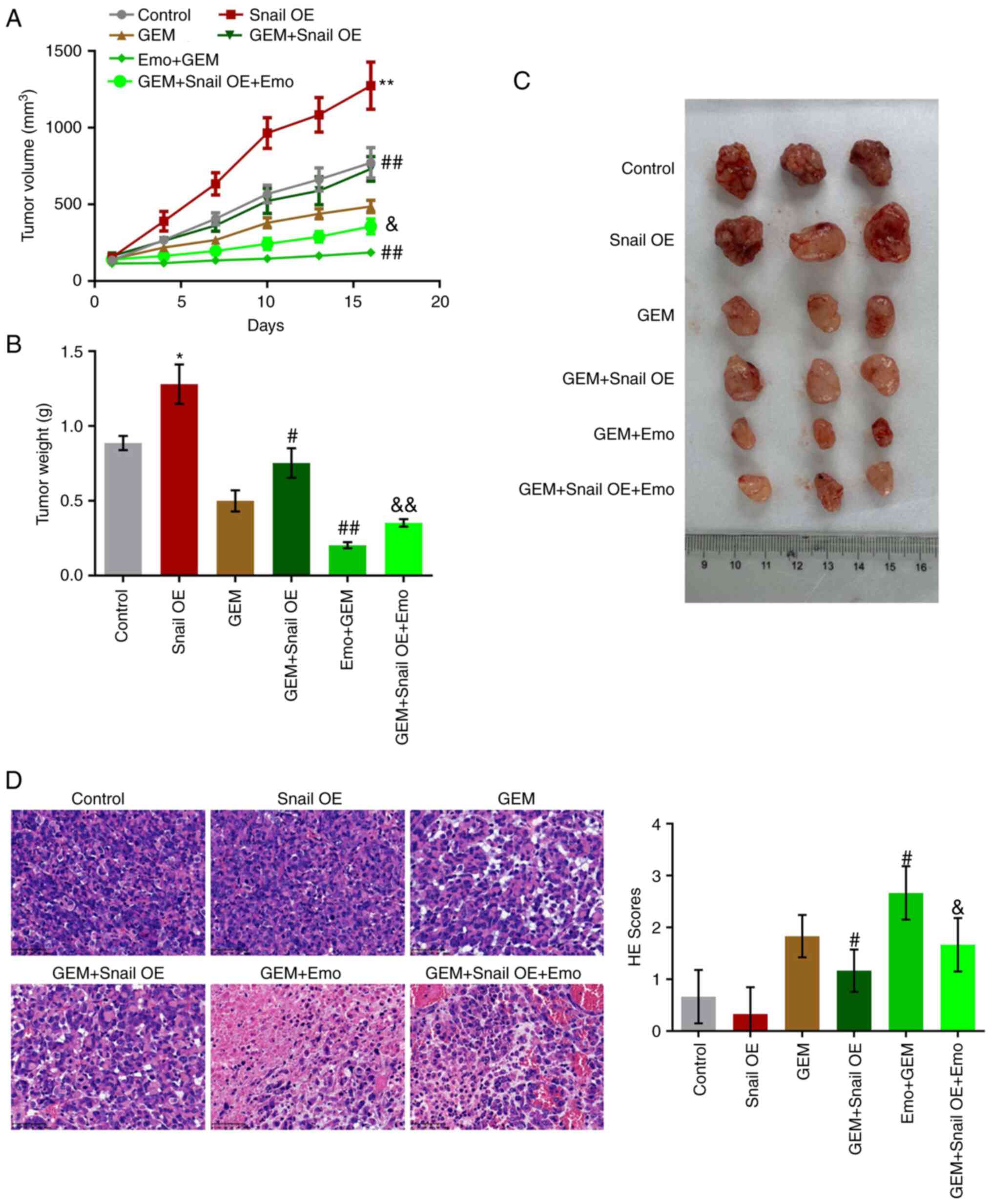
Snail overexpression abolishes the
enhancement of Emo on the anti-tumor efficacy of GEM on SW1990/GZ
xenograft model
To verify the effects of Emo on the anti-tumor
efficacy of GEM against GEM-resistant pancreatic cancer, a
xenograft model in nude mice with SW1990/GZ cells or
Snail-overexpressing SW1990/GZ cells was established. After 16 days
of treatments, tumor volume and weight were significantly increased
in the Snail OE group compared with control (Fig. 6A-C). Compared with the GEM group,
tumor volume and weight were significantly increased in the GEM +
Snail OE group and decreased in the Emo + GEM group, which were
significantly reversed in the Emo + Snail OE + GEM group.
Furthermore, H&E staining (Fig.
6D) was used to evaluate the pathological state of the tumor
tissues. No significant differences were observed between the
control and Snail OE groups. However, compared with the GEM group,
the H&E score was greatly reduced in the GEM + Snail OE group
and elevated in the Emo + GEM group, which was dramatically rescued
in the Emo + Snail OE + GEM group. (P<0.05 vs. control;
P<0.01 vs. control; P<0.05 vs. GEM; P<0.01 vs. GEM;
P<0.05 vs. Emo + GEM; P<0.01 vs. Emo + GEM).
Snail overexpression abolishes the
inhibitory effect of Emo against stem cell growth and EMT
progression in SW1990/GZ xenograft model
Tumor tissues were homogenized for western blotting
analysis and tissue sections were utilized for immunohistochemical
assays. Compared with the control, the expression levels of CD44,
ALDH1, Nanog and Snail were significantly elevated in the Snail OE
group (P<0.05 vs. control; Fig.
7A and B). Compared with the
GEM group, CD44, ALDH1, Nanog and Snail were significantly
upregulated in the GEM + Snail OE group and downregulated in the
Emo + GEM group, which was reversed in the Emo + Snail OE + GEM
group (P<0.05 and P<0.01 vs. GEM; P<0.05 vs. Emo + GEM;
P<0.01 vs. Emo + GEM). These data indicate that the stem-like
symptoms in SW1990/GZ tumors were significantly alleviated by Emo
by inactivating Snail. In addition, compared with the control, the
expression level of E-cadherin was decreased and that of vimentin
was increased in the Snail OE group (P<0.05; Fig. 7C and D). Compared with the GEM group,
E-cadherin was downregulated and vimentin was upregulated in the
GEM + Snail OE group and E-cadherin was upregulated and vimentin
was downregulated in the Emo + GEM group, while it was
significantly reversed in the Emo + Snail OE + GEM group compared
to the Emo + GEM group (P<0.05 and P<0.01 vs. GEM; P<0.05
and P<0.01 vs. Emo + GEM).
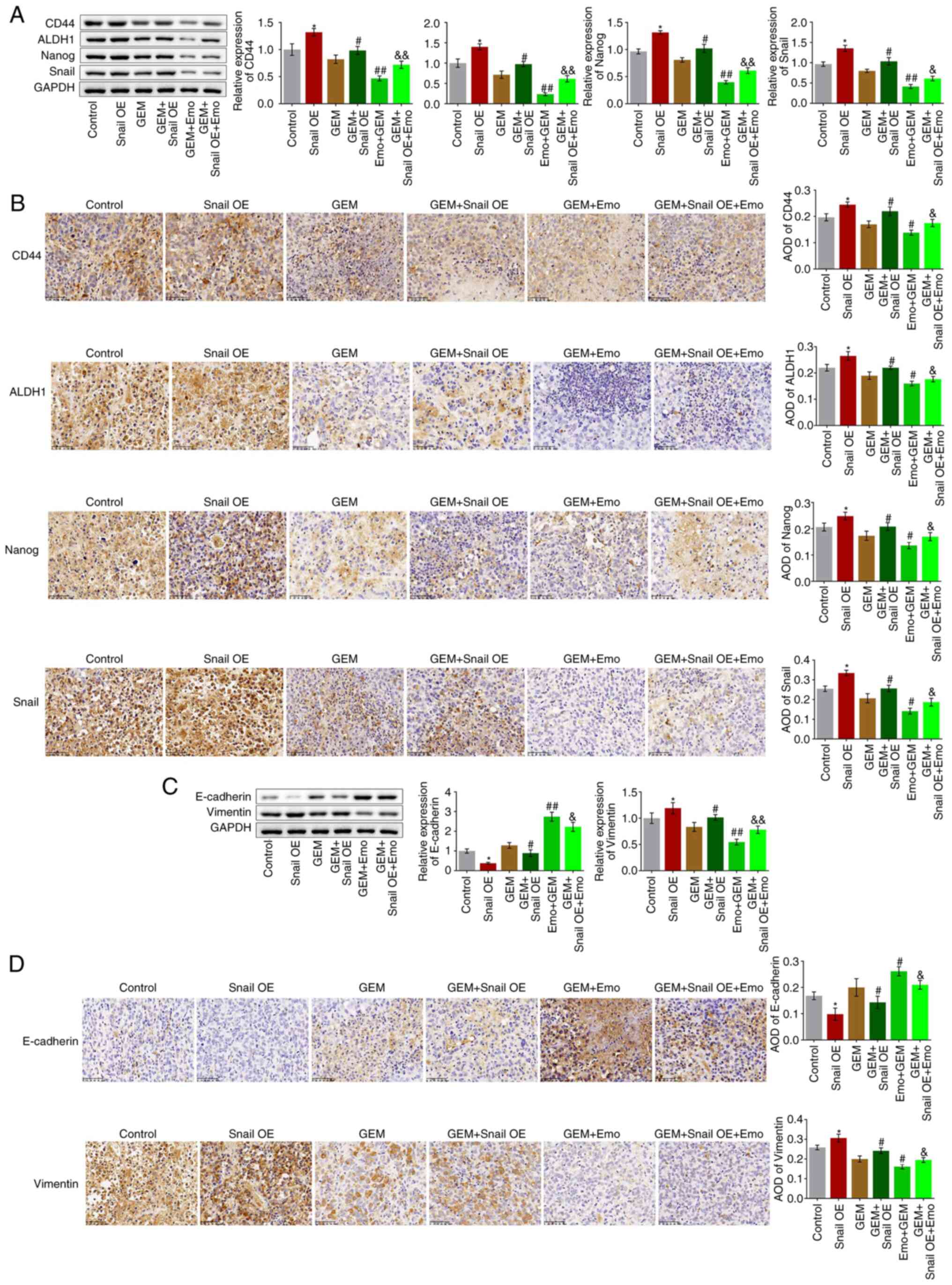 | Figure 7Growth of stem cells and EMT
progression in SW1990/GZ xenograft model is inhibited by Emo by
inactivating Snail. (A) Expression levels of CD44, ALDH1, Nanog and
Snail as determined via western blotting. (B) Expression level of
CD44, ALDH1, Nanog and Snail in tumor tissues measured via
immunohistochemistry (x200 magnification). (C) Expression levels of
vimentin and E-cadherin as determined via western blotting. (D)
Expression levels of vimentin and E-cadherin in tumor tissues
measured via immunohistochemistry (x200 magnification).
*P<0.05 vs. control. #P<0.05 vs. GEM.
##P<0.01 vs. GEM. &P<0.05 vs. Emo +
GEM. &&P<0.01 vs. Emo + GEM. Emo, emodin;
GEM, gemcitabine; SW1990/GZ, GEM-resistant SW1990 cells; Snail,
Snail family transcriptional repressor 1 gene; EMT,
epithelial-mesenchymal transition; ALDH1, Aldehyde dehydrogenase 1;
Snail, Snail family transcriptional repressor 1 gene; Snail OE,
Snail overexpression. |
Discussion
Resistance to chemotherapy is an important barrier
in the clinical treatment of pancreatic cancer (24). CSC-like characteristics in
pancreatic tumor cells, which contribute to chemotherapy
resistance, tumorigenesis, tumor relapse and metastasis have been
shown in multiple reports (25-27).
In addition, similar biofunctions are observed between CSCs and
tumor cells with EMT phenotypes, which are reported to be involved
in the development and progression of chemotherapy resistance,
tumor relapse and metastasis (7).
In the present study, GEM-resistant pancreatic tumor cells were
established in SW1990 cells by successively doubling the
concentration of GEM, which was verified by the increased
IC50 against GEM in a 3,000-fold manner. In addition,
the resistance of SW1990/GZ cells to GEM was accompanied by
enhanced self-renewal and EMT progression, which was consistent
with a previous report (28).
Pancreatic circulating tumor cells enter the
circulatory system to achieve EMT phenotypes and their migration is
significantly enhanced. Rhim et al (29) report that in pancreatic circulating
tumor cells, the number of CD24+CD44+ cells
is ~100-fold higher compared with that in pancreatic tumor tissues,
indicating that stem-like characteristics are observed in tumor
cells with EMT phenotypes. It has been reported that in isolated
CD24+CD44+ pancreatic tumor cells,
significant resistance against GEM is observed, accompanied by
upregulation of EMT phenotypes (25). Voon et al (30) induced EMT phenotypes in GIF-14
rodent gastric epithelial cells using TGF-β1 by activating the
EGFR/RAS pathway and found that the expression level of the stem
cell biomarker leucine-rich repeat-containing G-protein coupled
receptor 5 was significantly elevated, accompanied by enhanced
sphere and colony-formation. In the present study, Emo treatment
significantly enhanced the inhibitory effect of GEM against the
proliferation and migration of SW1990/GZ cells, accompanied by
suppressed stem-like and EMT phenotypes, indicating that Emo might
reverse the GEM resistance of SW1990/GZ cells by inhibiting the
formation of CSCs and the progression of EMT. In vivo
experiments further confirmed the role of Emo as enhancer of the
anti-tumor effect of GEM against GEM-resistant SW1990/GZ
xenografts, accompanied by the inhibition of stem-like and EMT
phenotypes in tumor tissues, which was consistent with the results
observed in the in vitro assays.
In the progression of EMT, the expression of
biomarkers is regulated by several transcriptional factors, such as
Snail, Twist and Zeb, among which Snail is an important
transcriptional factor originally discovered in Drosophila
melanogaster and proved to be the basis for mesoderm formation
(31). By directly binding to the
E-box sequence on the promoter of E-cadherin, Snail suppresses EMT
progression by inhibiting E-cadherin transcription (32). Xiong et al (33) claimed a significant role in
regulating EMT progression in pancreatic CSCs (33). In the present study,
Snail-overexpressing SW1990/GZ cells were established. These cells
showed enhanced proliferation, migration, self-renewal and EMT
progression, in agreement with previous results (34,35).
By comparing the results of Emo-treated SW1990/GZ cells and
Emo-treated Snail-overexpressing SW1990/GZ cells it was found that
the reverse effects of Emo against GEM-resistant pancreatic cancer
were significantly abolished by the overexpression of Snail in
vitro and in vivo, indicating that Emo enhanced the
anti-tumor efficacy of GEM against GEM-resistant pancreatic cancer
by downregulating Snail. However, some findings in the present
study require further investigation. For example, compared with the
control, the cell proliferation and migration were significantly
enhanced in the Snail-overexpressing group. However, no significant
differences were observed between the GEM and GEM + Snail OE
groups. Moreover, the establishment of Snail-overexpressed cells
was not as successful as expected. The relatively low transfection
efficiency of the transfection reagent might be accountable for
this. In future studies, a more successful establishment of
Snail-overexpressed cells will be explored to verify the data
achieved in the present study. In addition, further investigation
need to be performed to explore the underlying molecular mechanism
of the regulatory effect of Emo on the expression level of Snail,
such as screening differentially expressed miRNAs by using gene
chips.
In the present study, promising effects of Emo
against GEM-resistance in pancreatic cancer were observed. However,
in developing Emo as a drug, its limitations should be considered,
including poor oral bioavailability (36) and rapid elimination (37). In further research, developing
formulation methods to improve the oral bioavailability of Emo,
such as introducing a solubility enhancer or crystallization
inhibitor, is necessary. Furthermore, hepatotoxicity,
nephrotoxicity and reproductive toxicity of Emo have been reported
(38), which significantly limits
the its further development as therapeutic drug. Further research
is needed to investigate structure-function relationships to
explore the potential of optimizing the structure of Emo to reduce
off-target toxicity and maintain on-target activity.
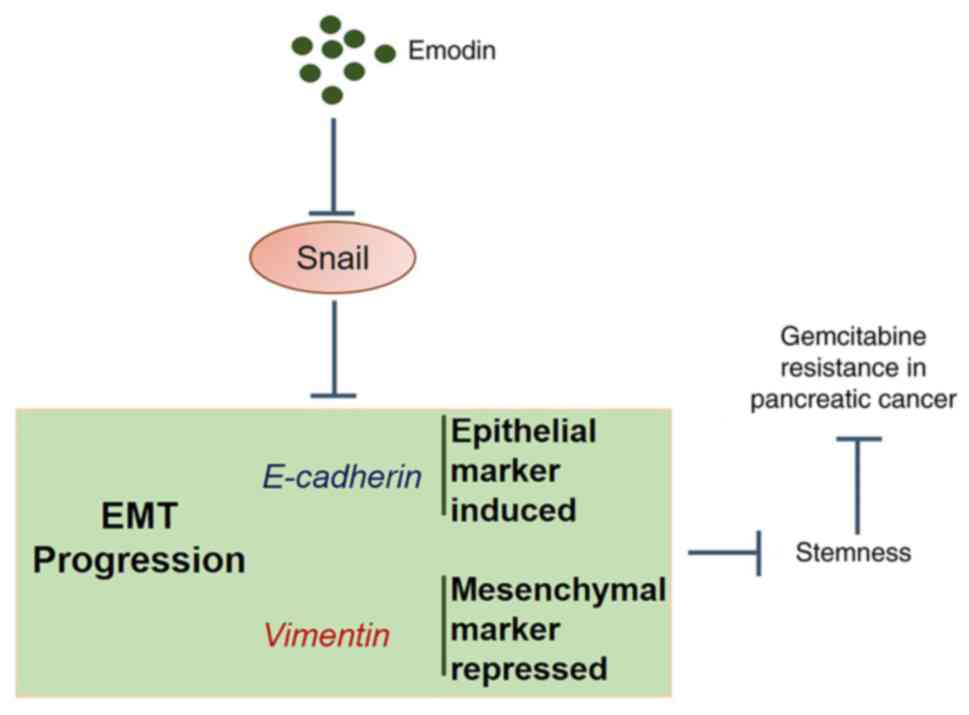
The data from the present study revealed that Emo
may reverse GEM-resistance in pancreatic cancer by suppressing
stemness through the regulation of EMT progression (Fig. 8).
Supplementary Material
Emo represses the self-renewal and EMT
progression of SW1990/GZ cells. (A) Cell sphere-forming rate
evaluated using the sphere formation assay (x100 magnification).
(B) Number of spheres measured using the colony-formation assay
(x200 magnification). (C) Expression levels of CD44, ALDH1, Nanog
and Snail as determined via western blotting. (D) Flow cytometry
used to detect the apoptosis of cells. (E) Cell migration measured
via Transwell assay (scale bar, 50 μm). (F) Expression
levels of vimentin and E-cadherin via western blotting.
*P<0.05 vs. SW1990/GZ. **P<0.01 vs.
SW1990/GZ. Emo, emodin; GEM, gemcitabine; SW1990/GZ, GEM-resistant
SW1990 cells; Snail, Snail family transcriptional repressor 1 gene;
EMT, epithelialmesenchymal transition; ALDH1, Aldehyde
dehydrogenase 1; Snail, Snail family transcriptional repressor 1
gene.
Snail-overexpressed SW1990/GZ cells.
Expression level of Snail was determined via western blotting.
*P<0.01 vs. control. SW1990/GZ, GEM-resistant SW1990
cells; Snail, Snail family transcriptional repressor 1 gene; NC,
negative control.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Zhejiang Province (grant no. LQ20H310001).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and WW were responsible for the conception and
design of the research. JW was responsible for the acquisition,
analysis and interpretation of the data. YH performed the
statistical analysis. JL obtained funding for the study. WW and SC
drafted the manuscript. SC performed the experiments and revised
the manuscript for important intellectual content. JL and WW
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments performed in this study were
authorized by the ethical committee of Zhejiang Cancer Hospital
(approval no. 2019-07-006) and conducted according to the
guidelines for care and use of laboratory animals and the
principles of laboratory animal care and protection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Park W, Chawla A and O'Reilly EM:
Pancreatic cancer: A review. JAMA. 326:851–862. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sarvepalli D, Rashid MU, Rahman AU, Ullah
W, Hussain I, Hasan B, Jehanzeb S, Khan AK, Jain AG, Khetpal N and
Ahmad S: Gemcitabine: A review of chemoresistance in pancreatic
cancer. Crit Rev Oncog. 24:199–212. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang L, Dong P, Wang W, Huang M and Tian
B: Gemcitabine treatment causes resistance and malignancy of
pancreatic cancer stem-like cells via induction of lncRNA HOTAIR.
Exp Ther Med. 14:4773–4780. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
de Sousa Cavalcante L and Monteiro G:
Gemcitabine: Metabolism and molecular mechanisms of action,
sensitivity and chemoresistance in pancreatic cancer. Eur J
Pharmacol. 741:8–16. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gangemi R, Paleari L, Orengo AM, Cesario
A, Chessa L, Ferrini S and Russo P: Cancer stem cells: A new
paradigm for understanding tumor growth and progression and drug
resistance. Curr Med Chem. 16:1688–1703. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Du Z, Qin R, Wei C, Wang M, Shi C, Tian R
and Peng C: Pancreatic cancer cells resistant to chemoradiotherapy
rich in ‘stem-cell-like’ tumor cells. Dig Dis Sci. 56:741–750.
2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Li Y, Kong D, Ahmad A, Bao B and Sarkar
FH: Pancreatic cancer stem cells: Emerging target for designing
novel therapy. Cancer Lett. 338:94–100. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Abel EV and Simeone DM: Biology and
clinical applications of pancreatic cancer stem cells.
Gastroenterology. 144:1241–1248. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sancho P, Alcala S, Usachov V, Hermann PC
and Sainz B Jr: The ever-changing landscape of pancreatic cancer
stem cells. Pancreatology. 16:489–496. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Shah AN, Summy JM, Zhang J, Park SI,
Parikh NU and Gallick GE: Development and characterization of
gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol.
14:3629–3637. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Iwatsuki M, Mimori K, Yokobori T, Ishi H,
Beppu T, Nakamori S, Baba H and Mori M: Epithelial-mesenchymal
transition in cancer development and its clinical significance.
Cancer Sci. 101:293–299. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Babaei G, Aziz SG and Jaghi NZZ: EMT,
cancer stem cells and autophagy; the three main axes of metastasis.
Biomed Pharmacother. 133(110909)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Reiman JM, Knutson KL and Radisky DC:
Immune promotion of epithelial-mesenchymal transition and
generation of breast cancer stem cells. Cancer Res. 70:3005–3008.
2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tian K, Zhang H, Chen X and Hu Z:
Determination of five anthraquinones in medicinal plants by
capillary zone electrophoresis with beta-cyclodextrin addition. J
Chromatogr A. 1123:134–137. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shrimali D, Shanmugam MK, Kumar AP, Zhang
J, Tan BK, Ahn KS and Sethi G: Targeted abrogation of diverse
signal transduction cascades by emodin for the treatment of
inflammatory disorders and cancer. Cancer Lett. 341:139–149.
2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li J, Liu P, Mao H, Wanga A and Zhang X:
Emodin sensitizes paclitaxel-resistant human ovarian cancer cells
to paclitaxel-induced apoptosis in vitro. Oncol Rep.
21:1605–1610. 2009.PubMed/NCBI
|
|
20
|
Way TD, Huang JT, Chou CH, Huang CH, Yang
MH and Ho CT: Emodin represses TWIST1-induced
epithelial-mesenchymal transitions in head and neck squamous cell
carcinoma cells by inhibiting the β-catenin and Akt pathways. Eur J
Cancer. 50:366–378. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Thacker PC and Karunagaran D: Curcumin and
emodin down-regulate TGF-β signaling pathway in human cervical
cancer cells. PLoS One. 10(e0120045)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pooja T and Karunagaran D: Emodin
suppresses Wnt signaling in human colorectal cancer cells SW480 and
SW620. Eur J Pharmacol. 742:55–64. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bai J, Wu J, Tang R, Sun C, Ji J, Yin Z,
Ma G and Yang W: Emodin, a natural anthraquinone, suppresses liver
cancer in vitro and in vivo by regulating VEGFR2 and
miR-34a. Invest New Drugs. 38:229–245. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zeng S, Pöttler M, Lan B, Grützmann R,
Pilarsky C and Yang H: Chemoresistance in pancreatic cancer. Int J
Mol Sci. 20(4504)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yin T, Wei H, Gou S, Shi P, Yang Z, Zhao G
and Wang C: Cancer stem-like cells enriched in Panc-1 spheres
possess increased migration ability and resistance to gemcitabine.
Int J Mol Sci. 12:1595–1604. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323.
2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rao CV and Mohammed A: New insights into
pancreatic cancer stem cells. World J Stem Cells. 7:547–555.
2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Izumiya M, Kabashima A, Higuchi H,
Igarashi T, Sakai G, Iizuka H, Nakamura S, Adachi M, Hamamoto Y,
Funakoshi S, et al: Chemoresistance is associated with cancer stem
cell-like properties and epithelial-to-mesenchymal transition in
pancreatic cancer cells. Anticancer Res. 32:3847–3853.
2012.PubMed/NCBI
|
|
29
|
Rhim AD, Mirek ET, Aiello NM, Maitra A,
Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK,
Vonderheide RH, et al: EMT and dissemination precede pancreatic
tumor formation. Cell. 148:349–361. 2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Voon DC, Wang H, Koo JK, Chai JH, Hor YT,
Tan TZ, Chu YS, Mori S and Ito Y: EMT-induced stemness and
tumorigenicity are fueled by the EGFR/Ras pathway. PLoS One.
8(e70427)2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Grau Y, Carteret C and Simpson P:
Mutations and chromosomal rearrangements affecting the expression
of snail, a gene involved in embryonic patterning in Drosophila
melanogaster. Genetics. 108:347–360. 1984.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xiong Y, Wang Y, Wang L, Huang Y, Xu Y, Xu
L, Guo Y, Lu J, Li X, Zhu M and Qian H: MicroRNA-30b targets Snail
to impede epithelial-mesenchymal transition in pancreatic cancer
stem cells. J Cancer. 9:2147–2159. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang H, Wang HS, Zhou BH, Li CL, Zhang F,
Wang XF, Zhang G, Bu XZ, Cai SH and Du J: Epithelial-mesenchymal
transition (EMT) induced by TNF-α requires AKT/GSK-3β-mediated
stabilization of snail in colorectal cancer. PLoS One.
8(e56664)2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hwang WL, Yang MH, Tsai ML, Lan HY, Su SH,
Chang SC, Teng HW, Yang SH, Lan YT, Chiou SH and Wang HW: SNAIL
regulates interleukin-8 expression, stem cell-like activity, and
tumorigenicity of human colorectal carcinoma cells.
Gastroenterology. 141:279–291, 291.e1-e5. 2011.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu W, Feng Q, Li Y, Ye L, Hu M and Liu Z:
Coupling of UDP-glucuronosyltransferases and multidrug
resistance-associated proteins is responsible for the intestinal
disposition and poor bioavailability of emodin. Toxicol Appl
Pharmacol. 265:316–324. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chen J, Li S, Liu M, Lam CWK, Li Z, Xu X,
Chen Z, Zhang W and Yao M: Bioconcentration and metabolism of
emodin in zebrafish eleutheroembryos. Front Pharmacol.
8(453)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Dong X, Fu J, Yin X, Cao S, Li X and Lin
L: Huyiligeqi and Ni J. Emodin: A review of its pharmacology,
toxicity and pharmacokinetics. Phytother Res. 30:1207–1218.
2016.PubMed/NCBI View Article : Google Scholar
|