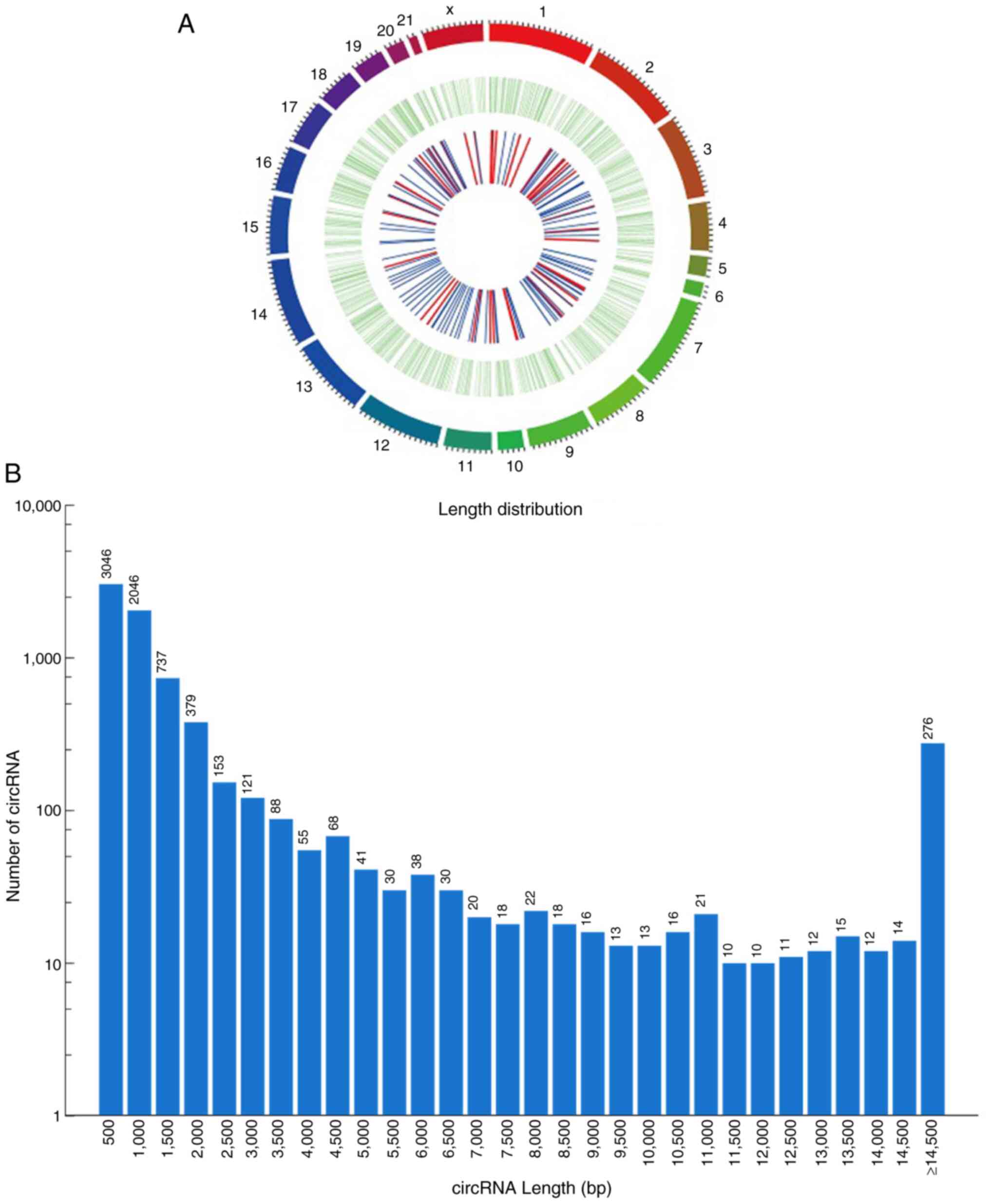
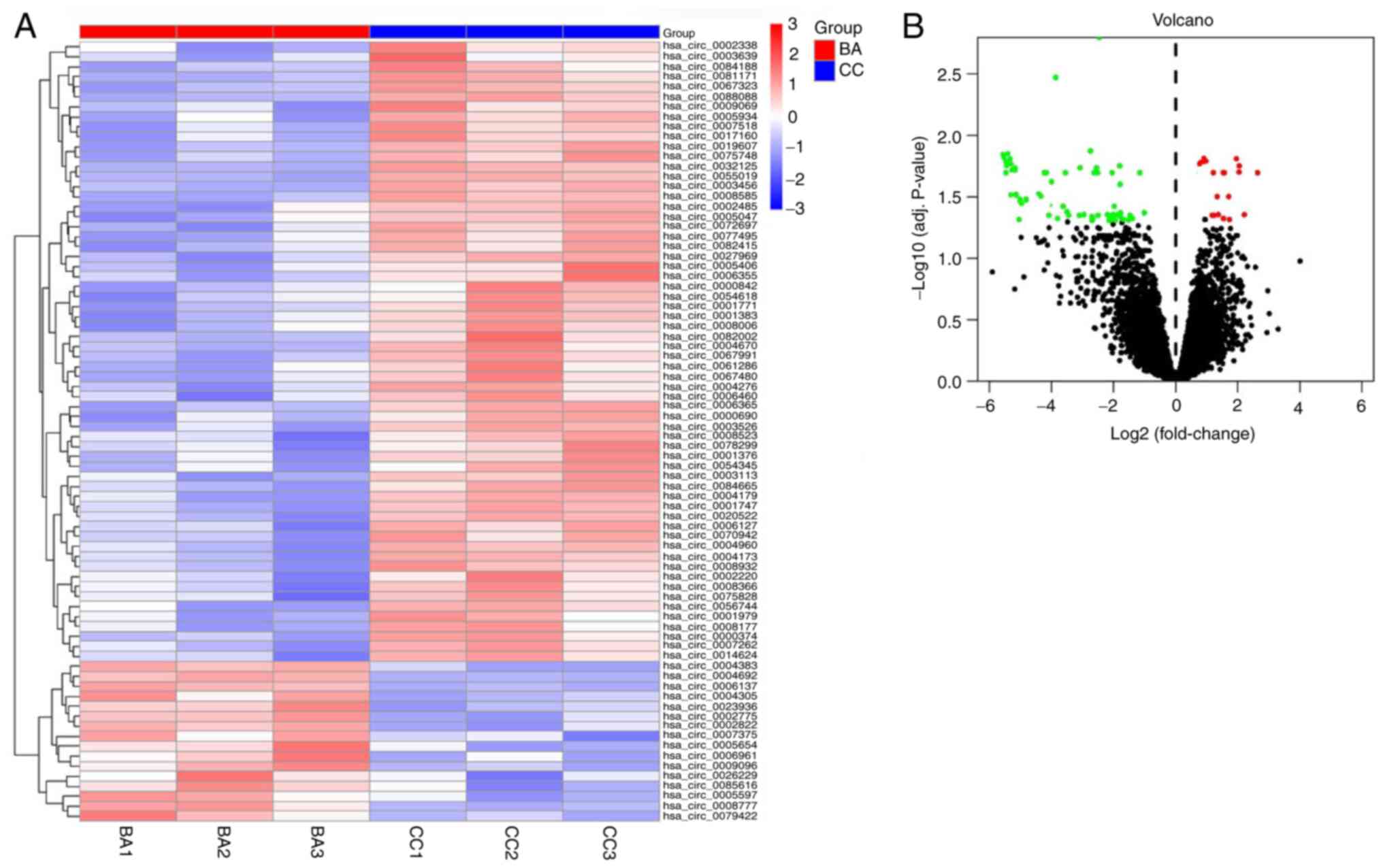
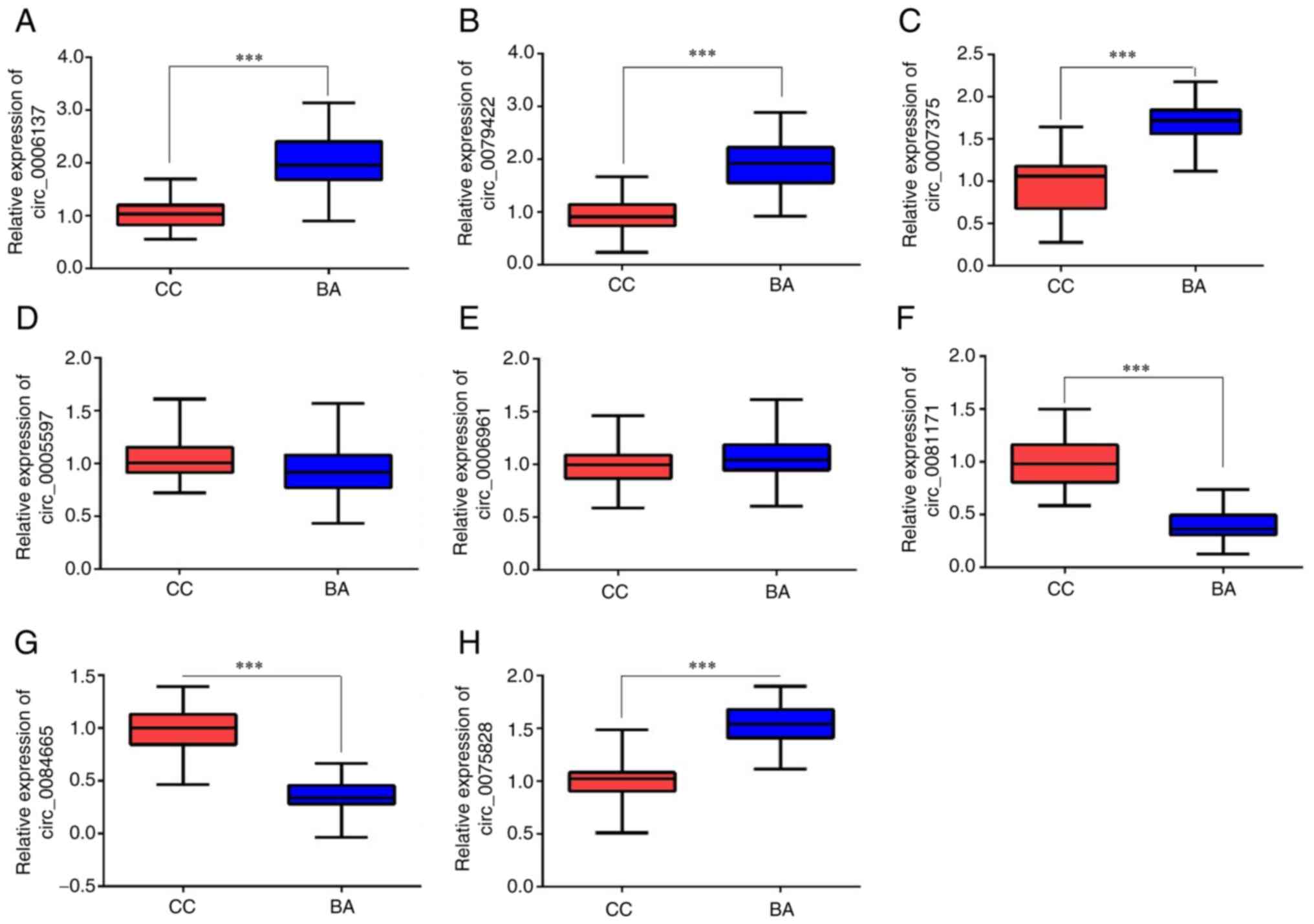
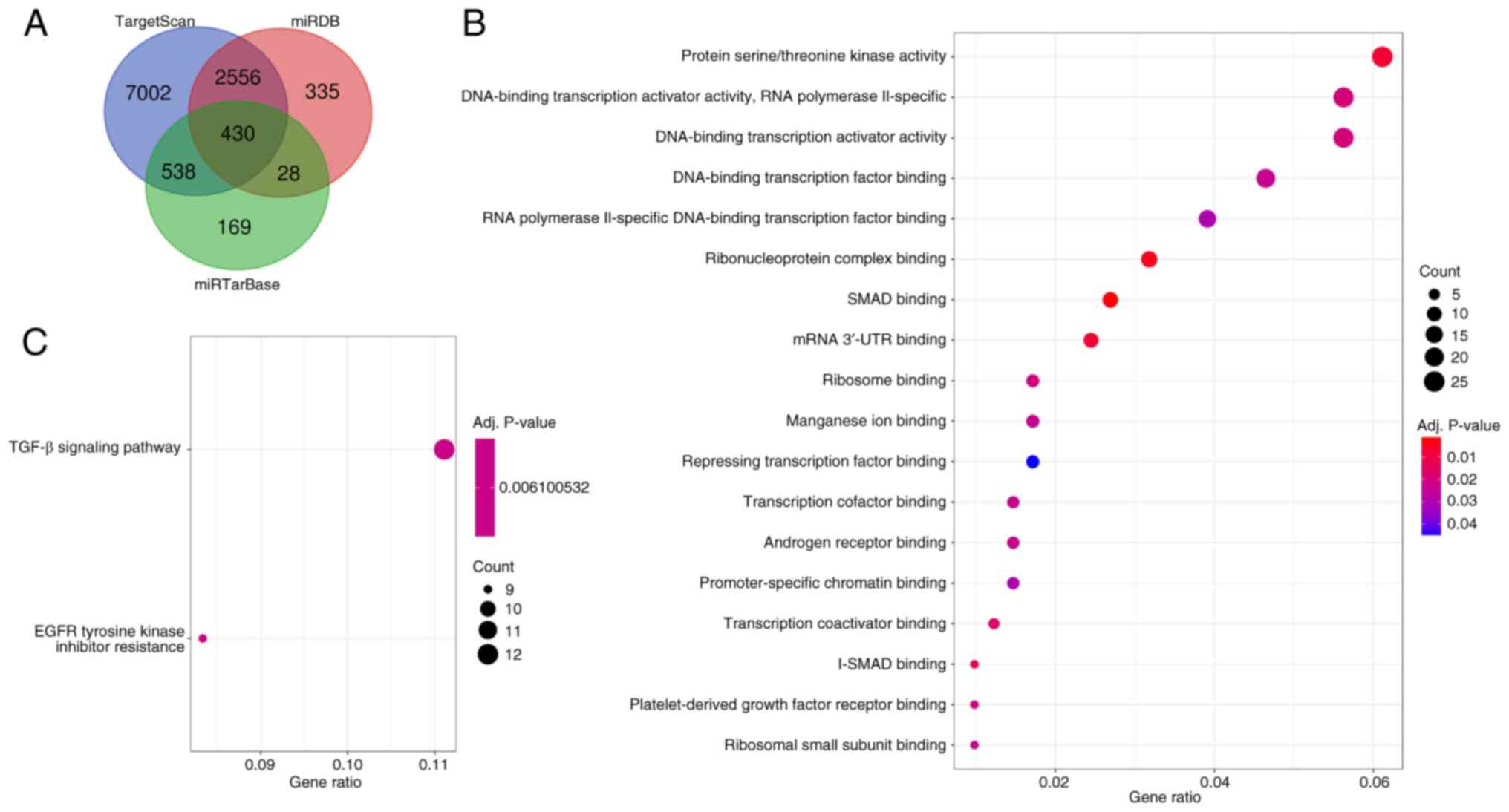
|
1
|
Kawano Y, Yoshimaru K, Uchida Y, Kajihara
K, Toriigahara Y, Shirai T, Takahashi Y and Matsuura T: Biliary
atresia in a preterm and extremely low birth weight infant: A case
report and literature review. Surg Case Rep. 6(321)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bezerra JA, Wells RG, Mack CL, Karpen SJ,
Hoofnagle JH, Doo E and Sokol RJ: Biliary atresia: Clinical and
research challenges for the twenty-first century. Hepatology.
68:1163–1173. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hsiao CH, Chang MH, Chen HL, Lee HC, Wu
TC, Lin CC, Yang YJ, Chen AC, Tiao MM, Lau BH, et al: Universal
screening for biliary atresia using an infant stool color card in
Taiwan. Hepatology. 47:1233–1240. 2008.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hartley JL, Davenport M and Kelly DA:
Biliary atresia. Lancet. 374:1704–1713. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Davenport M, Tizzard SA, Underhill J,
Mieli-Vergani G, Portmann B and Hadzić N: The biliary atresia
splenic malformation syndrome: A 28-year single-center
retrospective study. J Pediatr. 149:393–400. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hartley JL, O'Callaghan C, Rossetti S,
Consugar M, Ward CJ, Kelly DA and Harris PC: Investigation of
primary cilia in the pathogenesis of biliary atresia. J Pediatr
Gastroenterol Nutr. 52:485–488. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Fabris L, Cadamuro M, Guido M, Spirli C,
Fiorotto R, Colledan M, Torre G, Alberti D, Sonzogni A, Okolicsanyi
L and Strazzabosco M: Analysis of liver repair mechanisms in
Alagille syndrome and biliary atresia reveals a role for notch
signaling. Am J Pathol. 171:641–653. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Muraji T: Biliary atresia: New lessons
learned from the past. J Pediatr Gastroenterol Nutr. 53:586–587.
2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Edom PT, Meurer L, da Silveira TR, Matte U
and dos Santos JL: Immunolocalization of VEGF A and its receptors,
VEGFR1 and VEGFR2, in the liver from patients with biliary atresia.
Appl Immunohistochem Mol Morphol. 19:360–368. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bolha L, Ravnik-Glavač M and Glavač D:
Circular RNAs: Biogenesis, function, and a role as possible cancer
biomarkers. Int J Genomics. 2017(6218353)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zeng X, Lin W, Guo M and Zou Q: A
comprehensive overview and evaluation of circular RNA detection
tools. PLoS Comput Biol. 13(e1005420)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Verduci L, Strano S, Yarden Y and Blandino
G: The circRNA-microRNA code: Emerging implications for cancer
diagnosis and treatment. Mol Oncol. 13:669–680. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Miao Q, Zhong Z, Jiang Z, Lin Y, Ni B,
Yang W and Tang J: RNA-seq of circular RNAs identified circPTPN22
as a potential new activity indicator in systemic lupus
erythematosus. Lupus. 28:520–528. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li LJ, Zhu ZW, Zhao W, Tao SS, Li BZ, Xu
SZ, Wang JB, Zhang MY, Wu J, Leng RX, et al: Circular RNA
expression profile and potential function of hsa_circ_0045272 in
systemic lupus erythematosus. Immunology. 155:137–149.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu
X, Li L, Yang B, Chen J, Chen S, et al: Identification of circular
RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for
coronary artery disease. Atherosclerosis. 286:88–96.
2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19(30)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Floris G, Zhang L, Follesa P and Sun T:
Regulatory role of circular RNAs and neurological disorders. Mol
Neurobiol. 54:5156–5165. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang X, Lu N, Wang L, Wang Y, Li M, Zhou
Y, Yan H, Cui M, Zhang M and Zhang L: Circular RNAs and esophageal
cancer. Cancer Cell Int. 20(362)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen X, Zhu S, Li HD, Wang JN, Sun LJ, Xu
JJ, Hui YR, Li XF, Li LY, Zhao YX, et al:
N6-methyladenosine-modified circIRF2, identified by
YTHDF2, suppresses liver fibrosis via facilitating FOXO3 nuclear
translocation. Int J Biol Macromol. 248(125811)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang Q, Long Z, Zhu F, Li H, Xiang Z,
Liang H, Wu Y, Dai X and Zhu Z: Integrated analysis of
lncRNA/circRNA-miRNA-mRNA in the proliferative phase of liver
regeneration in mice with liver fibrosis. BMC Genomics.
24(417)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42 (Database Issue):D68–D73. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42 (Database Issue):D92–D97. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47 (D1):D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12(77)2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xu X, Song B, Zhang Q, Qi W and Xu Y:
Hsa_circ_0022383 promote non-small cell lung cancer tumorigenesis
through regulating the miR-495-3p/KPNA2 axis. Cancer Cell Int.
23(282)2023.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang C and He W: Circ_0020014 mediates
CTSB expression and participates in IL-1β-prompted chondrocyte
injury via interacting with miR-24-3p. J Orthop Surg Res.
18(877)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Karreth FA, Reschke M, Ruocco A, Ng C,
Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al:
The BRAF pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. Cell. 161:319–332. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lakshminarayanan B and Davenport M:
Biliary atresia: A comprehensive review. J Autoimmun. 73:1–9.
2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Girard M and Panasyuk G: Genetics in
biliary atresia. Curr Opin Gastroenterol. 35:73–81. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Debray D, Corvol H and Housset C: Modifier
genes in cystic fibrosis-related liver disease. Curr Opin
Gastroenterol. 35:88–92. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Costa FF: Non-coding RNAs, epigenetics and
complexity. Gene. 410:9–17. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Calvopina DA, Coleman MA, Lewindon PJ and
Ramm GA: Function and regulation of microRNAs and their potential
as biomarkers in paediatric liver disease. Int J Mol Sci.
17(1795)2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Vicens Q and Westhof E: Biogenesis of
circular RNAs. Cell. 159:13–14. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li H, Zheng X, Gao J, Leung KS, Wong MH,
Yang S, Liu Y, Dong M, Bai H, Ye X and Cheng L: Whole transcriptome
analysis reveals non-coding RNA's competing endogenous gene pairs
as novel form of motifs in serous ovarian cancer. Comput Biol Med.
148(105881)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wu S, Wu Y, Deng S, Lei X and Yang X:
Emerging roles of noncoding RNAs in human cancers. Discov Oncol.
14(128)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ramljak S, Schmitz M, Repond C, Zerr I and
Pellerin L: Altered mRNA and protein expression of monocarboxylate
transporter MCT1 in the cerebral cortex and cerebellum of prion
protein knockout mice. Int J Mol Sci. 22(1566)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Acalovschi M: Gallstones in patients with
liver cirrhosis: Incidence, etiology, clinical and therapeutical
aspects. World J Gastroenterol. 20:7277–7285. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Konyn P, Alshuwaykh O, Dennis BB,
Cholankeril G, Ahmed A and Kim D: Gallstone disease and its
association with nonalcoholic fatty liver disease, all-cause and
cause-specific mortality. Clin Gastroenterol Hepatol.
21:940–948.e2. 2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Li M, Xiao Y, Liu M, Ning Q, Xiang Z,
Zheng X, Tang S and Mo Z: MiR-26a-5p regulates proliferation,
apoptosis, migration and invasion via inhibiting hydroxysteroid
dehydrogenase like-2 in cervical cancer cell. BMC Cancer.
22(876)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Xing X, Guo S, Zhang G, Liu Y, Bi S, Wang
X and Lu Q: miR-26a-5p protects against myocardial
ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT
signaling pathway. Braz J Med Biol Res. 53(e9106)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ye Y, Li Z, Feng Q, Chen Z, Wu Z, Wang J,
Ye X, Zhang D, Liu L, Gao W, et al: Downregulation of microRNA-145
may contribute to liver fibrosis in biliary atresia by targeting
ADD3. PLoS One. 12(e0180896)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhang C, Zhou H, Yuan K, Xie R and Chen C:
Overexpression of hsa_circ_0136666 predicts poor prognosis and
initiates osteosarcoma tumorigenesis through miR-593-3p/ZEB2
pathway. Aging (Albany NY). 12:10488–10496. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Abd El-Aziz A, El-Desouky MA, Shafei A,
Elnakib M and Abdelmoniem AM: Influence of pentoxifylline on gene
expression of PAG1/miR-1206/SNHG14 in ischemic heart disease.
Biochem Biophys Rep. 25(100911)2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhang Y, Wang D, Zhu T, Yu J, Wu X, Lin W,
Zhu M, Dai Y and Zhu J: CircPUM1 promotes hepatocellular carcinoma
progression through the miR-1208/MAP3K2 axis. J Cell Mol Med.
25:600–612. 2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Roskoski R Jr: Small molecule inhibitors
targeting the EGFR/ErbB family of protein-tyrosine kinases in human
cancers. Pharmacol Res. 139:395–411. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Tzavlaki K and Moustakas A: TGF-β
signaling. Biomolecules. 10(487)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chung-Davidson YW, Ren J, Yeh CY, Bussy U,
Huerta B, Davidson PJ, Whyard S and Li W: TGF-β signaling plays a
pivotal role during developmental biliary atresia in sea lamprey
(petromyzon marinus). Hepatol Commun. 4:219–234. 2019.PubMed/NCBI View Article : Google Scholar
|