Introduction
Hypercholesterolemia is a metabolic disease with
dyslipidemia and can be divided into two categories: One is based
on genetics including familial and polygenic hypercholesterolemia;
the second is based on elevated lipoproteins (1), which are mainly manifested by
abnormally elevated serum or plasma levels of total cholesterol
(TC) and low-density lipoprotein cholesterol (LDL-C) (2). Obesity and diet are important factors
in causing elevated lipoproteins, especially foods that are high in
saturated fatty acids and high in saturated cholesterol (3). Statins are commonly used drugs for
the treatment of hypercholesterolemia (4) but are accompanied by serious adverse
reactions (5), such as liver
dysfunction, rhabdomyolysis and increased susceptibility to
diabetes (6). Therefore, it is
urgent to find other lipid-regulating drugs to reduce the
occurrence of adverse drug reactions.
The Chinese Herbal Medicine Gynostemma
pentaphyllum (Thunb.) Makino is the dried whole herb of
Gynostemma pentaphyllum, family Cucurbitaceae. Gypenosides
(GP), the main active ingredient of G. pentaphyllum, can
significantly reduce blood lipid levels (7). Megalli et al (8) found that following 12 days of oral
administration the gypenosides extract (250 mg/kg) reduced TC and
TG levels in rats and no adverse effects were found with long-term
oral administration of large doses (9,10).
Gypenosides possess a core structure of Dammaran-type triterpenes,
similar to the core structure of endogenous bile acids (7). Bile acids are the main product of
cholesterol metabolism. Excess cholesterol in the liver is
essential for hepatic protection by converting it into bile acids
that enter the bile ducts, maintain cholesterol homeostasis and
prevent cholesterol accumulation in the liver (11). Cholesterol metabolism is closely
related to the processes of bile acid synthesis, metabolism and
transport, which are influenced by proteins and genes related to
the bile acid pathway.
Bile acids are found mainly in the liver and are
mediated by CYP7A1, CYP8B1 and various enzymes for hepatic
cholesterol to cholic acid (CA) and chenodeoxycholic acid (CDCA)
(12), coupled with taurine or
glycine to form bound bile acids taurine/glycine-conjugated cholic
acid (T/G-CA), taurine/glycine-conjugated chenodeoxycholic acid
(T/G-CDCA) (13-15).
The bile acid efflux transporter bile salt export pump (ABCB11) is
responsible for mediating the transport of free and conjugated bile
acids from hepatocytes to the gallbladder. solute carrier organic
anion transporter family member (SLCO) 1A1 is expressed in the
basement membrane of hepatocytes in the portal vein, where it is
thought to be a bidirectional transporter protein that transports
free saprophytic acid and organic anions into the blood (16,17).
SLCO1A1 is expressed on the basal hepatocytes of the hepatic
sinusoids and is responsible for the translocation and reabsorption
of free bile acids from the blood into the liver (18), where they are eventually excreted
through the body's circulation. In addition, the nuclear receptor
farnesoid X receptor (FXR) acts as a bile acid receptor with a role
in regulating glucose metabolism, lipid metabolism and energy
metabolism (19). Bile acids
maintain cholesterol-bile acids homeostasis by activating hepatic
FXR to induce feedback inhibition of CYP7A1 expression by small
heterodimer partner (SHP, NR0B2), thereby inhibiting cholesterol
metabolism and bile acids synthesis (20,21).
NR4A1 is also a key transcription factor for glucose lipid
homeostasis and its overexpression attenuates hepatic triglyceride
production and regulates a variety of key genes involved in lipid
metabolism (22), accelerating the
initial phase of lipogenesis by regulating mitosis (23). Hypercholesterolemia upregulates
NR4A1expression (24) and NR4A1
may be the upstream molecule of the SHP upstream molecule, which
indirectly regulates lipid metabolic processes by regulating
transcription factors (25,26).
The present study analyzed the regulatory effects of
GP on bile acid pathway and the mechanism of hepatic cholesterol
lowering in mice stimulated by HFD through a combination of
metabolomics, proteomics and transcriptomics.
Materials and methods
Plant and Materials
Gypenosides (purity >98% assayed by UV) were
purchased from Zhongxin Biotechnology (Zhejiang) Co., Ltd. The
purity of gypenosides >98% was verified using high-performance
liquid chromatography with diode-array detection (HPLC-DAD) as in a
previous study (7). The standard
reference material of bile acids was bought from MilliporeSigma.
TRIzol® was purchased from Thermo Fisher Scientific,
Inc. Trypsin-EDTA solution, 0.25% (without phenol red) was
purchased from Promega Corporation. Acetonitrile was purchased from
Tedia Co., Inc. Trypsin inhibitors were purchased from Calbiochem
(Merck KGaA). Iodoacetamide (IAA), 1,4-dithiothreitol (DTT) and
urea were purchased from MilliporeSigma. Isopropanol, methanol and
trichloromethane were purchased from Chengdu Kelong Chemical
Reagent Factory.
Animal experiments
The experimental protocol was approved by the
Institutional Animal Care and Use Committee at Zunyi Medical
University (approval no. 2-557). Male C57BL/6J mice weighing 23-25
g, 6-8 weeks old, n=90 were kept in a controlled animal room on the
SPF level, and were purchased from Beijing Huafukang Biotechnology,
approval number: SCXK 2014-0004. Temperature was 21-23˚C and the
humidity 50-60%. The animals consumed food and drank water freely
and a 12-h light/dark cycle was used. Following an acclimation
period of 1 week, the mice were randomly grouped into 3 groups (n=5
per group). One group was fed a normal diet (ND; Research Diets,
Inc.; cat. no. D12450B;10% Kcal fat; energy density: 3.82 kcal/g)
and the others were fed a high-fat diet (HFD) purchased from
Research Diets, Inc.; cat. no. D12492. The processed formula is:
Casein 25.84%, L-cystine 0.39%, maltodextrin 16.15%, sucrose 8.90%,
cellulose 6.46%, soybean oil 3.23%, lard 31.66%, mineral mixture
1.29%, dicalcium phosphate 1.68%, calcium carbonate 0.71%,
potassium citrate 2.13%, vitamin mixture 1.29%, choline Tartaric
acid salt 0.26% and dye 0.01%, respectively. According to the
previous research of the authors, the hypercholesterolemia model
could be built at 16 weeks of HFD (27). One of the HFD groups was treated
under intragastric administration with GP (HFD + GP, 250 mg/kg)
from weeks 17 to 38, while mice in ND and the other HFD group was
treated under intragastric administration with 0.1% of
Carboxymethyl-Na solution (medium used to suspend gypenosides in
reverses osmosis water, once per/d). After 18, 20, 26, 32 and 38
weeks, mice were injected intraperitoneally using 1.5 g/kg of 20%
urethane, then whole blood and liver were taken following
anaesthesia. Blood was centrifuged at 4˚C 4,500 g for 10 min after
settling at 4˚C for 60 min to isolate serum. All of the other
samples were transferred to liquid nitrogen immediately for quick
freezing, then moved to a -80˚C refrigerator until testing and
analysis.
Lipid assessment and hematoxylin and
eosin (HE) staining
The kits for TC and LDL-C assays were purchased from
Nanjing Jiancheng Bioengineering Institute and were used according
to the manufacturer's instructions. The liver was fixed in 10%
neutral formaldehyde at room temperature for 24 h and rinsed under
running water. Dehydration was carried out by immersion in ethanol
solutions of different concentrations (70, 80, 90, 95 and 100%) for
30 min, respectively. Then, it was soaked in xylene solution and
wax in turn (60 min for soft wax, 120 min for soft wax, 120 min for
hard wax). It was embedded in a mold and placed on a freezing table
and left to cool and solidify rapidly for about 10 min and then
removed (modular tissue embed- ding centre; cat. no. EG1150; Leica
Microsystems GmbH). Then the blocks were sectioned at 3-5 µm using
a Leica RM2245 Biosystems (Leica Microsystems GmbH). Tissue
sections were dewaxed and redehydrated before staining with
hematoxylin solution for 5 min, rinsed with running water, stained
with 0.5% eosin staining solution for 1 min and rinsed with tap
water at room temperature. Images were captured at x400
magnification on a light microscope Olympus BX43 (Olympus
Corporation).
Metabolomics of GP regulating
molecular levels of bile acid in mice
Liver was homogenized and then centrifuged at 4˚C at
12,000 g for 15 min. 250 µl of supernatant was removed and blown
dry on a nitrogen blower at room temperature. Rehydrated with 50%
methanol in water, centrifuged under the above conditions and 50 µl
was taken as the sample to be tested.
A chromatographic column, ACQUITY UPLC BEH
C18 (1.7 µm, 3.0x150 mm), was used for the separation of
bile acids. The flow rate was 0.3 ml/min. The injection volume was
10 µl and the column temperature was 45˚C. Mobile phase A was 20%
acetonitrile (containing 5 mmol/l ammonium acetate) and mobile
phase B was 80% acetonitrile (containing 5 mmol/l ammonium acetate)
with gradient elution. The elution conditions and the rest of the
chromatographic conditions are referred to in a previous study
(28) Table I.
 | Table IConditions of gradient elution. |
Table I
Conditions of gradient elution.
| Time (min) | 20% Acetonitrile +
5 mM Ammonium acetate (%) | 80% Acetonitrile +
5 mM Ammonium acetate (%) |
|---|
| 0 | 95 | 5 |
| 5 | 95 | 5 |
| 14 | 86 | 14 |
| 14.5 | 75 | 25 |
| 17.5 | 75 | 25 |
| 18 | 50 | 50 |
| 22 | 50 | 50 |
| 22.5 | 20 | 80 |
| 24.5 | 20 | 80 |
| 25 | 0 | 100 |
| 27 | 0 | 100 |
| 28 | 95 | 5 |
| 33 | 95 | 5 |
The single ion monitoring (SIM) mode was adopted to
capture the [M-H]- ion of expected bile acids (BAs).
Peak areas were used for comparison and statistical analysis. The
mass spectrometer used an electrospray ionization source (ESI) in
negative ion. The ion source parameters were set as gas temp:
326˚C, gas flow: 12 l/min, nebulizer pressure: 55 psi and capillary
voltage: 3.5 kV. Detection ion: Taurocholate acid (TCA)
[M-H]- m/z 514.1, Tauroursodeoxycholic acid
(TUDCA), taurohyodeoxycholic acid (THDCA), Taurochenodeoxycholic
acid (TCDCA), tauroursodeoxycholic acid (TDCA) [M-H]-
m/z 498.2, taurolithocholic acid (TLCA) [M-H]-
m/z 482.1, glycocholic acid (GCA) [M-H]-
m/z 464.6, Glycochenodeoxycholic acid (GCDCA),
glycodeoxycholic acid (GDCA), glycoursodeoxycholic acid (GUDCA)
[M-H]- m/z 448.2, cholic acid (CA)
[M-H]- m/z 407.6, ursodesoxycholic acid (UDCA),
hyodeoxycholic acid (HDCA), chenodeoxycholic acid (CDCA),
deoxycholic acid (DCA) [M-H]- m/z 391.5 and
lithocholic acid (LCA) [M-H]- m/z 375.2.
Proteomics of GP regulating bile acid
pathway in mouse liver
Samples were removed from -80˚C, added to four times
the volume of lysis buffer (1% Triton X-100 and 1% protease
inhibitor) and lysed by ultrasonics (25 KHz, ultrasound 5 sec, 10
ec interval, 100 repetitions, samples were placed on ice.) before
being centrifuged at 12,000 g for 10 min at 4˚C. Protein
concentration was determined using a BCA kit (Beijing Solarbio
Science & Technology Co., Ltd.).
Each sample protein was digested by adding an equal
amount of standard protein, then adding DTT to a final
concentration of 5 mM and reducing at 56˚C for 30 min. Afterwards,
IAA was added to a final concentration of 11 mM and incubated for
15 min at room temperature and protected from light. Finally,
triethylammonium bicarbonate buffer (TEAB) was added to dilute urea
to ensure that the concentration was <2 M. Trypsin was added at
a ratio of trypsin: protein=1:50 and digestion performed at 37˚C
overnight. The next day, trypsin was added again in the ratio of
trypsin: protein=1:100 and the trypsin digestion were continued for
4 h.
The peptides were dissolved with liquid
chromatography mobile phase A and then separated using a NanoElute
ultra performance liquid phase system. Mobile phase A was 0.1%
formic acid solution and mobile phase B was acetonitrile
(containing 0.1% formic acid). Liquid phase gradient settings: 0-70
min, 6-24%B; 70-84 min, 24-35%B; 84-87 min, 35-80%B; 87-90 min,
80%B, with the flow rate maintained at 400 nl/min.
The peptides were separated by the Ultra-HPLC system
and then injected into the capillary ion source for ionization and
then into the TOF Pro mass spectrometer for analysis. The ion
source voltage was set to 1.6 kV and the peptide parent ions and
their secondary fragments were detected and analyzed using time of
flight (TOF). The secondary mass spectrometry scan range was m/z
100-1,700. The data acquisition mode was used in parallel
accumulated serial fragmentation (PASEF) mode. A primary mass
spectrum acquisition was followed by 10 PASEF mode acquisitions of
secondary spectra with parent ion charge numbers in the range of
0-5. The dynamic exclusion time of the tandem mass spectrometry
scan was set to 30 sec to avoid repeated scanning of the parent
ions.
Secondary mass spectrometry data were retrieved
using Maxquant (v1.6.6.0; Max Planck Institute of Biochemistry).
The search parameters were set: the database was SwissProt Mouse
(17,032 sequences), an inverse library was added to calculate the
false positive rate (FDR) due to random matching. A common
contamination library was added to the database to eliminate the
effect of contaminated proteins in the identification results; the
digestion mode was set to Trypsin/P; the number of missed cut sites
was set to 2; the first mass error tolerance of the primary parent
ion was set to 20 and 20 ppm for search and main search,
respectively and the mass error tolerance of the second fragment
ion was 0.02 Da. The cysteine alkylation was set to fixed
modification, the variable modification to oxidation of methionine
and the acetylation of protein N-terminal. The FDR for
protein identification and PSM identification were set to 1%.
The proteomics KEGG database was searched for its
pathway map (PATHWAY: map00120) using Bile acid as a keyword, and
proteins associated with bile acid/cholesterol were searched in the
NCBI database, resulting in 45 proteins for analysis.
Transcriptomics of GP regulating bile
acid pathway in mouse liver
Briefly, liver tissues were lysed using
TRIzol® (Thermo Fisher Scientific, Inc.) RNA was
extracted with trichloromethane, centrifuged at 12,000 g for 15 min
at 4˚C. The supernatant was enriched with isopropyl alcohol and
centrifuged at 12,000 g for 15 min at 4˚C. The precipitate was
washed with 75% ethanol and centrifuged at 7,500 g for 15 min at
4˚C. The RNA was resolubilized using the appropriate amount of DEPC
water and detected by NanoDrop. The concentration and OD 260/280
ratio of total RNA samples were measured by NanoDrop ultra-micro
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.).
After the samples had been extracted, the RNA
concentration purity and RNA integrity values were checked with an
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.); the screened
RNA was reverse transcribed into cDNA for library construction and
sequenced and analyzed by using a BGISEQ-500RS RNA-Seq platform
(BGI Biotechnology (Wuhan) Co., Ltd.); GRCm38/mm10 was used for
comparison and annotation and fragments per kilobase million (FPKM)
values were used as the standardized value output matrix for post
data analysis.
The FPKM values of Fxr, Shp, and
Nr4a1 were selected to form a matrix based on the
transcriptomics FPKM data matrix. Correlation analysis of data in R
language by ggplot2 and ggpbur packages, where x-axis is set to
Shp expression levels (FPKM) and y-axis is set to Fxr
expression levels (FPKM) and Nr4a1 expression levels (FPKM)
respectively. Finally, the correlation between Fxr,
Nr4a1 and Shp is determined based on the R value,
where the closer the absolute value of R is to 1, the stronger the
linear correlation between the two variables.
Expression levels of genes involved in
bile acids regulation
Mouse liver RNA extraction is the same as in
transcriptomics. Reverse transcription kits were purchased from
Bio-Rad Laboratories and reverse transcription was performed in a
multifunctional PCR instrument according to the instructions. The
reverse transcription conditions were: preheating at 25˚C for 10
min, reverse transcription at 37˚C for 2 h, reverse transcriptase
inactivation at 85˚C for 5 min, and storage at 4˚C. Real Time
Quantitative PCR (RT-qPCR) was performed by using 2*SYBR Green
Supermix (Bio-Rad Laboratories, Inc.) on a CFX96 RT-PCR System
(C1000 Touch; Bio-Rad Laboratories, Inc.). The RT-PCR parameters
were as follows: 3 min, 95˚C and 1 cycle; 10 sec at 95˚C plus 45
sec at 60˚C, 40 cycles. The melting curve analysis consisted of 5
sec at 55˚C followed by heating up to 95˚C with a ramp rate of
0.5˚C/5 sec. Names and corresponding primer sequences of genes
checked in the present study are in Table II. Expression levels of GAPDH in
each sample were used as internal reference.
 | Table IIPrimers used for reverse
transcription-quantitative PCR. |
Table II
Primers used for reverse
transcription-quantitative PCR.
| Gene | Forward primer (5'
to 3') | Reverse primer (5'
to 3') |
|---|
| Cyp7a1 |
GAGCCCTGAAGCAATGAAAG |
GCTGTCCGGATATTCAAGGA |
| Cyp8b1 |
GGACAGCCTATCCTTGGTGA |
GACGGAACTTCCTGAACAGC |
| Fxr |
TTCCTCAAGTTCAGCCACAG |
TCGCCTGAGTTCATAGATGC |
| Shp |
GGAGTCTTTCTGGAGCCTTG |
ATCTGGGTTGAAGAGGATCG |
| Gapdh |
TGTGTCCGTCGTGGATCTGA |
CCTGCTTCACCACCTTCTTGA |
Liver (0.2 g) was ground into fine powder in liquid
nitrogen, 1.1% formaldehyde solution was cross-linked for 15 min
and 1.25 M glycine for 5 min terminated the cross-linking in the
ice, the supernatant was centrifuged and resuspended twice in
pre-cooled PBS. Then, 2 ml of frozen chromatin immunoprecipitation
(ChIP) cell lysis buffer (containing 20 µl Protease inhibitors and
20 µl PMSF) was resuspended, ground 20 times on ice in a glass
homogenizer, incubated on ice for 15 min and the supernatant was
discarded by centrifugation at 4˚C, 1,000 x g, 5 min. Then, 1 ml of
ChIP nuclear lysis buffer (containing 10 µl Protease inhibitors and
10 µl PMSF) was resuspended and incubated on ice for 5 min.
Protease inhibitors and 10 µl PMSF) and incubate for 5 min on ice.
The samples were sonicated on moist ice at 20% power for 2 sec with
a 4 sec gap for a total of 6 min. The supernatant was divided into
120 µl tubes after centrifugation at 4˚C, 1,000 x g, 5 min and
stored at -80˚C.
50 µl of ultrasound chromatin was added to 450 µl of
ChIP dilution buffer (containing 2.25 µl Protease Inhibitor
Cocktail II), and 5 µl (1%) was taken as Input and stored at 4˚C. 1
µg/µl IgG antibody (Millipore, 12-371) and 20 µl of mixed Protein G
Magnetic Beads (Millipore, 16-662) were added to the negative
control group. Add 2 µg/µl FXR antibody (Santa Cruz Biotechnology,
sc-25309x) and 20 µl of well-mixed Protein G Magnetic Beads to the
experimental group. The magnetic beads were separated from the
liquid using a magnetic holder, and the supernatant was carefully
pipetted into Low salt buffer, High salt buffer, and LiCl Wash
buffer, respectively. For each sample (including Input samples),
add 100 µl of freshly prepared Elution Buffer and 1 µl of Rnase A
(10 mg/ml Solarbio, Beijing) in 37˚C water bath for 30 min with a
shaking frequency of 300 rpm. Add 1 µl of proteinase K and shake at
300 rpm for 2 h. Incubate at 95˚C for 10 min and cool to room
temperature. Separate the magnetic beads from the supernatant using
a magnetic holder and carefully pipette the supernatant to a new
1.5 ml centrifuge tube. Purify the eluted DNA using the MinElute
PCR Purification Kit (Qiagen, Germany 28006). The purified DNA was
examined by ultra-micro spectrophotometer with 2 µl samples for DNA
concentration, and 10 µl samples were examined by 1.2% agarose gel
electrophoresis for chromatin fragments. Fluorescent quantitative
PCR was performed using 2*SYBR Green Supermix. Primers are shown in
Table III. pre-denaturation at
95˚C for 3 min, denaturation at 95˚C for 10 sec, extension at 60˚C
for 45 sec, 40 cycles, and lysis curves were plotted. The results
were compared with Input and then statistical analysis was
performed with the expression value of IgG as the reference
value.
 | Table IIIPrimers used for chromatin
immunoprecipitation-reverse transcription-quantitative PCR. |
Table III
Primers used for chromatin
immunoprecipitation-reverse transcription-quantitative PCR.
| Gene | Forward primer (5'
to 3') | Reverse primer (5'
to 3') |
|---|
| Abcb11 |
TGCGTGGGGACCTTCTGAG |
AGAGTCGGGCCTCTCACCA |
Statistical analysis
All data were shown as mean ± standard error of the
mean. Principal component analysis (PCA) was performed using
the ‘pca’ function in the mixOmics package of the R program
(http://mixomics.org/). Statistical significance
levels tested by two-way ANOVA followed by a Tukey's test, which
were performed by using the base functions ‘aov’ and ‘TukeyHSD’ in
R program. The statistical significance levels of data were tested
by the Student's t test, which was performed by using the base
function ‘t.test’ in R program. P<0.05 was considered to
indicate a statistically significant difference. The raw sequence
reads generated in the article have been uploaded to the NCBI
BioProject database (https://www.ncbi.nlm.nih.gov/sra/) under accession
number PRJNA885754.
Results
Biochemical index testing and HE
staining
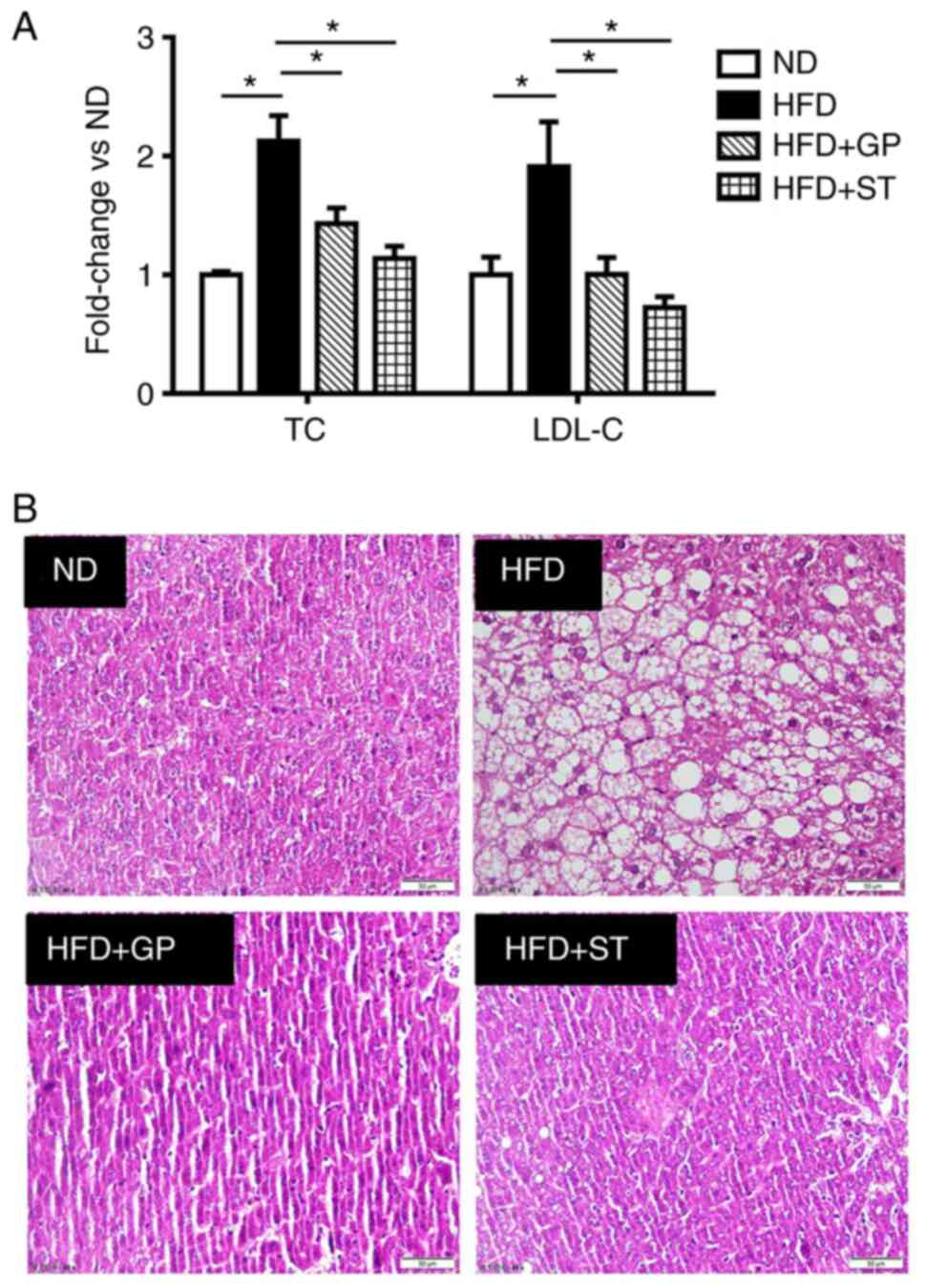
Metabolic disorders caused by HFD are accompanied by
hyperglycemia, hypercholesterolemia and hyperlipidemia.
Hypercholesterolemia is mainly manifested by abnormally high serum
levels of TC and LDL-C. These two indicators were tested and it was
shown that HFD significantly increased the expression levels of TC
and LDL-C and the administration of GP treatment downregulated the
expression of TC and LDL-C (Fig.
1A). HE staining showed that the HFD caused vacuolar-like
lesions and irregular arrangement of hepatocytes. By contrast, the
administration of GP and simvastatin (positive control drug)
reduced the hepatocyte pathology-like lesions and neatly aligned
cells (Fig. 1B), it was shown that
the high-fat model was successfully constructed.
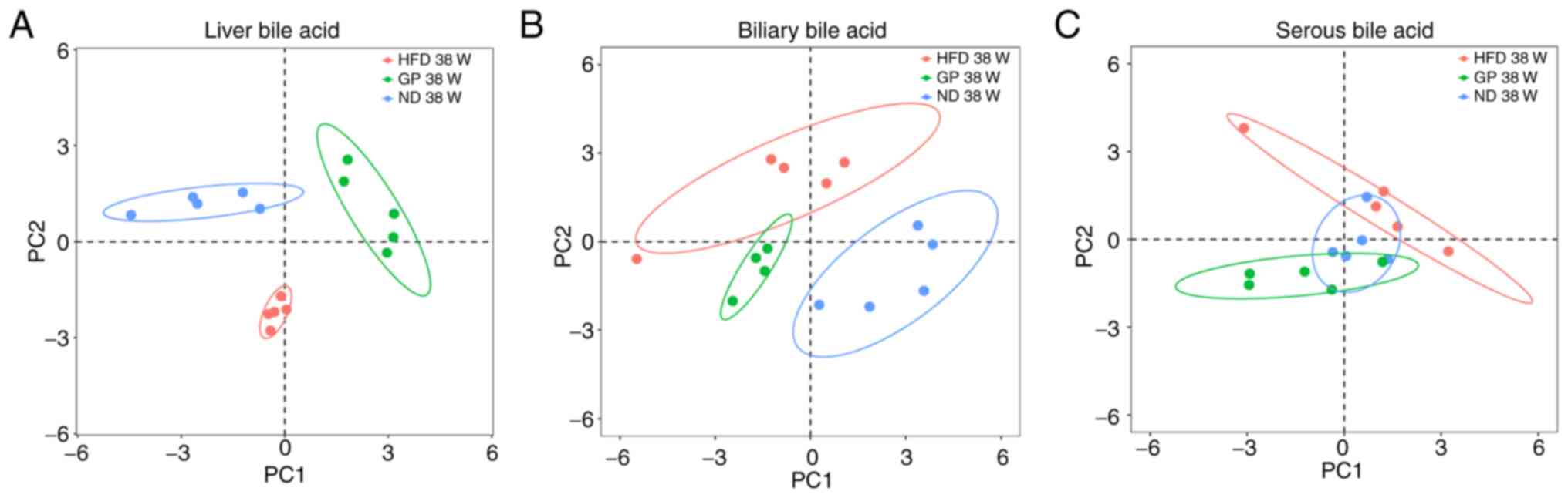
Metabolomics analysis of the effect of
GP on the molecular level of bile acid
The bile acid profile in the liver, bile and serum
of mice in the ND, HFD and GP groups was significantly different
and the HFD-induced changes in the bile acids profile in the liver,
bile and serum of mice. The bile acids profile of the liver,
biliary and serum also changed following GP administration,
ameliorating the changes caused by the HFD (Fig. 2) This suggested that GP has a
therapeutic effect on changes in bile acids in mice caused by
HFD.
In a further study, 16 bile acids were analyzed
according to their bile acids structure into free bile acids,
taurine-conjugated bile acids and glycine-conjugated bile acids.
Based on our previous study (7),
gypenosides decreased the CA/CDCA ratio which is positively related
to cholesterol absorption and increasing CDCA levels suggested that
gypenosides may accelerate hepatic free bile acid synthesis by
promoting the key bile acid synthase. The changes in bile acids
levels of 16 bile acids in bile and serum were further investigated
(Fig. 3) and in serum, GP
significantly altered the levels of DCA and LCA-free bile acids
(Fig. 3A and D). LCA and DCA expression in the
intestine is a key function of the intestinal microbiota (29), indicating that GP promotes the
entry of bile acids into the blood and their absorption and
expression in the intestine. Analysis of the levels of
taurine-conjugated bile acids in bile and serum (Fig. 3B and E) showed that the results of changes in
the levels of taurine-conjugated bile acids in the liver were
similar to those in bile, with no significant differences in serum.
The opposite expression was observed for the levels of
glycine-conjugated bile acids in bile; presumably, GP promotes the
transport of glycine-conjugated bile acids to the gallbladder. As
glycine-conjugated bile acids were not detected in serum, only the
effect of GP on the levels of glycine-conjugated bile acids in the
liver and bile was analyzed in the present study (Fig. 3C). The increase in CDCA levels
following the action of GP suggests that GP may increase the
production of conjugated bile acids and facilitate the
translocation of conjugated bile acids to the gallbladder by
promoting the key bile acid synthase and the significant elevation
of LCA and DCA in serum also demonstrates that GP also promote the
entry of bile acids into the blood and their absorption and
expression in the intestine.
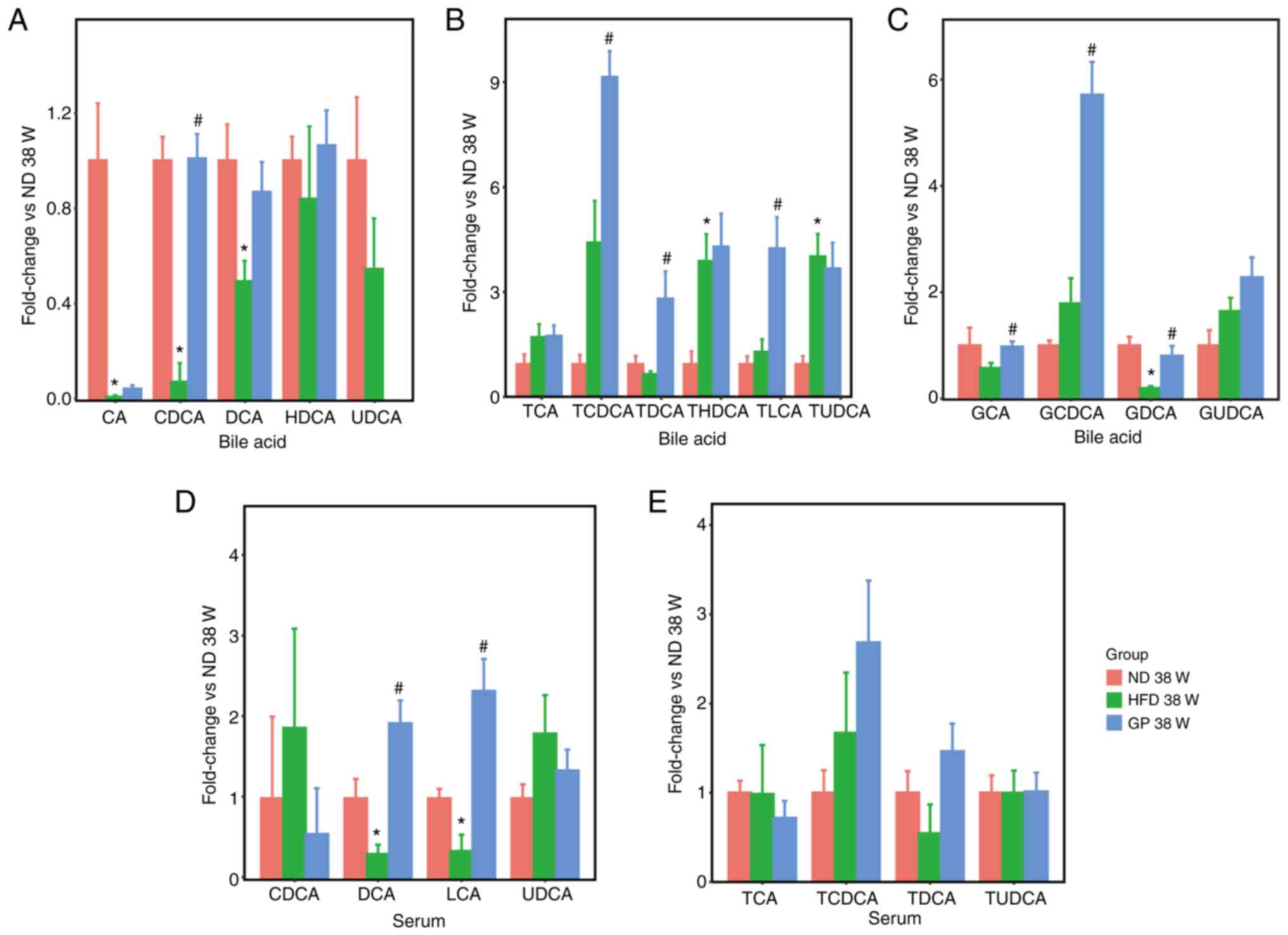 | Figure 3The effects of GP on the levels of
bile acids with different structures in bile and serum. (A-C) The
expression levels of free bile acids, taurine-conjugated bile
acids, and glycine-conjugated bile acids in bile in bile acid,
respectively. (D, E) The expression levels of free bile acids and
taurine-conjugated bile acids in serum, respectively. Data
represent means ± standard error of the mean. n=5,
*P<0.05 HFD 38 W vs. ND 38 W, #P<0.05
GP 38 W vs. HFD 38 W. GP, gypenosides/GP treatment; HFD, high fat
diet; W, weeks. |
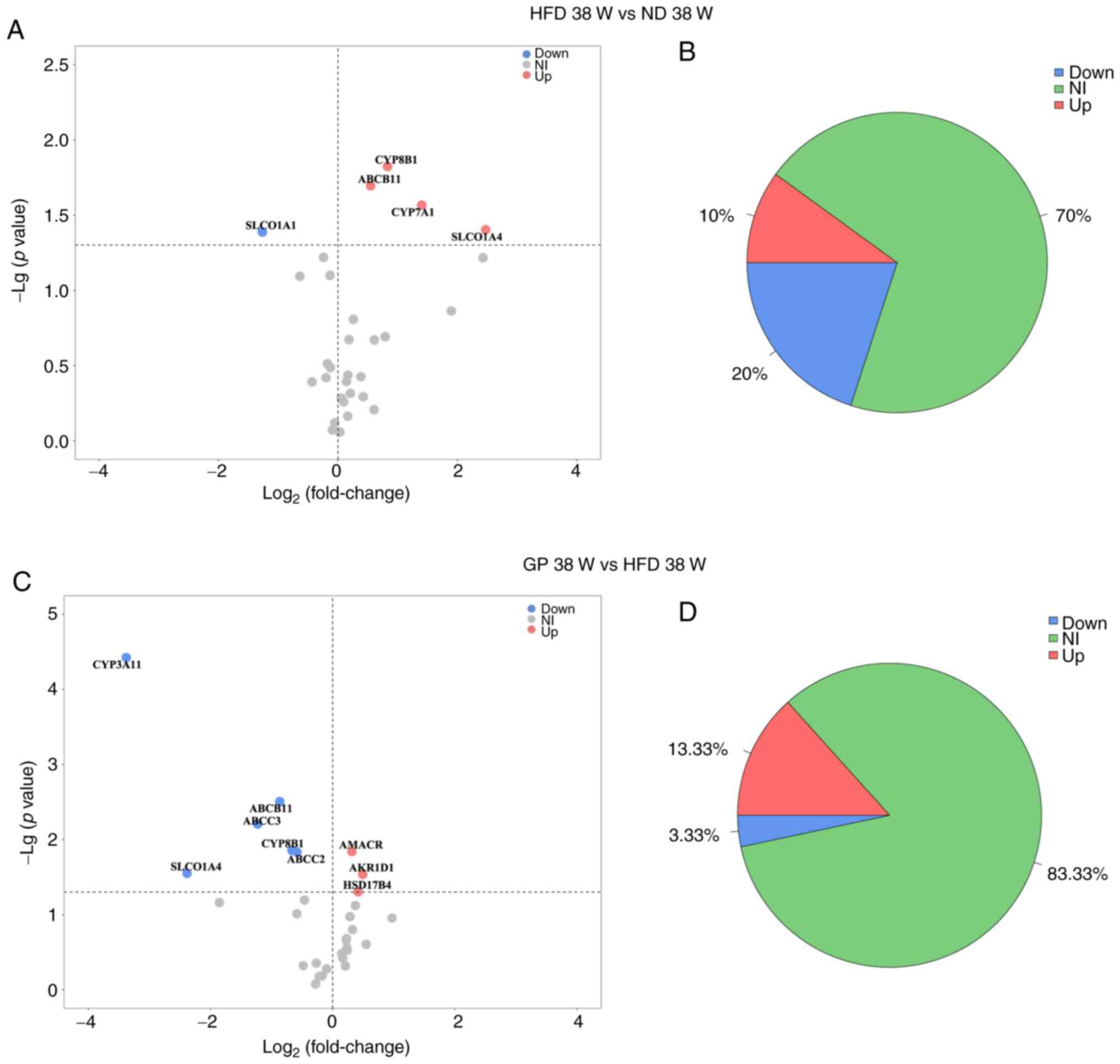
Proteomics analysis of the effect of
GP on the expression of bile acid pathway-related proteins
The effect of GP on the bile acids pathway in mouse
liver was further explored by proteomics. In the present study, the
bile acids-related proteins were queried by KEGG and NCBI databases
and then a literature search conducted to screen 45 proteins
closely related to the bile acids synthesis, metabolism and
transport pathways for analysis. The present study obtained 31 bile
acids pathway proteins by high-resolution mass spectrometry and
analyzed the expression of mouse liver bile acids pathway-related
proteins after the effects of HFD and GP using volcano plots
(Fig. 4). HSD17B4, AKR1D1 and
AMACR were upregulated in response to HFD stress. CYP3A11, ABCB11,
ABCC3, CYP8B1, ABCC2 and SLCO1A4 were significantly downregulated
(Fig. 4A). It was hypothesized
that HFD may cause hypercholesterolemia in mice through processes
such as inhibition of bile acids synthesis and bile acids transport
to the gallbladder. Following treatment with GP, mouse bile acids
pathway-related proteins CYP8B1, ABCB11, CYP7A1 and SLCO1A4 were
significantly upregulated and SLCO1A1 was significantly
downregulated (Fig. 4C). SLCO1A1
is responsible for the translocation and reabsorption of free bile
acids from the blood into the liver (30,31),
suggesting that GP may regulate hypercholesterolemia by promoting
bile acids synthesis, further translocation of bile acids to the
gallbladder and inhibition of bile acids reabsorption into the
liver. Under HFD stress, significantly upregulated proteins
accounted for 10% of the total bile acids pathway proteins and
significantly downregulated proteins accounted for 20% of the total
bile acids pathway proteins; following administration of GP
(Fig. 4B), significantly
upregulated proteins accounted for 13% of the total bile acids
pathway proteins and significantly downregulated proteins accounted
for 3% of the total bile acids pathway proteins (Fig. 4D). Thus it was shown that GP had a
significant effect on bile acids pathway proteins in mice with the
hypercholesterolemia model.
The present study performed association analysis for
proteins that underwent significant changes (Fig. 5A). The horizontal coordinate
represents the change in protein expression after the effect of a
HFD and the vertical coordinate represents the change in protein
expression after the effect of GP. The results showed that GP
significantly downregulated the expression of SLCO1A4, CYP3A11,
CYP8B1 and ABCB11 induced by HFD. HFD had significant effects on
ABCC3, ABCC2, HSD17B4, AMACR, AKR1D1, CYP7A1 and SLCO1A1, which
were also significantly regulated by GP. Selected proteins where GP
significantly modulated the changes induced by a HFD were analyzed
(Fig. 5C-H) and the results showed
that the expression levels of CYP8B1, CYP3A11, ABCB11 and SLCO1A4
were significantly downregulated by HFD and the expression levels
of CYP7A1, CYP8B1, ABCB11, SLCO1A4 were significantly upregulated,
while the expression level of SLCO1A1 was significantly decreased.
This is further evidence that GP promoted the expression level of
CYP7A1, CYP8B1 and ABCB11 and increased bile acids synthesis and
secretion to the bile duct. It was observed that GP inhibited the
expression of SLCO1A1 and promoted the expression of CLCO1A4,
indicating that GP increased the level of free bile acids in the
blood and accelerated the excretion of free bile acids in the blood
to the kidneys, thereby maintaining bile acids homeostasis.
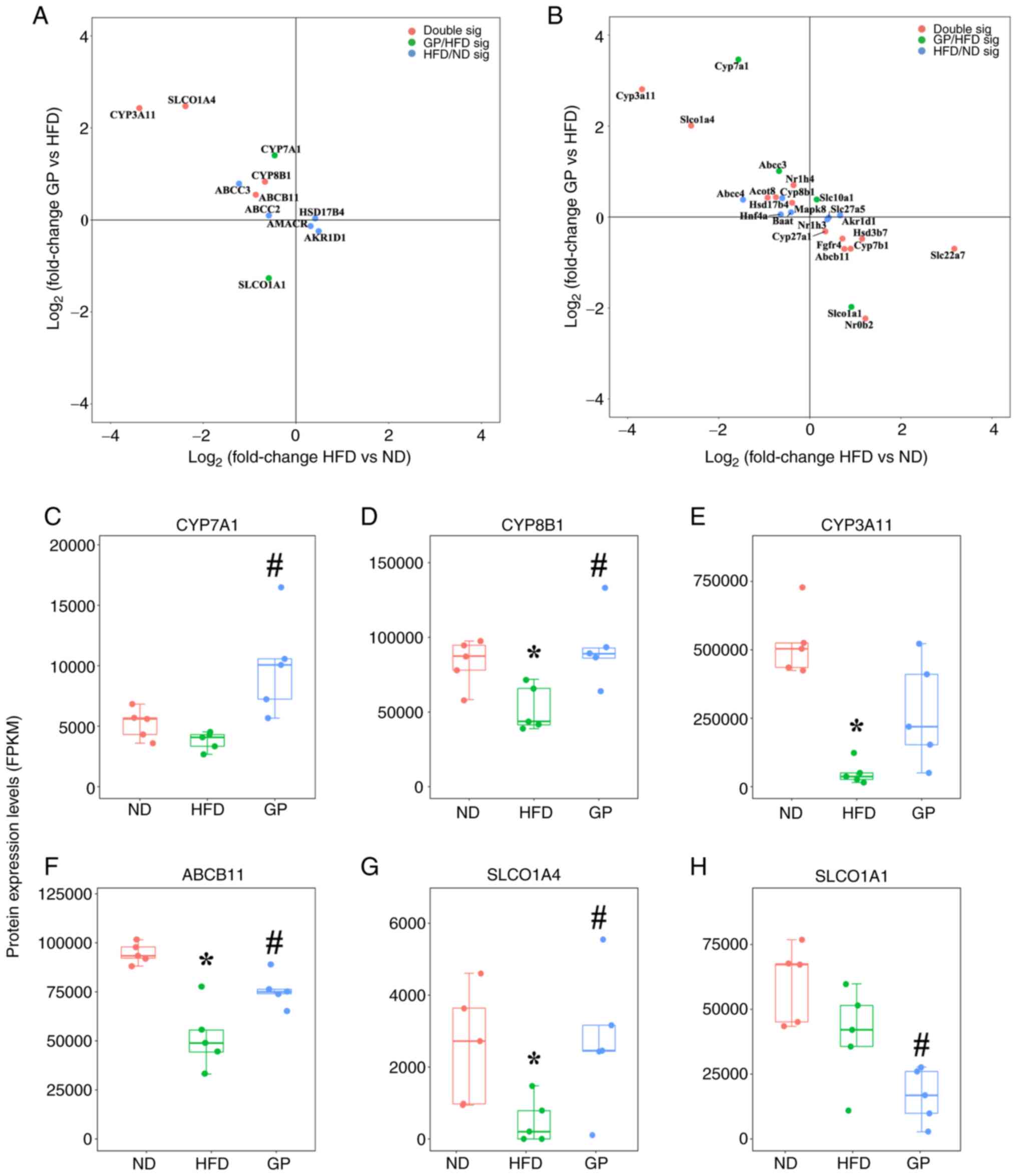 | Figure 5Differential analysis of bile acid
proteins and genes by GP and the back-regulation of key proteins of
the pathway. (A) Differential analysis of proteins in bile acid
pathway on GP. (B) Differential analysis of genes in bile acid
pathway on GP. (C-H) Regulation of CYP7A1, CYP8B1, CYP3A11, ABCB11,
SLCO1A4, and CLCO1A1 key pathway proteins by GP. Data represent
means ± standard error of the mean. n=5, *P<0.05 HFD
38 W vs. ND 38 W, #P<0.05 GP 38 W vs. HFD 38 W. Red
represents 38 W of ND; Green represents 38 W of HFD; Blue
represents 38 W of GP. GP, gypenosides/GP treatment; HFD, high fat
diet; ND, normal diet; W, weeks; FPKM, fragments per kilobase
million. |
Transcriptomics analysis of the effect
of GP on the expression of bile acids-related genes
Proteomic analysis found that GP ameliorated the
abnormal changes in proteins responsible for bile acids synthesis,
metabolism and transport in the liver of mice affected by a HFD.
The effect of GP on the expression of 45 bile acid pathway-related
genes in mouse liver was further verified. These genes come from
KEGG and NCBI databases. A total of 26 differentially expressed
genes were screened using the volcano plots (Fig. S1) and association analysis was
performed for these genes (Fig.
5B), with the horizontal coordinates representing gene
expression changes after the effect of HFD and the vertical
coordinates representing gene expression changes following GP, with
the horizontal coordinates representing the changes in gene
expression following the effects of HFD and the vertical
coordinates representing the changes in gene expression following
the effects of GP. Under HFD stress, GP caused significant
upregulation of mRNA for Cyp3a11, Slco1a4, Nr1h4, Acot8,
Hsd17b4 and Mapk8 and caused significant downregulation
of mRNA for Cyp27a1, Fgfr4, Abcb11, Cyp7a1, Hsd3b7, Slc22a7
and Nr0b2. In genes encoding bile acids pathway-related
proteins, GP significantly reversed the mRNA expression of
Cyp7a1, Cyp3a11, Slco1a4, Abcb11 and Slco1a1
(Fig. 6A-H) and tended to
upregulate Cyp8b1, which was similar to the proteomic
result. Further PCR validation of key genes on Fxr pathway
showed that the expression of Cyp7a1, Cyp8b1, Fxr, Aacb11
was significantly downregulated following GP treatment compared
with HFD, but the expression of Shp was significantly
decreased following GP treatment, which may be related to the
treatment cycle of Shp (Fig.
7).
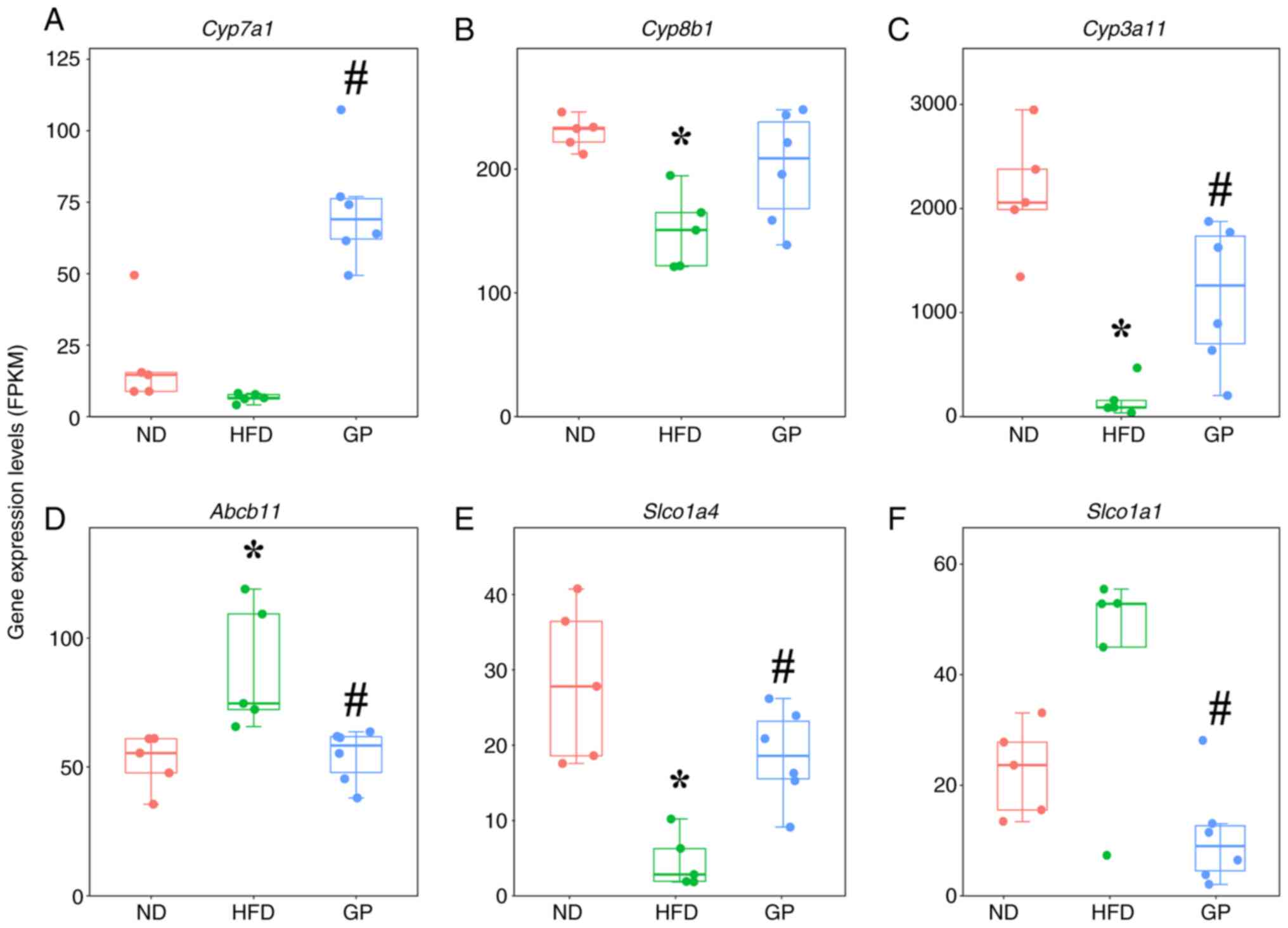 | Figure 6Differential analysis of gene in bile
acids pathway on gypenosides. (A-F) Regulation of Cyp7a1,
Cyp8b1, Cyp3a11, Abcb11, Slco1a4, and
Slco1a1 key pathway genes by gypenosides. Data represent
means ± standard error of the mean. n=5, *P<0.05 HFD
38 W vs. ND 38 W, #P<0.05 GP 38 W vs. HFD 38 W. Red
represents 38 W of ND; green represents 38 W of HFD; blue
represents 38 W of GP. GP, gypenosides/GP treatment; HFD, high fat
diet; ND, normal diet; W, weeks; FPKM, fragments per kilobase
million. |
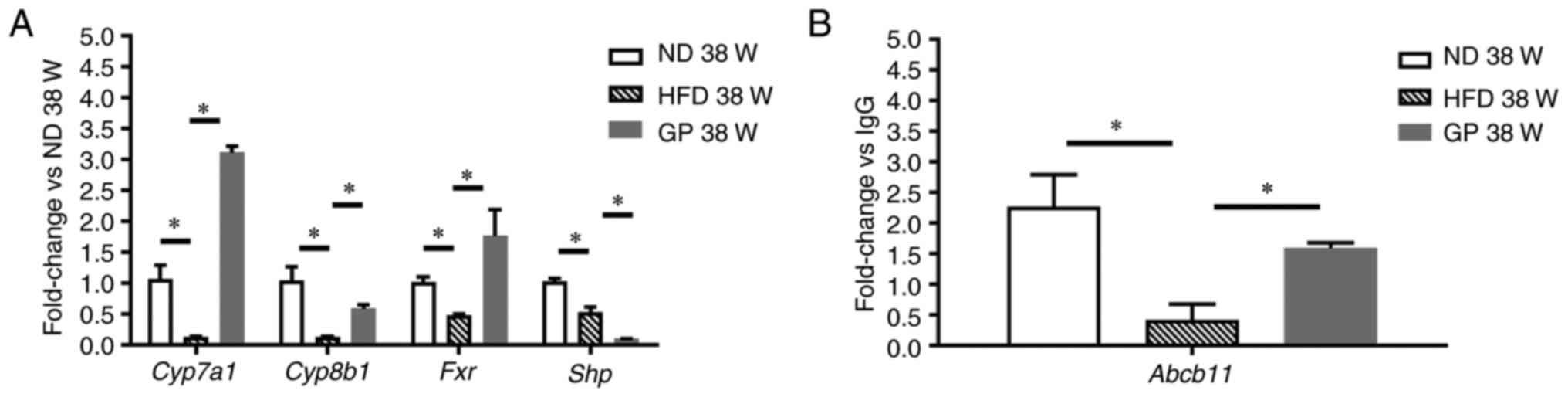 | Figure 7Expression of genes involved in the
Fxr pathway. (A) Cyp7a1, Cyp8b1, Fxr,
and Shp were detected by RT-qPCR, and (B) Abcb11 gene
was detected by CHiP-qPCR. Data represent means ± standard error of
the mean. n=3, *P<0.05; RT-qPCR, reverse
transcription-quantitative PCR; ChIP, chromatin
immunoprecipitation; ND, normal diet; HFD, high fat diet; GP, GP
treatment. |
Effects of GP administration time on
key genes and proteins of the FXR pathway
In addition to cholesterol metabolism via the bile
acids pathway, cholesterol is also required for synthesis in the
body, including steroid hormones, cell membranes and vitamin D.
Therefore, to ensure that cholesterol is available for the rest of
the physiological functions, the body may initiate feedback
inhibition of Cyp7a1 expression by the hepatic Fxr
pathway, thus maintaining bile acids homeostasis. By studying the
regulatory effects of GP on key genes and proteins such as
Fxr, Shp, Cyp7a1 and Nr4a1 the effect
of GP on the feedback regulation of bile acids in the liver
Fxr pathway can be revealed (Fig. 8). Compared with the HFD group, the
mRNA expression of Cyp7a1 was significantly upregulated at
38 weeks and the expression level of this encoded protein was
significantly upregulated at 18 and 38 weeks; the mRNA expression
of Fxr was significantly downregulated at 18 weeks and
significantly upregulated after 38 weeks and the expression level
of its encoded protein was significantly upregulated at 32 weeks.
However, at 26 and 32 weeks, it was significantly downregulated,
probably due to organismal self-regulation; the mRNA expression of
Shp was significantly downregulated after 38 weeks and the
mRNA expression of Nr4a1 was significantly downregulated at
both 26 and 38 weeks. Since the protein expression of Shp
and Nr4a1 was not examined (possibly due to the low
concentration of this protein in the group), only the transcript
levels of Shp and Nr4a1 were analyzed in the present
study. Further correlation analysis of genes Fxr,
Nr4a1 and Shp (Fig.
8G and H) showed a significant
negative correlation between Fxr and Shp (R=-0.633),
while Nr4a1 showed a significant positive correlation with
Shp (R=0.800). GP may regulate cholesterol metabolism
through Nr4a1-mediated FXR pathway.
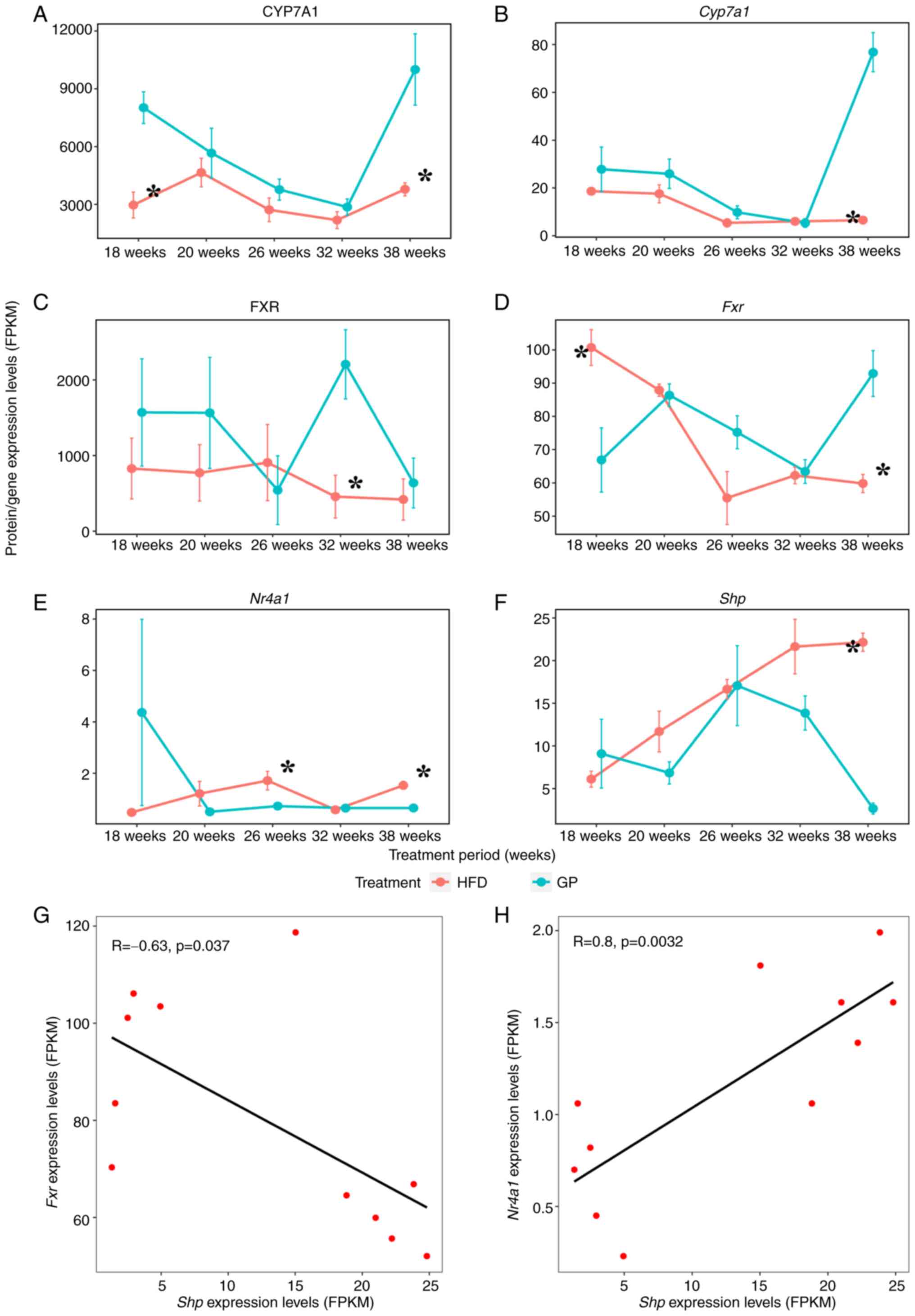 | Figure 8Effects of administration time on FXR
pathway and correlation comparison. (A-F) Time course of key gene
and protein expression levels in the FXR pathway at each identical
time point GP vs. HFD for CYP7A1, Cyp7a1, FXR, Fxr,
Nr4a1, Shp. Data represent means ± standard error of
the mean. n=5, *P<0.05 HFD 18 W vs. GP 18 W,
*P<0.05 HFD 20 W vs. GP 20 W, *P<0.05
HFD 26 W vs. GP 26 W, *P<0.05 HFD 32 W vs. GP 32 W,
*P<0.05 HFD 38 W vs. GP 38 W. (G, H) Correlation
analysis of mRNA expression of Fxr, Nr4a1, and
Shp. FXR, farnesoid X receptor; HFD, high fat diet; GP, GP
treatment; HFD, high fat diet; CYP7A1, cholesterol 7α-hydroxylase;
FPKM, fragments per kilobase million. |
Discussion
GP has the effect of regulating blood lipid and
blood sugar levels and protecting the liver, which is of great
clinical value in the treatment of obesity, fatty liver and
metabolic diseases (32). The
present study, using a hypercholesterolemic model mouse as the
research object and using metabolomics, proteomics, transcriptomics
and other multi-omics research tools, found that GP may accelerate
the synthesis of bile acids by promoting the expression of key bile
acids synthesis enzymes and increasing the expression of bile acids
metabolizing enzymes to reduce the damage of bile acids to the
liver. GP may also promote the expression of bile acids efflux
transporters to accelerate the efflux of bile acids from the liver
and inhibit the expression of bile acids uptake transporter to
reduce bile acids reabsorption to the liver, which in turn promotes
the excretion of bile acids into the kidney for detoxification and
promotes the conversion of cholesterol to bile acids to maintain
the homeostasis of the cholesterol-bile acids internal environment
(Fig. 9). Unfortunately, the
experiments were not performed for groups treated with GP on ND,
but it will be included in future studies.
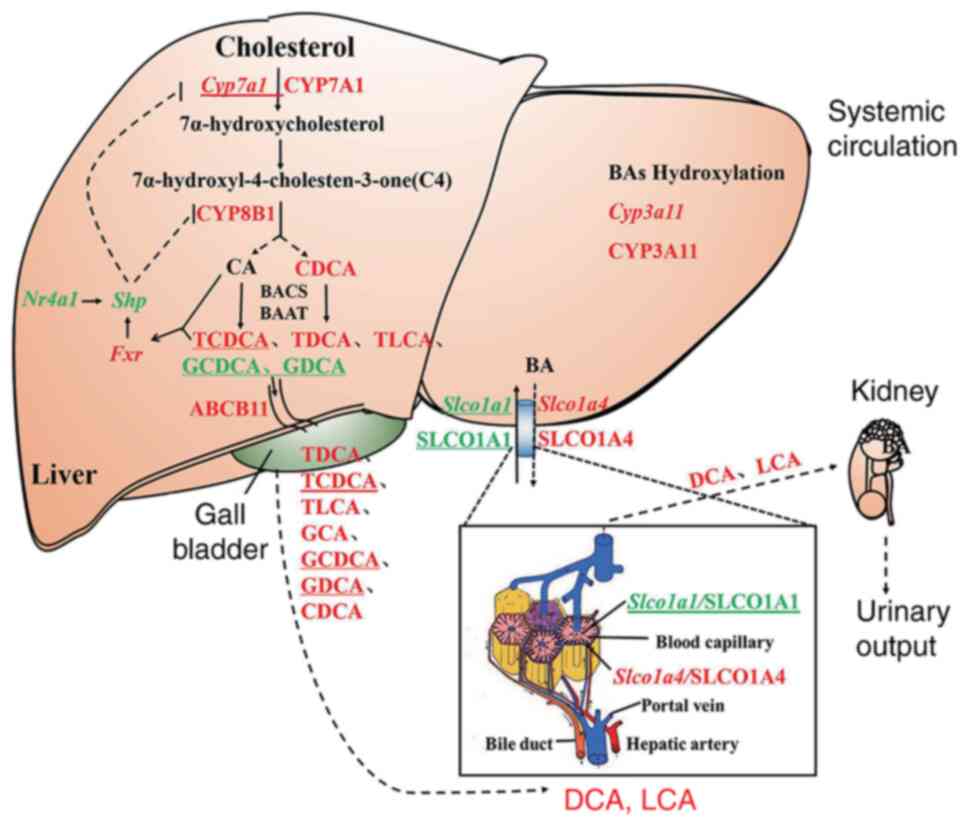 | Figure 9The regulation of GP on genes and
proteins expression in bile acid pathway in mice liver. Capital
letters represent proteins, lowercase italics represent genes; red
represents the expression is significantly downregulated by
high-fat diet and upregulated by GP; green represents the
expression is significantly upregulated by high-fat diet and
downregulated by GP; the underline represents no significant change
in high-fat diet and GP significantly modulates; black represents
no change in either. GP, gypenosides/GP treatment; CYP7A1,
Cholesterol 7α-hydroxylase; CYP8B1, Oxysterol-7α-hydroxylase; CA,
cholic acid; BACS, bile acid-CoA synthase; BAAT, amino acid
N-acyltransferase; TCDCA, taurochenodeoxycholic acid; TDCA,
tauroursodeoxycholic acid; TLCA, taurolithocholic acid; GCDCA,
glycochenodeoxycholic acid; GDCA, glycodeoxycholic acid; ABCB11,
bile salt export pump; GCA, glycocholic acid; CDCA,
chenodeoxycholic acid; SLCO1A1, solute carrier organic anion
transporter family member 1A1; SLCO1A4, solute carrier organic
anion transporter family, member 1A4; DCA, deoxycholic acid; LCA,
lithocholic acid. |
A combination of time course line (Fig. 8A-F) and PCR data used to analyze
high-fat diet in the early stages: Low Fxr expression, low
Shp expression and normal bile acid synthesis by
Cyp7a1. In the late hyperlipidemic phase, bile acid
accumulation activates Fxr, which increases Shp
expression and thus inhibits Cyp7a1, limiting cholesterol
catabolism and thus causing hypercholesterolemia. Significant
increase in Fxr gene expression followed GP administration
(Fig. 7). The significant decrease
in Shp expression may be due to the fact that GP treatment
is a slow process and the gene expression level of Shp had
not recovered at the time point the material was collected
(Fig. 9).
In the liver, CYP7A1 and CYP8B1 protein expression
levels, as well as mRNA expression of the genes encoding these
proteins, were downregulated under HFD stress, consistent with
literature reports (33-35),
suggesting that a HFD inhibits liver metabolism and causes the
development of hypercholesterolemia (36). The upregulation of the expression
levels of CYP7A1 and CYP8B1 and the mRNA expression of the genes
encoding these proteins following the action of GP may cause an
increase in the concentration of 7α-hydroxycholesterol, a catalytic
reaction product of 7α-hydroxylase and induce the processing
modification of enzymes such as CYP8B1, thereby promoting liver
cholesterol metabolism and producing primary bile acids. It was
also observed by UPLC-MS analysis that the levels of free bile
acids such as CDCA, UDCA and DCA in mouse liver increased following
the administration of GP, indicating that one of the mechanisms by
which GP lowers hepatic cholesterol may be through promoting the
expression of CYP7A1 and CYP8B1, the key bile acid synthesis
enzymes, to accelerate the synthesis of free bile acids and thus
promote the metabolism of hepatic cholesterol. Nr1h3
expression was significantly increased under HFD (Fig. 5B) and HFD can cause metabolic
disorders with hypercholesterolemia. Unfortunately, a limitation of
the present study was that it did search for alterations in SREBP
levels.
The bile acids efflux transporter protein ABCB11 is
the main transporter protein for hepatic secretion of bile salts
and ABCB11 is the rate-limiting enzyme of the entire enterohepatic
cycle (37), which efficiently
transports bound bile acids across the hepatocyte parietal membrane
and pumps them through the bile ducts into the gallbladder by
stimulating the formation of bile salt-dependent bile flow
(17). HFD decreases ABCB11
expression (38) and in humans
ABCB11 mutations lead to severe intrahepatic cholestasis. ABCB11
protein expression is significantly reduced in patients with
nonalcoholic steatohepatitis (39), consistent with Abcb11 mRNA
experimental results in the present study (Fig. 7B). In the present study, GP could
alleviate HFD-induced liver injury by promoting the expression of
ABCB11 and accelerating the secretion of hepatic conjugated bile
acids into the bile ducts. Meanwhile, the analysis of UPLC-MS
results showed that the levels of GCDCA and GDCA in liver were
significantly decreased following the administration of GP, while
the levels of GCA, GCDCA and GDCA in bile were significantly
increased, indicating that GP could alleviate the liver injury
caused by HFD by promoting the expression level of ABCB11 protein
and accelerating the secretion of liver-bound bile acids into the
bile ducts. In addition, the present study found that ABCB11 was
inconsistently expressed at the transcriptional and protein levels,
possibly due to GP indirectly regulating the expression of
ABCB11.
Bile acids transporters SLCO1A1 and SLCO1A4 are
members of the OATP family. SLCO1A1 is responsible for transporting
free bile acids from the blood for reabsorption back to hepatocytes
and SLCO1A4 transports free bile acids from hepatocytes into the
blood (40). In the present study,
the expression of SLCO1A1 protein as well as its encoding gene was
significantly reduced in the hypercholesterolemic model mice
following GP intervention, while the expression of SLCO1A4 protein
as well as its encoding gene was significantly increased,
suggesting a reduction in free bile acids reabsorption into the
liver and facilitation of free bile acids transport to the body
circulation in the liver. In the present study, bile acids
reabsorption was inhibited, therefore, it was hypothesized that the
bile acids entering the kidney through blood circulation was
increased, thus preventing bile acids reabsorption into the liver
and causing bile stasis and reducing the toxic effect on liver
cells. It is worth mentioning that previous studies found that
increased fecal bile acids were accompanied by increased hepatic
bile acids synthesis (41,42). By inhibiting SLCO1A1 and promoting
SLCO1A4 expression, GP reduced the reabsorption of hepatic free
bile acids and accelerated the entry of hepatic free bile acids
into the bloodstream for excretion into the kidney through blood
circulation, which may be another mechanism to promote bile acids
metabolism and lower hepatic cholesterol.
CYP3A11, homologous to human CYP3A4 and a bile acids
metabolic enzyme, is mainly responsible for converting hydrophobic
bile acids such as LCA, DCA and CDCA into hydrophilic bile acids by
hydroxylation (43), enhancing
their water solubility for excretion and thus maintaining the
internal environment homeostasis of bile acids. The results of the
present study showed that a HFD induced a decrease in CYP3A11
protein expression levels and mRNA expression levels of its
encoding gene. Following the administration of GP, CYP3A11 protein
expression levels and mRNA expression levels of its encoding gene
were increased, resulting in increased water solubility of bile
acids and accelerated excretion of bile acids. Thus, GP avoids
biliary stasis and liver injury by promoting hepatic bile acids
hydroxylation, which in turn promotes bile acids metabolism and
excretion.
GP promotes CYP7A1-catalyzed hepatic cholesterol
metabolism for the synthesis of bile acids, and hepatic cholesterol
levels decrease (44). In addition
to cholesterol metabolism through the bile acids pathway, besides
being metabolized by the bile acid pathway, cholesterol is also
required for the synthesis of steroid hormones, cell membranes, and
vitamin D in the body (45) and to
ensure that cholesterol can meet the needs of the remaining
physiological functions, the body may initiate feedback inhibition
of the hepatic FXR pathway for CYP7A1 expression, thus maintaining
the homeostasis of the cholesterol-bile acids internal environment.
Under normal physiological conditions, bile acids are the catalytic
products of CYP7A1 and have a negative feedback regulation on their
expression (46,47). Endogenous bile acid CDCA is the
most potent ligand of FXR (48,49),
which increases the expression of SHP by activating FXR and
inhibiting bile acids synthesis (50,51).
Activation of FXR increases the expression of SHP
and NR4A1, another nuclear receptor in the body, which can
indirectly regulate glycolipid metabolism by regulating the
transcription factor SHP (52). In
the present study, the correlation analysis of Fxr and
Nr4a1 with Shp using transcriptomic data revealed
that the regulatory effect of Nr4a1 on Shp was
stronger than that of Fxr on Shp. GP may further
inhibit the expression of Shp by suppressing the expression
of Nr4a1, while upregulating the expression levels of
Cyp7a1, as well as its encoded protein, to promote hepatic
cholesterol metabolism. Synthesis of CDCA and TCDCA levels
increased, which in turn promoted Fxr expression. GP may
improve bile acids metabolism and maintain the dynamic
cholesterol-bile acids balance by regulating the
Nr4a1-mediated bile acids metabolic pathway. Nr4a1
expression was reduced following GP treatment and Shp
expression was reduced (Fig. 8E
and F), which in turn further
accelerated Cyp7a1 catabolism of cholesterol and alleviated
hypercholesterolemia. The bile acid pathway mediated by
Nr4a1, another nuclear receptor in the liver, may be another
mechanism to maintain the homeostasis of bile acids.
Research has shown that glycine-conjugated bile
acids are predominant in human serum. Taurine-conjugated bile acids
predominate in the serum of odontocetes (53). Through extensive literature search
none was found stating that glycine-conjugated bile acids are not
present in the liver and bile. Glycine-conjugated bile acids are
not detected in serum probably because they are hydrolyzed in the
intestine by bile salt hydrolases (BSHs), which hydrolyze the amide
bond and release the glycine/taurine molecule from the steroid via
the intestinal microbiota (54).
Studies have shown that BSHs preferentially hydrolyze
glycine-conjugated bile acids (55,56),
so glycine-conjugated bile acids are hydrolyzed in the intestine,
leading to a decrease in their absorption into the blood, which may
be one of the reasons why it was not possible to measure
glycine-conjugated bile acids in the serum of mice. Conjugated bile
acids of the glycine/taurine conjugated bile acids exhibit a
different dynamic balance in the circulatory system (57).
In summary, GP mainly regulates the synthesis,
metabolism and transport of bile acids and hepatic NR4A1-mediated
bile acids pathway by regulating the expression of key bile acids
synthases CYP7A1 and CYP8B1, bile acids metabolizing enzyme
CYP3A11, major bile acids efflux and uptake transporters ABCB11,
SLCO1A4 and SLCO1A1 at the transcriptional or protein level to
maintain internal environment homeostasis for bile acids, which in
turn regulates the bile acids pathway, promotes hepatic cholesterol
metabolism and lowers hepatic cholesterol.
Supplementary Material
The effect of GP on expressions of
bile acid pathway genes in mice. Volcano plots show the
significantly regulated genes. (A) In high-fat diet vs. normal diet
with volcano plot of differential genes. (B) In GP diet vs. HFD
with volcano plot of differentially expressed genes. The horizontal
dashed line represents P<0.05. GP, gypenosides/GP treatment;
HFD, high fat diet; ND, normal diet; W, weeks.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Nature
Science Foundation of China (grant nos. 82260843 and 82060649),
Guizhou Provincial Postgraduate Research Fund Project [grant no.
QJH YJSKYJJ (2021)186], the Science and Technology Foundation of
Guizhou Province of China [grant nos. QKHPTRC (2017)5733-060 and
QKHPTRC (2017)5733-062] and the Doctoral Research Start-up Fund
Project of Zunyi Medical University (grant no. F-938).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CCF wrote the manuscript and analyzed data. YPY and
AJL established the mice model and collected tissue samples. LQ,
DPT, YLL and YQH designed the present study, contributed to
interpretation of the data and revised the manuscript. CCF and YPY
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All procedures involving the use of laboratory
animals were in accordance with the requirements of Animal
Experiment Ethics Committee of Zunyi Medical University (approval
no. 2-557).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu JN, Cunningham SR, Thouin T, Gurvich D
and Liu D: Hyperlipidemia. Prim Care. 27:541–587. 2000.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bunnoy A, Saenphet K, Lumyong S, Saenphet
S and Chomdej S: Monascus purpureus-fermented Thai glutinous rice
reduces blood and hepatic cholesterol and hepatic steatosis
concentrations in diet-induced hypercholesterolemic rats. BMC
Complement Altern Med. 15(88)2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Karr S: Epidemiology and management of
hyperlipidemia. Am J Manag Care. 23:139–148. 2017.PubMed/NCBI
|
|
4
|
Collins R, Reith C, Emberson J, Armitage
J, Baigent C, Blackwell L, Blumenthal R, Danesh J, Smith GD, DeMets
D, et al: Interpretation of the evidence for the efficacy and
safety of statin therapy. Lancet. 19:2532–2561. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sessa M, Rafaniello C, Scavone C, Mascolo
A, di Mauro G, Fucile A, Rossi F, Sportiello L and Capuano A:
Preventable statin adverse reactions and therapy discontinuation.
What can we learn from the spontaneous reporting system? Expert
Opin Drug Saf. 17:457–465. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Livingstone SJ, Looker HC, Akbar T,
Betteridge DJ, Durrington PN, Hitman GA, Neil HA, Fuller JH and
Colhoun HM: Effect of atorvastatin on glycaemia progression in
patients with diabetes: An analysis from the collaborative
atorvastatin in diabetes trial (CARDS). Diabetologia. 59:299–306.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lu Y, Du Y, Qin L, Wu D, Wang W, Ling L,
Ma F, Ling H, Yang L, Wang C, et al: Gypenosides altered hepatic
bile acids homeostasis in mice treated with high fat diet. Evid
Based Complement Alternat Med. 2018(8098059)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Megalli S, Aktan F, Davies NM and
Roufogalis BD: Phytopreventative anti-hyperlipidemic effects of
gynostemma pentaphyllum in rats. J Pharm Pharm Sci. 8:507–515.
2005.PubMed/NCBI
|
|
9
|
Attawish A, Chivapat S, Phadungpat S,
Bansiddhi J, Techadamrongsin Y, Mitrijit O, Chaorai B and
Chavalittumrong P: Chronic toxicity of Gynostemma pentaphyllum.
Fitoterapia. 75:539–551. 2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chiranthanut N, Teekachunhatean S,
Panthong A, Khonsung P, Kanjanapothi D and Lertprasertsuk N:
Toxicity evaluation of standardized extract of Gynostemma
pentaphyllum Makino. J Ethnopharmacol. 149:228–234. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lazarević S, Đanić M, Goločorbin-Kon S,
Al-Salami H and Mikov M: Semisynthetic bile acids: A new
therapeutic option for metabolic syndrome. Pharmacol Res.
146(104333)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Staley C, Weingarden AR, Khoruts A and
Sadowsky MJ: Interaction of gut microbiota with bile acid
metabolism and its influence on disease states. Appl Microbiol
Biotechnol. 101:47–64. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vallim TQdA, Tarling EJ and Edwards PA:
Pleiotropic roles of bile acids in metabolism. Cell Metab.
17:657–669. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Malhi H and Camilleri M: Modulating bile
acid pathways and TGR5 receptors for treating liver and GI
diseases. Curr Opin Pharmacol. 37:80–86. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Trauner M, Fuchs D, Halilbasic E and
Paumgartner G: New therapeutic concepts in bile acid transport and
signaling for management of cholestasis. Hepatology. 65:1393–1404.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang L, Wang Q, Liu W, Liu F, Ji A and Li
Y: The orphan nuclear receptor 4A1: A potential new therapeutic
target for metabolic diseases. J Diabetes Res.
2018(9363461)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Dawson PA and Oelkers P: Bile acid
transporters. Curr Opin Lipidol. 6:109–114. 1995.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Meier PJ, Eckhardt U, Schroeder A,
Hagenbuch B and Stieger B: Substrate specificity of sinusoidal bile
acid and organic anion uptake systems in rat and human liver.
Hepatology. 26:1667–1677. 1997.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Modica S, Gadaleta RM and Moschetta A:
Deciphering the nuclear bile acid receptor FXR paradigm. Nucl
Recept Signal. 8(e005)2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Goodwin B, Jones SA, Price RR, Watson MA,
McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al:
A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1
represses bile acid biosynthesis. Mol Cell. 6:517–526.
2000.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chiang JY, Kimmel R, Weinberger C and
Stroup D: Farnesoid X receptor responds to bile acids and represses
cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J Biol
Chem. 275:10918–10924. 2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hu YW, Zhang P, Yang JY, Huang JL, Ma X,
Li SF, Zhao JY, Hu YR, Wang YC, Gao JJ, et al: Nur77 decreases
atherosclerosis progression in apoE(-/-) mice fed a
high-fat/high-cholesterol diet. PLoS One. 9(e87313)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jung YS, Lee HS, Cho HR, Kim KJ, Kim JH,
Safe S and Lee SO: Dual targeting of Nur77 and AMPKα by
isoalantolactone inhibits adipogenesis in vitro and decreases body
fat mass in vivo. Int J Obes (Lond). 43:952–962. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kudo T, Nakayama E, Suzuki S, Akiyama M
and Shibata S: Cholesterol diet enhances daily rhythm of Pai-1 mRNA
in the mouse liver. Am J Physiol Endocrinol Metab. 287:E644–E651.
2004.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Abdou HS, Robert NM and Tremblay JJ:
Calcium-dependent Nr4a1 expression in mouse Leydig cells requires
distinct AP1/CRE and MEF2 elements. J Mol Endocrinol. 56:151–161.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
De Fabiani E, Mitro N, Anzulovich AC,
Pinelli A, Galli G and Crestani M: The negative effects of bile
acids and tumor necrosis factor-alpha on the transcription of
cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic
nuclear factor-4: A novel mechanism of feedback regulation of bile
acid synthesis mediated by nuclear receptors. J Biol Chem.
276:30708–30716. 2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
He Y, Yang T, Du Y, Qin L, Ma F, Wu Z,
Ling H, Yang L, Wang Z, Zhou Q, et al: High fat diet significantly
changed the global gene expression profile involved in hepatic drug
metabolism and pharmacokinetic system in mice. Nutr Metab (Lond).
17(37)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang Y and Klaassen CD: Effects of
feeding bile acids and a bile acid sequestrant on hepatic bile acid
composition in mice. J Lipid Res. 51:3230–3242. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kakiyama G, Pandak WM, Gillevet PM,
Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon
JM, et al: Modulation of the fecal bile acid profile by gut
microbiota in cirrhosis. J Hepatol. 58:949–955. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wood M, Ananthanarayanan M, Jones B,
Wooton-Kee R, Hoffman T, Suchy FJ and Vore M: Hormonal regulation
of hepatic organic anion transporting polypeptides. Mol Pharmacol.
68:218–225. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Miyazaki H, Sekine T and Endou H: The
multispecific organic anion transporter family: Properties and
pharmacological significance. Trends Pharmacol Sci. 25:654–662.
2004.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang H, Chen X, Zong B, Yuan H, Wang Z,
Wei Y, Wang X, Liu G, Zhang J, Li S, et al: Gypenosides improve
diabetic cardiomyopathy by inhibiting ROS-mediated NLRP3
inflammasome activation. J Cell Mol Med. 22:4437–4448.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
He X, Zheng N, He J, Liu C, Feng J, Jia W
and Li H: Gut microbiota modulation attenuated the hypolipidemic
effect of simvastatin in High-Fat/cholesterol-diet fed mice. J
Proteome Res. 16:1900–1910. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yu L, Lu H, Yang X, Li R, Shi J, Yu Y, Ma
C, Sun F, Zhang S and Zhang F: Diosgenin alleviates
hypercholesterolemia via SRB1/CES-1/CYP7A1/FXR pathway in high-fat
diet-fed rats. Toxicol App Pharmacol. 412(115388)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gillard J, Clerbaux LA, Nachit M, Sempoux
C, Staels B, Bindels LB, Tailleux A and Leclercq IA: Bile acids
contribute to the development of non-alcoholic steatohepatitis in
mice. JHEP Rep. 4(100387)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Gryn SE and Hegele RA: Ezetimibe plus
simvastatin for the treatment of hypercholesterolemia. Expert Opin
Pharmacother. 16:1255–1262. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ren T, Pang L, Dai W, Wu S and Kong J:
Regulatory mechanisms of the bile salt export pump (BSEP/ABCB11)
and its role in related diseases. Clin Res Hepatol Gastroenterol.
45(101641)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Okushin K, Tsutsumi T, Ikeuchi K, Kado A,
Enooku K, Fujinaga H, Yamauchi N, Ushiku T, Moriya K, Yotsuyanagi H
and Koike K: Heterozygous knockout of Bile salt export pump
ameliorates liver steatosis in mice fed a high-fat diet. PLoS One.
15(e0234750)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Okushin K, Tsutsumi T, Enooku K, Fujinaga
H, Kado A, Shibahara J, Fukayama M, Moriya K, Yotsuyanagi H and
Koike K: The intrahepatic expression levels of bile acid
transporters are inversely correlated with the histological
progression of nonalcoholic fatty liver disease. J Gastroenterol.
51:808–818. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kalliokoski A and Niemi M: Impact of OATP
transporters on pharmacokinetics. Br J Pharmacol. 158:693–705.
2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Herrema H, Meissner M, Dijk TH, Brufa G,
Boverhof R, Oosterveer MH, Reijngoud DJ, Müller M, Stellaard F,
Groen AK and Kuipers F: Bile salt sequestration induces hepatic de
novo lipogenesis through farnesoid X receptor- and liver X receptor
alpha-controlled metabolic pathways in mice. Hepatology.
51:806–816. 2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Out C, Hageman J, Bloks VW, Gerrits H,
Gelpke MDS, Bos T, Smit MJ, Kuipers F and Groen AK: Liver receptor
homolog-1 is critical for adequate up-regulation of Cyp7a1 gene
transcription and bile salt synthesis during bile salt
sequestration. Hepatology. 53:2075–2085. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chen J, Zhao KN and Chen C: The role of
CYP3A4 in the biotransformation of bile acids and therapeutic
implication for cholestasis. Ann Transl Med. 2(7)2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Cao K, Zhang K, Ma M, Ma J, Tian J and Jin
Y: Lactobacillus mediates the expression of NPC1L1, CYP7A1, and
ABCG5 genes to regulate cholesterol. Food Sci Nutr. 9:6882–6891.
2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Maekawa M: Domain 4 (D4) of perfringolysin
O to visualize cholesterol in cellular membranes-the update.
Sensors (Basel). 17:504–518. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lorbek G, Lewinska M and Rozman D:
Cytochrome P450s in the synthesis of cholesterol and bile
acids-from mouse models to human diseases. FEBS J. 279:1516–1533.
2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chiang JYL: Bile acids: Regulation of
synthesis. J Lipid Res. 50:1955–1966. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Makishima M, Okamoto AY, Repa JJ, Tu H,
Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ and Shan B:
Identification of a nuclear receptor for bile acids. Science.
284:1362–1365. 1999.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li G and Guo GL: Farnesoid X receptor, the
bile acid sensing nuclear receptor, in liver regeneration. Acta
Pharm Sin B. 5:93–98. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xiang D, Yang J, Liu Y, He W, Zhang S, Li
X, Zhang C and Liu D: Calculus bovis sativus improves bile acid
homeostasis via Farnesoid X receptor-mediated signaling in rats
with estrogen-induced cholestasis. Front Pharmacol.
10(48)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang Y, Jackson JP, St Claire RL III,
Freeman K, Brouwer KR and Edwards JE: Obeticholic acid, a selective
farnesoid X receptor agonist, regulates bile acid homeostasis in
sandwich-cultured human hepatocytes. Pharmacol Res Perspect.
5:329–340. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Miao L, Yang Y, Liu Y, Lai L, Wang L, Zhan
Y, Yin R, Yu M, Li C, Yang X and Ge C: Glycerol kinase interacts
with nuclear receptor NR4A1 and regulates glucose metabolism in the
liver. FASEB J. 33:6736–6747. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wahlström A, Sayin SI, Marschall HU and
Bäckhed F: Intestinal crosstalk between bile acids and microbiota
and its impact on host metabolism. Cell Metab. 24:41–50.
2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Begley M, Hill C and Gahan CG: Bile salt
hydrolase activity in probiotics. App Environs Microbiol.
72:1729–1738. 2006.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Tanaka H, Hashiba H, Kok J and Mierau I:
Bile salt hydrolase of Bifidobacterium longum-biochemical and
genetic characterization. App Environ Microbiol. 66:2502–2512.
2000.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Kim GB, Miyamoto CM, Meighen EA and Lee
BH: Cloning and characterization of the bile salt hydrolase genes
(bsh) from Bifidobacterium bifidum strains. App Environ Microbiol.
70:5603–5612. 2004.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Yang T, Shu T, Liu G, Mei H, Zhu X, Huang
X, Zhang L and Jiang Z: Quantitative profiling of 19 bile acids in
rat plasma, liver, bile and different intestinal section contents
to investigate bile acid homeostasis and the application of
temporal variation of endogenous bile acids. J Steroid Biochem Mol
Biol. 172:69–78. 2017.PubMed/NCBI View Article : Google Scholar
|