Introduction
Carbapenems used to be antibiotics reserved for the
treatment of patients with infections caused by extended-spectrum
β-lactamase (ESBL)-producing Enterobacteriaceae. In recent
years, the emergence of carbapenem-resistant strains has been
increasing worldwide and as such also in Romania (1,2).
Based on the Ambler classification of β-lactamases, according to
their molecular structure, these enzymes with versatile hydrolytic
capacities, known as carbapenemases (3), are divided into class A (clavulanic
acid inhibitory enzymes), B (metallo-β-lactamases) and D
(oxacillinases) (4).
Class A β-lactamases include the Klebsiella
pneumoniae carbapenemase (KPC), the first reported
carbapenem-hydrolysing β-lactamase. KPC was initially extracted
from a carbapenem-resistant (CR) strain of Klebsiella
pneumoniae in a North Carolina hospital in 2001(5). KPC-producing organisms are usually
resistant to penicillin, cephalosporins and carbapenems. Moreover,
their growth is not inhibited by clavulanic acid or other common
β-lactamase inhibitors such as sulbactam and tazobactam. Thus, only
a few therapeutic options remain, namely colistin, polymyxin B and
tigecycline (6).
The New Delhi metallo-β-lactamase (NDM) emerged from
the Indian subcontinent as a major cause of carbapenem resistance
and has since been identified all over the world (7). NDM strains are usually resistant to
most antibiotics except for tigecycline and colistin. Regarding NDM
resistance, of great concern is its frequent association with
Escherichia coli, a ubiquitous and non-nosocomial microbe,
making it extremely difficult to contain community transmission
(2). Other MBLs are the Verona
integron-encoded metallo-β-lactamase (VIM) and the imipenemase
(IMP). Both were reported worldwide, with a higher prevalence in
southern Europe and Asia (8).
Class D β-lactamases include oxacilinase-48
(OXA-48), a carbapenem-hydrolysing β-lactamase, initially
identified in Europe, which has been increasingly reported in
various species of Enterobacteriaceae. Though colistin and
tigecycline are most likely to be active against OXA-48 producer
strains, some resistance has been reported (9). Fosfomycin as part of combination
therapy could be used as a last resort, though the emergence of
resistance is highly likely (10).
Point-of-care (POC) tests for the identification of
carbapenemases were developed during the second half of the last
century and gradually gained a foothold as an aid in the fast and
accurate diagnosis of CR strains. They are easy to perform, fast,
inexpensive and allow for on-site application. The technologies
used in POC tests include particle agglutination assays, monoclonal
antibody-based immunodot tests, immunochromatography and
immunofiltration (11). Most
immunochromatographic tests (ICT) used in microbiological
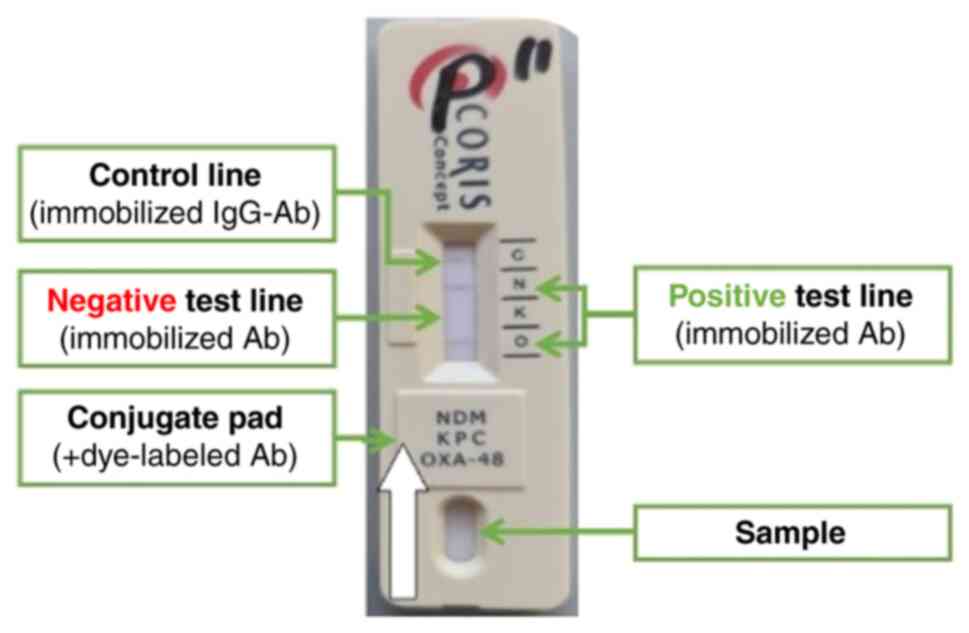
laboratories are lateral flow double-antibody sandwich assays.
Antigens from the biological sample migrate along a nitrocellulose
membrane by capillarity. In doing so, they encounter the conjugate
pad which contains dye-labelled antibodies, forming an
antigen-antibody complex. A second wave of antibodies is
immobilized at the level of the test line; when the
antigen-antibody complex encounters the test line antibodies a
colour reaction occurs and produces a visible band (positive
result). The control line contains immobilized anti-IgG antibodies
aimed at the excess dye-labelled antibodies (with or without the
antigen) and is used to confirm that the sample migrated along the
nitrocellulose membrane (Fig. 1)
(4).
In terms of post-mortem use, POC tests are already
in use for the diagnosis of acute coronary syndrome (rapid troponin
T test) (12), sepsis (a
semi-quantitative procalcitonin test) (13) as well as several antibody-based
tests for the diagnosis of hepatitis virus C and HIV (14,15).
More recently, the post-mortem application of a SARS-COV-2 rapid
antigen test was assessed on nasopharyngeal exudate samples
collected from 30 autopsy cases in a pilot study with promising
results; this showed an overall lower sensitivity compared with
reverse transcription-quantitative PCR considered to be due to
viral loads below the threshold of cultivability in cases with
false-negative results (16).
Rapid diagnostic tests are currently commercially
available for the detection of carbapenem resistance. The
O.K.N.V.I. RESIST-5 (Coris BioConcept) targets KPC, NDM, VIM, IMP
and OXA-48 types of carbapenemases and is performed on pure
microbial cultures, in laboratories, without the use of specialized
equipment (17). The detection
limit, as provided by the manufacturer, varies according to
carbapenemase and was determined using purified recombinant
proteins obtained from KPC (0.5 ng/ml), NDM (0.0625 ng/ml), VIM
(0.23 ng/ml), IMP (0.781 ng/ml) and OXA-48-like carbapenemases
(0.25 ng/ml) (18). To the best of
our knowledge, currently, there are no ICT or POC tests available
for the identification of CR strains directly from clinical samples
(without prior isolation), as the sensitivity of these assays is
not high enough (19). However,
simplified procedures are under research in both laboratory and
clinical settings, aiming to concentrate sample antigens and
eliminate any potential interference with impurities. Simplified
procedures have been tested on blood cultures (NG-Test Carba 5
assay) (20), rectal swabs
(RESIST-4 O.K.N.V. K-SeT test) (21,22)
and urine samples using a dedicated device (19).
Not yet available for routine clinical practice,
rapid tests for the detection of antimicrobial resistance using
immunochromatographic analysis would guide the diagnosis and
management of infections with CR strains. Some of its potential
applications lie in the field of legal medicine/forensic pathology,
where other rapid tests have already proven useful [troponin
(12), C reactive protein
(23), HIV (15) etc.]. Post-mortem sampling offers
the advantage of targeted access to biological samples,
irrespective of the area of interest, by allowing sampling of fluid
collections, including pus, as well as profound structures, such as
the distal respiratory tract. Moreover, the post-mortem dehydration
results in a concentrated sample of antigens.
In Romania, autopsy-performing facilities are
dependent on external laboratories for microbiological assessment;
thus, sampling is strictly case-oriented and not routinely
performed. A cheaper, faster and easier method of assessing
antibiotic resistance would potentially redefine the incidence and
prevalence of ESBL-producing microorganisms. For patients who die
during hospitalization where the issue of malpractice arises,
medico-legal practice would benefit from rapid tests validated for
post-mortem samples. Thus, detection and differentiation of the
five targeted carbapenemases, KPC, NDM, VIM, IMP and OXA-48, is
useful in guiding sampling for external microbiological assessment
and could also be an asset from an epidemiological standpoint.
The current study aimed to test the performance of
the O.K.N.V.I. RESIST-5 test on impure post-mortem biological
samples compared with a standard consisting of pure microbial
cultures obtained from the initial sample through inoculation and
incubation.
Materials and methods
The present study was a prospective, single-centre
pilot study conducted at Mina Minovici National Institute of Legal
Medicine (Bucharest, Romania) aimed to compare CR identifying rapid
test results obtained from post-mortem samples either on-site or as
pure microbial cultures after inoculation, isolation and
identification of the causative agents.
Sample collection
Bronchial swabs were preferred to tracheal swabs due
to agonal aspiration of gastric content. The trachea was dissected
just above the bronchial bifurcation and four sterile swabs were
inserted distally into the bronchi. For other biological samples,
four swabs per site were collected through a sterile technique
(using sterile gloves and dissection instruments, immediately after
removing the wound dressings or opening the anatomical cavities,
prior to other extensive manipulation of body or organs, wound
swabbing, collection of meningeal fluid etc.). Following sample
collection, two swabs were used for O.K.N.V.I. RESIST-5 testing and
the other two were sent to a third party laboratory for further
testing the results obtained from the latter were outside the scope
of the study.
Patient inclusion
A total of 200 autopsies were performed at The Mina
Minovici National Institute of Legal Medicine between June and July
2022. Patient inclusion criteria were: Hospitalization over 3 days
and a carbapenem-resistant (CR) strain identified during
hospitalization.
Autopsies that did not have post-mortem bacteriology
results available for review were excluded from the present study.
A total of 21 medico-legal autopsies met the inclusion criteria and
rapid on-site tests were performed in 14 cases out of the 21. The
location of post-mortem microbiological sampling was selected in a
case-oriented manner, guided by the anatomical area or biological
sample from which a positive culture for CR strains was identified
during hospitalization, thereby suspected as a possible cause of
death.
Examination technique and
O.K.N.V.I
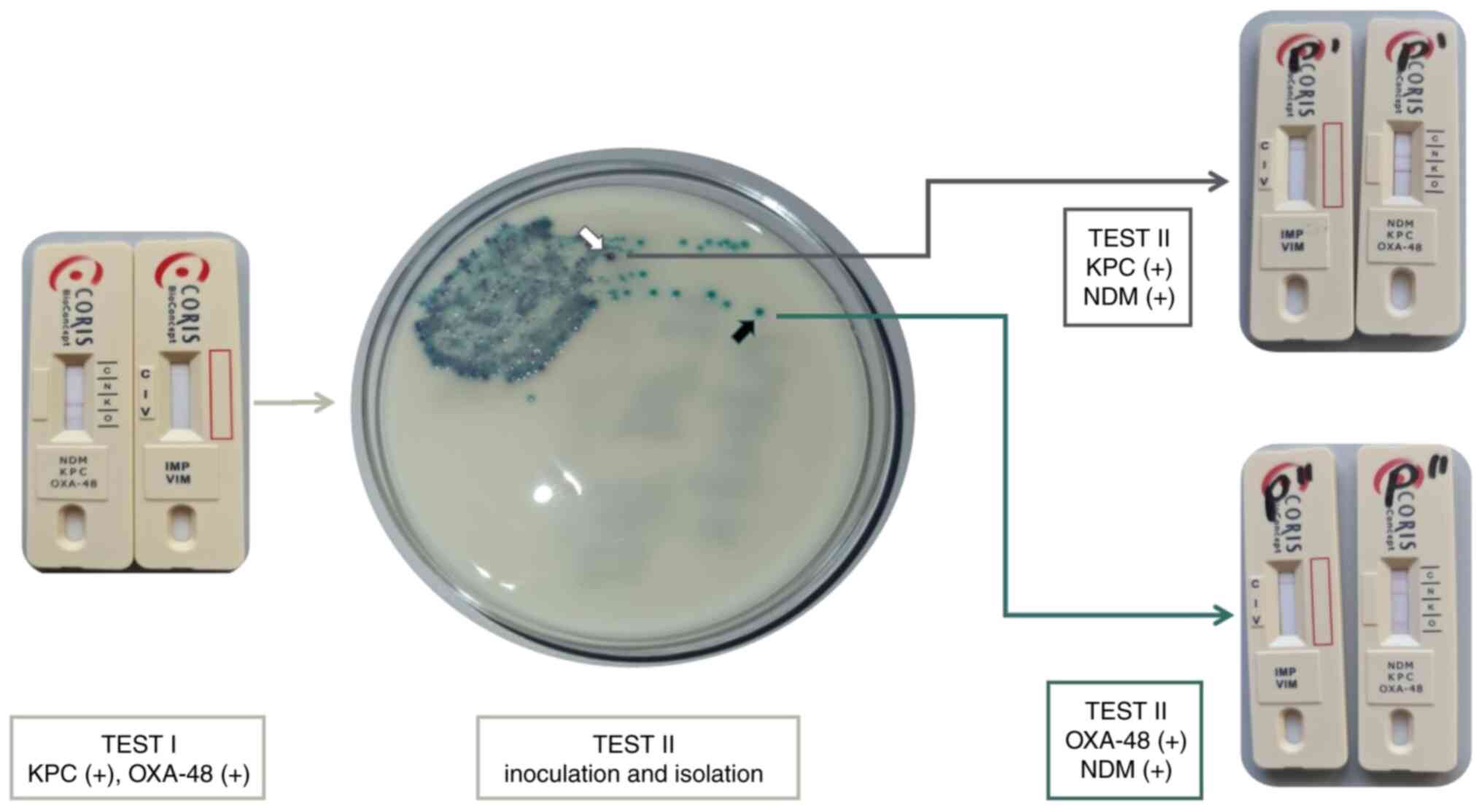
RESIST-5 testing. A total of two sets of
O.K.N.V.I. RESIST-5 tests (Test I and II) were performed for each
collected specimen. Test I was performed on-site from post-mortem
samples, while Test II was performed on pure cultures obtained
after isolation and served as standard, as described below:
The swab for Test I was tested on-site, immediately
after post-mortem sampling. The swab was immersed into a semi-rigid
tube containing 12 drops of buffer (LY-D) provided by the
manufacturer of the rapid diagnostic kits (Coris BioConcept), mixed
thoroughly, and the diluted sample transferred using a pipette onto
each of the two cassettes of O.K.N.V.I. RESIST-5. The results were
read within 15 min of sampling (Fig.
2; bottom green arrow, Test I).
The swab for Test II was tested locally. The swab
was used to inoculate Brilliance™ ESBL Agar (Oxoid, Ltd.; Thermo
Fisher Scientific, Inc. Fisher Scientific), a chromogenic screening
plate with a selective medium for the detection of
extended-spectrum β-lactamase-producing strains. The Brilliance™
ESBL Agar plates were examined after incubation at 36±2˚C, for
18-24 h. If bacterial growth showed mixed populations, an isolation
plate was performed for further differentiation, by aseptically
transferring a small amount of an isolated colony using a smear
loop onto CLED Agar plate (Oxoid, Ltd.; Thermo Fisher Scientific,
Inc.), followed by incubation at 36±2˚C for 18-24 h. Once pure
cultures were obtained, O.K.N.V.I. RESIST-5 tests were performed
according to manufacturer instructions (Fig. 2; top green arrow, Test II). Results
were obtained within 1-2 days of sampling depending on culture
growth rate and incubation facilities.
Statistical analysis
Categorical variables were reported as counts or
frequencies (%). Continuous data are presented as mean ± standard
deviation for normally distributed data, while non-normally
distributed data are presented as median and interquartile range
(IR) or range (R). P<0.05 was considered to indicate a
statistically significant difference. The statistical analysis was
performed using IBM® SPSS® Statistics
software for Windows, version 24.0 (IBM Corp.). Sensitivity,
specificity, and positive and negative predictive values were
assessed using crosstabs column and row percentages with syntax for
95% CIs. The effect of postmortem interval upon concordance of the
two sets of test for each type of carbapenemase was analyzed by
dividing the samples into two groups and since >20% of the cells
in the contingency table contained an expected count of five or
less, Fisher's exact test was used (Table SII).
Ethics
This study was approved by the Institutional
Research Ethics Committee from Mina Minovici National Institute of
Legal Medicine, Bucharest (approval no. 5152/2022).
Results
Descriptives of included cases,
sampling and rapid testing
The present study included four female (28.8%) and
10 male (71.4%) patients with a median age of 63 years (IR,
48.7-77.0 years). Patients had a hospital stay of 43.5 days (IR,
23.5-58.2 days) with a maximum of 107 days. The post-mortem
interval (PMI), i.e. time to autopsy and sample collection, was 3
days (IR, 1.7-4.0 days), the longest being 6 days. A total of 10
patients (71.4%) were hospitalized due to violent circumstances
such as traumatic brain injury, polytrauma (motor vehicle
collision) or thermal burn injuries, of whom six (60.0%) had
pre-existing conditions (Table I).
Patient characteristics and additional data extracted from medical
records obtained before death are available in Table I.
 | Table IStudy patient characteristics and
additional data extracted from medical records obtained before
death. |
Table I
Study patient characteristics and
additional data extracted from medical records obtained before
death.
| Patient no. | Age, years/sex | Cause of
hospitalization | Pre-existing
conditions | Hospital stay,
days | Post-mortem interval,
days | Empirical
antimicrobial treatment (duration, days) | Positive microbiology
during hospitalization | Targeted
antimicrobial treatment (duration, (days) | Superimposed
infection during hospitalization | CR positive sample
location |
|---|
| 1 | 50/F | Spontaneous
pneumothorax | Chronic obstructive
pulmonary disease; Obesity | 59 | 4 | Ceftriaxone (5);
Moxifloxacin (5); Meropenem (7) | Acinetobacter
baumannii (MDR); Klebsiella pneumoniae | Colistin (14);
Tigecycline (10); TZP (13) | Providencia
stuartii | Respiratory
tract |
| 2 | 83/M | Traumatic brain
injury | Epilepsy; HTN | 31 | 2 | Ceftriaxone (12) | Klebsiella
pneumoniae (CR) | Colistin (14);
Tigecycline (10) | Pseudomonas
aeruginosa; Providencia stuartii | Respiratory
tract |
| 3 | 45/M | Perforated colonic
diverticulitis | | 38 | 3 | Colistin (7) | Klebsiella
pneumoniae (CR) | Recarbrio (21);
Linezolid (14) | Acinetobacter
baumannii | Respiratory
tract |
| 4 | 88/F | Thermal burns | T2DM; Heart
failure | 4 | 1 | | Klebsiella
pneumoniae (CR) | Colistin (3);
Tigecycline (2) | - | Wound swab |
| 5 | 57/M | Traumatic brain
injury | | 27 | 1 | Ceftriaxone
(14) | Klebsiella
pneumoniae (CR) | Recarbrio (5);
Vancomycin (2) | - | Respiratory
tract |
| 6 | 40/M | Septic arthritis of
the hip | Heroin addiction;
Hepatitis C virus infection | 11 | 3 | Ceftriaxone
(1) | Escherichia
coli ESBL; Klebsiella pneumoniae (CR) | Colistin (10);
tigecycline (10) | - | Respiratory tract,
wound swab |
| 7 | 74/M | Traumatic brain
injury | Alcoholism;
HTN | 53 | 2 | Ceftriaxone
(7) | Klebsiella
pneumoniae (CR) | Colistin (14)
tigecycline (14) | Burkholderia
cepacia complex | Respiratory
tract |
| 8 | 69/M | Polytrauma | | 31 | 4 | Ceftriaxone
(6) | Acinetobacter
baumannii (MDR); Klebsiella pneumoniae | Colistin (10);
tigecycline (10); TZP (14) | Pseudomonas
aeruginosa | Respiratory
tract |
| 9 | 37/F | Thermal burns | | 58 | 6 | Ceftriaxone
(6) | Escherichia
coli (ESBL); Acinetobacter baumannii (MDR) | Colistin (10);
tigecycline (14); TZP (8) | Pseudomonas
aeruginosa (MDR) | Wound swab |
| 10 | 89/M | Polytrauma | | 49 | 3 | Ceftriaxone (10);
Moxifloxacin (14) | Pseudomonas
aeruginosa (MDR); Acinetobacter baumannii | Colistin (10); TZP
(14) | Klebsiella
pneumoniae (CR) | Respiratory
tract |
| 11 | 54/M |
Meningoencephalitis | T2DM; HTN | 13 | 5 | Meropenem (3) | Salmonella
spp. | Meropenem (10);
vancomycin (10); levofloxacin (7) | Acinetobacter
baumannii | Respiratory
tract |
| 12 | 59/M | Thermal burns | T2DM; Obesity | 79 | 2 | Ceftriaxone
(14) | Pseudomonas
aeruginosa (MDR); Acinetobacter baumannii | TZP (14);
Vancomycin (10) | Klebsiella
pneumoniae (CR) A. baumannii | Respiratory
tract |
| 13 | 75/M | Traumatic brain
injury | Cirrhosis | 55 | 1 | Ciprofloxacin
(7) | Proteus
mirabilis (extended spectrum beta-lactamase) | Amoxicillin
clavulanate (14) | Providencia
stuartii (CR) | Respiratory
tract |
| 14 | 67/F | Smoke
inhalation | HTN | 107 | 3 | Doxycycline (10);
Levofloxacin (7); TZP (10) | Acinetobacter
baumannii Pseudomonas aeruginosa | TZP (14); Colistin
(14) | Klebsiella
pneumoniae (CR) | Respiratory
tract |
A total of 39 O.K.N.V.I. RESIST-5 rapid tests were
performed on 19 biological samples collected from 14 patients; one
sample per case and, in five cases, two samples. For two of the
samples, Test I was negative and Test II was not performed since
the 24 h incubation showed no microbial growth on the Oxoid
Brilliance ESBL Agar screening plate. The first negative sample
(Table I, patient no. 6) was a
tracheal swab collected from a patient who tested positive for CR
Klebsiella pneumoniae during hospitalization (wound swab).
They had been treated empirically with ceftriaxone for 1 day,
followed by combination therapy with colistin and tigecycline for
10 days. The second negative sample (Table I, patient no. 11) was a meningeal
swab collected from a patient with meningoencephalitis of unknown
origin. The patient had been treated empirically with meropenem for
3 days followed by combination therapy with meropenem, vancomycin
and levofloxacin.
Moreover, fourteen of the inoculated samples
generated monocultures, while three samples required transfer to a
second plate for microbial pure culture. An invalid result was
obtained from one rapid test performed on a microbial culture (Test
II); therefore a third test was performed. The following
Enterobacteriaceae species were identified in pure culture:
Klebsiella pneumoniae (16 cultures), Pseudomonas
aeruginosa (two cultures) and Escherichia coli (one
culture). For 10 of the pure cultures, the same CR strain showed
positive results for two different types of carbapenemases
(Fig. 3).
Antimicrobial therapy
All patients included in the present study had
positive microbiology results for carbapenem-resistant
microorganisms while in-hospital (Table I). All but one (13 patients)
received empirical antibacterial therapy with broad-spectrum
antibiotics. Of the 13 patients, 10 received single-agent
antimicrobial therapy as empirical treatment. Ceftriaxone was the
most popular option for empirical antibacterial therapy being used
in 10 of the 13 patients, with a median duration of treatment of 7
days (range, 1-14 days).
All 14 patients received targeted antibacterial
therapy for a variable number of days after CR strain
identification. Combination therapy regiments most often included
colistin (9/14 patients) for a mean duration of 10 days (range,
3-14 days), tigecycline (seven patients) for a mean duration of 10
days (range, 2-14 days), piperacillin/tazobactam (six patients) for
a mean duration of 14 days (range, 8-14 days), vancomycin (three
patients) for a mean duration of 10 days (range, 2-10 days) and
Recarbrio (imipenem/cilastatin/relebactam; Merck Sharp &
Dohme-Hoddesdon) in two patients for a mean duration of 13 days
(range, 5-21 days).
In the context of the present study, for any type of
carbapenemase, the results were interpreted as follows: A negative
result during Test I with a positive result during Test II as a
false negative; a positive result during both sets of tests as a
true positive; a negative result during both sets of tests as a
true negative; and a positive result during Test I with a negative
result during Test II as a false positive. The Test I results for
the two samples that showed no microbial growth were excluded from
the interpretation.
For tests performed on-site from post-mortem
biological samples (Test I results-interpretation), eight false
negative results were obtained: Five for NDM; two for OXA-48; and
one for VIM. Five false negative results were found in cases that
received CO for at least 10 days (two NDM false negative results
from cases with a combination of antibacterial therapy with TZP for
14 days; one NDM and one OXA-48 false negative result from cases
with a combination of antibacterial therapy with TZP for 13 days
and TG for 10 days). Two false negative results (one VIM and one
NDM) were encountered in a case that received RECARBRIO for 5 days
and Vanco for 2 days before death, and an OXA-48 false negative in
a case with the following antibacterial therapy: TZP for 14 days
and Vanco for 10 days. True negative results for all five
carbapenemases were found in four samples, three samples from cases
that received a combination of antibacterial therapy with CO and TG
for at least 10 days, with or without TZP, and one sample from a
case with RECARBRIO for 21 days (Table SI).
Time to sampling (PMI)
The present study aimed also to evaluate whether the
PMI had any influence on the results of the O.K.N.V.I. RESIST-5
performed on-site (Test I) or from a pure culture (Test II). Given
the small sample size consistent results were computed without
statistical tests for each carbapenemase type, the true negative
and true positive results were both considered as concordance of
tests. Due to the small sample size, we took into account the
length of time between death and biological sample collection
(postmortem interval-PMI) as a binary variable dividing the samples
into two groups (PMI under/over 24 h) (Table II) and used 2x5 contingency table
with Fisher's exact test (Table SII) to determine if there was a
significant association with concordance (C) for each type of
carbapenemase. The O.K.N.V.I. RESIST-5 test concordance for each
carbapenemase type does not seem to differ with PMI groups,
respectively with time elapsed from death to biospecimen collection
(P=0.989).
 | Table IICarbapenem-resistance is identified
through RESIST-5 O.K.N.V.I. performed on-site (Test I) and from a
pure culture (Test II). |
Table II
Carbapenem-resistance is identified
through RESIST-5 O.K.N.V.I. performed on-site (Test I) and from a
pure culture (Test II).
| CP | Imipenemase | Verona
integron-encoded metallo-β-lactamase | New Delhi
metallo-β-lactamase | Klebsiella
pneumoniae CP | Oxacilinase-48 |
|---|
| PMI groups | Test I | Test II | C | Test I | Test II | C | Test I | Test II | C | Test I | Test II | C | Test I | Test II | C |
|---|
| <24 h | | | | | | | | | | | | | | | |
| 1 | - | - | Yes | + | + | Yes | - | - | Yes | - | - | Yes | - | - | Yes |
| 2 | - | - | Yes | - | + | No | - | - | Yes | + | + | Yes | - | - | Yes |
| 3 | - | - | Yes | - | - | Yes | - | + | No | - | - | Yes | + | + | Yes |
| >24 h | | | | | | | | | | | | | | | |
| 1 | - | - | Yes | - | - | Yes | + | + | Yes | - | - | Yes | + | + | Yes |
| 2 | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes |
| 3 | - | - | Yes | - | - | Yes | + | + | Yes | - | - | Yes | + | + | Yes |
| 4 | - | - | Yes | - | - | Yes | + | + | Yes | - | - | Yes | - | + | No |
| 5 | - | - | Yes | - | - | Yes | - | + | No | - | - | Yes | - | + | No |
| 6 | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes |
| 7 | - | IC | N/A | - | IC | N/A | - | IC | N/A | - | IC | N/A | - | IC | N/A |
| 8 | - | - | Yes | - | - | Yes | - | + | No | - | - | Yes | + | + | Yes |
| 9 | - | - | Yes | - | - | Yes | + | + | Yes | - | - | Yes | + | - | No |
| 10 | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes |
| 11 | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes | - | - | Yes |
| 12 | - | - | Yes | - | - | Yes | - | + | No | + | + | Yes | + | - | No |
| 13 | - | - | Yes | - | - | Yes | - | + | No | + | - | No | + | + | Yes |
| 14 | - | - | Yes | - | - | Yes | - | - | Yes | + | + | Yes | - | - | Yes |
| 15 | - | IC | N/A | - | IC | N/A | - | IC | N/A | - | IC | N/A | - | IC | N/A |
| 16 | - | - | Yes | - | - | Yes | + | + | Yes | - | - | Yes | + | + | Yes |
| Test I/Test II | 17/17(100%) | 16/17 (94.1%) | 12/17 (70.6%) | 16/17 (94.1%) | 13/17 (76.5%) |
| C rates for CP | | | | | | | | | | | | | | | |
Sensitivity and Specificity of
O.K.N.V.I
RESIST-5 tests. The number of positive and
negative results for the O.K.N.V.I. RESIST-5 tests performed during
both Test I and II are shown in Table
II.
The sensitivity, specificity, positive predictive
value and negative predictive value of the O.K.N.V.I. RESIST-5
tests performed on-site from a biological sample were computed for
each of the types of carbapenemase (IMP, VIM, NDM, KPC and OXA-48)
and obtained via the positive and negative results of both sets of
O.K.N.V.I. RESIST-5 tests, Test II values serving as standard
(Table III). The O.K.N.V.I.
RESIST-5 tests performed on-site from biological samples (Test I)
showed an overall sensitivity of 92.3% with 100% specificity, 100%
positive predictive value and an 80.0% negative predictive
value.
 | Table IIIThe sensitivity, specificity,
positive predictive value, negative predictive value and accuracy
of the O.K.N.V.I. |
Table III
The sensitivity, specificity,
positive predictive value, negative predictive value and accuracy
of the O.K.N.V.I.
| Carbapenemase | Sensitivity, % (95%
CI) | Specificity, % (95%
CI) | Positive predictive
value, % (95% CI) | Negative predictive
value, % (95% CI) | Accuracy % (95%
CI) |
|---|
| IMP | N/A | N/A | - | - | - |
| VIM | 50.0
(1.3-98.7) | 100 (78.2-100) | 100 (2.5-100) | 93.8
(69.8-99.8) | 94.1
(71.3-99.9) |
| NDM | 50.0
(18.7-81.3) | 100 (59.0-100) | 100 (47.8-100) | 58.3
(34.8-93.3) | 70.6
(44.0-89.7) |
| KPC | 100 (39.8-100) | 92.9
(66.1-99.8) | 75.0
(29.0-100) | 100 (76.8-100) | 94.1
(71.3-99.9) |
| OXA-48 | 75.0
(44.0-97.5) | 77.8
(30.8-89.1) | 75.0
(54.1-100) | 77.8
(66.4-100) | 76.5
(50.1-93.2) |
| O.K.N.V.I.
RESIST-5 | 92.3
(63.9-99.81) | 100 (39.8-100) | 100 (73.5-100) | 80.0
(37.9-96.3) | 94.12
(71.3-99.9) |
Discussion
Carbapenem-resistant strains are increasingly
described in both hospital and community-acquired infections
(24) as well as in animal
reservoirs (25). Consequently,
medical negligence accusations focusing on healthcare-associated
infections that go unnoticed or untreated are becoming prominent
issues in forensic medicine. Thus, post-mortem bacteriology
analysis is of paramount importance both in establishing the cause
of death and, in a wider sense, identifying CR strain outbreaks in
hospitals.
Post-mortem bacteriology is rarely requested during
medico-legal autopsies at The Mina Minovici National Institute of
Legal Medicine where the present study was performed (26) due to its costly and time-consuming
nature. However, rapid diagnostic tests are emerging, some with
applications in forensic medicine (12-15,23).
The present study aimed to assess their performance in the
detection of carbapenem-resistant strains from post-mortem samples
(bronchial swabs, wound swabs and meningeal fluid) without the need
for incubation, isolation and identification. Blood cultures were
also collected for each case without being included in the
O.K.N.V.I. RESIST-5 testing sequence, due to the potentially lower
concentration of microorganisms. The overall sensibility and
sensitivity of the O.K.N.V.I. RESIST-5 tests performed on-site were
satisfactory.
The O.K.N.V.I. RESIST-5 tests we used in the current
study could identify up to five CR strains. However, as no
IMP-producing strains were recovered and only two VIM strains among
the 19 samples were collected, we hypothesise that less specialized
versions of multiplex tests like RESIST-4 OKNV (27) or RESIST-3 OKN (28) may be sufficient for post-mortem
screening of CR strains.
Traditionally, POC tests guide the course of
treatment. The present study proposes a legal-medicine-oriented
application by guiding the course of complementary examinations for
determining probable cause of death. For example, in medico-legal
cases with macroscopic signs of infection (with or without a
previously identified causative agent), positive screening for
carbapenem resistance could guide the pathologist towards
requesting a complete post-mortem microbiology assessment.
Although limited by the small sample size, the
present results showed a good correlation between the tests
performed on-site from biological samples (‘point of autopsy’) and
results from pure cultures after 24-48 h of incubation.
In medico-legal autopsies, the pathologist often
needs to answer questions regarding the contribution of
healthcare-associated infections to thanatogenesis. On the one
hand, a legislative framework provides specific case definitions
based on clinical and laboratory findings recorded during
hospitalization. On the other, post-mortem analysis is of
tantamount importance in determining if correct in-hospital
management was achieved regarding the diagnosis and treatment of
healthcare-associated infections.
Performing O.K.N.V.I. RESIST-5 tests on biological
samples collected on-site during medico-legal autopsy are easy,
fast and inexpensive. The results obtained through rapid tests
using post-mortem impure samples are comparable to results from
sample cultures with good sensitivity and specificity. Through
post-mortem screening for carbapenem resistance, it is possible to
improve the screening of cases that require further bacteriological
assessment. Nonetheless, larger studies are required to confirm the
present findings.
Acknowledgements
Publication of this paper was supported by the
University of Medicine and Pharmacy Carol Davila, through the
institutional program Publish not Perish.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ID conceptualized the study. ID, MH and AAK
discussed and designed the methodology. ID, MC and CD performed the
investigation. ID and MC confirm the authenticity of all the raw
data. ID provided the resources for the study. ID, MC, MH and LNC
performed the data validation. ID and LNC provided the data
curation. ID, MH and LNC performed the statistical analysis, figure
design and interpretation of the results. ID and MH prepared the
original draft. ID, MH and AAK worked on writing, reviewing and
English editing. MC and CD supervised the study. ID insured project
administration. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate.
Relevant ethical issues were identified and
discussed with the institutional ethical committee. All
medico-legal autopsies in Romania are mandated by the police or the
state prosecutor. Institutional access to data was obtained.
Informed consent of relatives for data and sample collection was
waived by the Institutional Research Ethics Committee of the Mina
Minovici National Institute of Legal Medicine, Bucharest,
considering that all microbiological samples were requested to
ascertain probable cause of death. The study was conducted in
accordance with the Declaration of Helsinki.
Patient consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nordmann P and Cornaglia G:
Carbapenemase-producing Enterobacteriaceae: A call for
action! Clin Microbiol Infect. 18:411–412. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nordmann P: Carbapenemase-producing
Enterobacteriaceae: Overview of a major public health
challenge. Med Mal Infect. 44:51–56. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Queenan AM and Bush K: Carbapenemases: The
versatile beta-lactamases. Clin Microbiol Rev. 20:440–458, table of
contents. 2007.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Noster J, Thelen P and Hamprecht A:
Detection of multidrug-resistant enterobacterales-from ESBLs to
carbapenemases. Antibiotics (Basel). 10(1140)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yigit H, Queenan AM, Anderson GJ,
Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K and
Tenover FC: Novel carbapenem-hydrolyzing beta-lactamase, KPC-1,
from a carbapenem-resistant strain of Klebsiella pneumoniae.
Antimicrob Agents Chemother. 45:1151–1161. 2001.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Paterson DL: 313-infections due to other
members of the Enterobacteriaceae, including management of
multidrug-resistant strains, Goldman L and Schafer AI (eds).
Goldman's Cecil Medicine (Twenty Fourth Edition), W.B. Saunders,
pp1874-1877, 2012. https://doi.org/10.1016/B978-1-4377-1604-7.00313-4.
|
|
7
|
Poirel L, Hombrouck-Alet C, Freneaux C,
Bernabeu S and Nordmann P: Global spread of New Delhi
metallo-β-lactamase 1. Lancet Infect Dis. 10(832)2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Nordmann P, Naas T and Poirel L: Global
spread of carbapenemase-producing Enterobacteriaceae. Emerg
Infect Dis. 17:1791–1798. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Stewart A, Harris P, Henderson A and
Paterson D: Treatment of infections by OXA-48-producing
Enterobacteriaceae. Antimicrob Agents Chemother.
62:e01195–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Poirel L, Potron A and Nordmann P:
OXA-48-like carbapenemases: The phantom menace. J Antimicrob
Chemother. 67:1597–1606. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Anfossi L, Giovannoli C and Baggiani C:
Introductory chapter: Rapid test-advances in design, formats, and
detection strategies [Internet]. Rapid Test-Advances in Design,
Format and Diagnostic Applications. InTech, 2018. Available from:
http://dx.doi.org/10.5772/intechopen.79805.
|
|
12
|
Cina SJ, Brown DK, Smialek JE and Collins
KA: A rapid postmortem cardiac troponin T assay: Laboratory
evidence of sudden cardiac death. Am J Forensic Med Pathol.
22:173–136. 2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ramsthaler F, Kettner M, Mall G and
Bratzke H: The use of rapid diagnostic test of procalcitonin serum
levels for the postmortem diagnosis of sepsis. Forensic Sci Int.
178:139–145. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Rose CE, Duncan L and Hawes AM: Validation
of OraQuick HCV rapid antibody test in postmortem specimens. Acad
Forensic Pathol. 10:81–86. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Karhunen PJ, Brummer-Korvenkontio H,
Leinikki P and Nyberg M: Stability of human immunodeficiency virus
(HIV) antibodies in post-mortem samples. J Forensic Sci.
39:129–135. 1994.PubMed/NCBI
|
|
16
|
Zacharias M, Stangl V, Thüringer A,
Loibner M, Wurm P, Wolfgruber S, Zatloukal K, Kashofer K and
Gorkiewicz G: Rapid antigen test for postmortem evaluation of
SARS-CoV-2 carriage. Emerg Infect Dis. 27:1734–1737.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hong J, Kang D and Kim D: Performance
evaluation of the newly developed in vitro rapid diagnostic test
for detecting OXA-48-Like, KPC-, NDM-, VIM- and IMP-type
carbapenemases: The RESIST-5 O.K.N.V.I. Multiplex lateral flow
assay. Antibiotics (Basel). 10(460)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
OKNVI RESIST-5, Coris BioConcept.
https://ifu.corisbio.com/corisbio/all?keycode=COR58R1104
(Visited on 02.06.2023).
|
|
19
|
Boutal H, Moguet C, Pommiès L, Simon S,
Naas T and Volland H: The revolution of lateral flow assay in the
field of AMR detection. Diagnostics (Basel).
12(1744)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Giordano L, Fiori B, D'Inzeo T, Parisi G,
Liotti FM, Menchinelli G, De Angelis G, De Maio F, Luzzaro F,
Sanguinetti M, et al: Simplified testing method for direct
detection of carbapenemase-producing organisms from positive blood
cultures using the NG-test carba 5 assay. Antimicrob Agents
Chemother. 63:e00550–19. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gallah S, Villageois-Tran K, Godmer A,
Arlet G, Rottman M, Benzerara Y and Garnier M: Four-hour
immunochromatographic detection of intestinal carriage of
carbapenemase-producing Enterobacteriaceae: A validation
study. J Clin Microbiol. 59:e02973–20. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Rescape & Resist-Bc, Coris BioConcept.
https://www.corisbio.com/pdf/Products/Coris-D-CUS-10-RES04-2023-02-V1.pdf
(Visited on 02.06.2023).
|
|
23
|
Soejima M and Koda Y: Evaluation of
point-of-care testing of C-reactive protein in forensic autopsy
cases. Forensic Sci Int. 237:27–29. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tilahun M, Kassa Y, Gedefie A and Ashagire
M: Emerging carbapenem-resistant Enterobacteriaceae
infection, its epidemiology and novel treatment options: A review.
Infect Drug Resist. 14:4363–4374. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Silva JMD, Menezes J, Marques C and Pomba
CF: Companion animals-an overlooked and misdiagnosed reservoir of
carbapenem resistance. Antibiotics (Basel). 11(533)2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Diac I, Keresztesi AA, Cerghizan AM,
Negrea M and Dogăroiu C: Postmortem bacteriology in forensic
autopsies-a single center retrospective study in Romania.
Diagnostics (Basel). 12(2024)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cointe A, Bonacorsi S, Truong J, Hobson C,
Doit C, Monjault A, Bidet P and Birgy A: Detection of
carbapenemase-producing Enterobacteriaceae in positive blood
culture using an immunochromatographic RESIST-4 O.K.N.V. Assay.
Antimicrob Agents Chemother. 62:e01828–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wareham DW and Abdul Momin MH: Rapid
detection of carbapenemases in Enterobacteriaceae:
Evaluation of the resist-3 O.K.N. (OXA-48, KPC, NDM) lateral flow
multiplexed assay. J Clin Microbiol. 55:1223–1225. 2017.PubMed/NCBI View Article : Google Scholar
|