In the face of social-economic development,
urbanization and the aging population, chronic diseases are
becoming a global public health issue that is associated with
premature death and disability (1). Chronic diseases are the leading cause
of mortalities worldwide and have led to a 16% increase in
childhood morbidity over the past few decades in the USA (1). Chronic diseases in children refer to
non-communicable diseases that last for >3 months and are caused
by factors associated with genetic metabolism, environment and an
unhealthy diet. Children with chronic diseases undergo long-term
medical treatment and inpatient care, have special dietary
requirements and experience psychological and physical changes as
well as learning difficulties owing to physical unfitness and
absenteeism (2). In the majority
of cases, conventional diagnostic methods can detect childhood
diseases at an advanced stage (3).
Given that microRNAs regulate most biological processes, they may
serve as efficient diagnostic and therapeutic tools for chronic
diseases in children (4).
MicroRNAs are small non-coding RNAs of 20-24
nucleotides in length that play an important role in regulating
gene expression (5-7).
Each microRNA can regulate several target genes simultaneously.
Alternatively, the combination of multiple microRNAs can regulate
the expression of multiple genes that of a single gene. A total of
~60% of human genes are regulated by microRNAs, which constitute
2-3% of the human genome (8).
However, abnormal expression of microRNAs in different cells or
tissues has been associated with the pathogenesis, progression and
severity of multiple chronic diseases, including cardiovascular,
neurodegenerative and endocrine diseases (9-13).
Highly specific microRNAs found in body fluids, such as blood,
saliva, urine and semen, may serve as important biomarkers and play
an essential role in developing treatment strategies for various
diseases (14). The present review
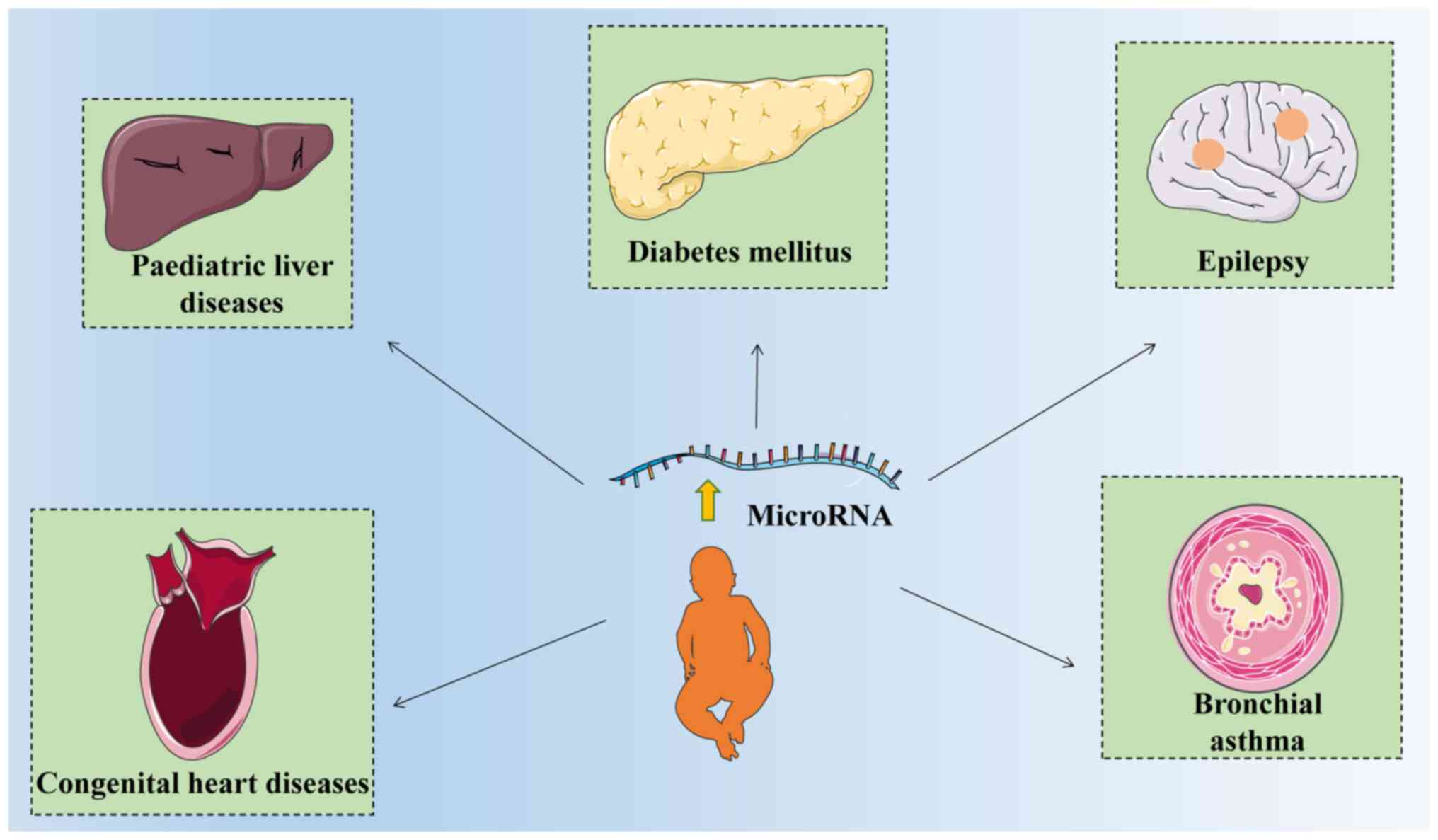
mainly summarizes the role of microRNAs in chronic diseases in
children, including diabetes mellitus, congenital heart disease
(CHD), liver diseases, bronchial asthma and epilepsy (Table I) (15-43).
MicroRNA biogenesis begins in the nucleus with the
synthesis of a long hairpin structure called primary microRNA
(Pri-microRNA) through RNA polymerase II-mediated transcription.
Subsequently, a splicing complex consisting of Drosha and DGCR8
cleaves the Pri-microRNA to form a smaller stem-loop structure
called precursor microRNA (Pre-microRNA). After the Pre-microRNA is
translocated into the cytoplasm via Exportin 5, the RNA-binding
protein and the RNase III endonuclease Dicer collectively produce a
mature double-stranded RNA structure. Decapping enzymes separate
the two RNA strands, resulting in the binding of the guide strand
to Argonaute-guided RNA-induced silencing complex 2. Finally, the
RNA-induced silencing complex-microRNA complexes recognize specific
mRNAs through sequence complementarity, leading to the degradation
of mRNAs or the inhibition of translation (44-49).
Diabetes mellitus is characterized by hyperglycemia
resulting from impaired secretion or function of insulin (50-52).
Prolonged hyperglycemia may cause structural and functional damage
to the eyes, kidneys, heart, blood vessels and nervous system
(53,54).
Studies have suggested that microRNA-124a2 and
microRNA-375 regulate the generation of pancreatic β-cells and are
required for the normal formation of vertebrate islets (55,56).
MicroRNA-375 is essential for the establishment and maintenance of
healthy pancreatic endocrine cells in mice postnatally (57), and its deficiency may result in
pancreatic cell defects and chronic hyperglycemia. In addition,
microRNAs have been reported to regulate various physiological
processes associated with diabetes mellitus, such as insulin
synthesis, secretion and sensitivity and energy homeostasis
(8,58). MicroRNA-15a reduces the levels of
endogenous uncoupling protein-2 to increase oxygen consumption and
decrease ATP production, thereby resulting in the positive
regulation of β-cell function and insulin biosynthesis (59). Additionally, the
microRNA-25/NEUROD1 axis prevents insulin biosynthesis by
regulating the transcription of cell-specific genes (60). MicroRNA-375 is specifically
expressed in pancreatic islets and regulates the secretion of
insulin from isolated pancreatic cells (8). Its overexpression inhibits the
translation of the cytoplasmic protein myotrophin to reduce insulin
secretion by inhibiting the exocytosis of insulin granules
(61). Mice lacking microRNA-375
have increased blood glucose levels and decreased pancreatic β-cell
volume owing to impaired proliferation (57).
CHD is the most common type of birth defect in
children, affecting 5.4-16.1 per 1,000 live births worldwide
(62). It refers to an anatomical
developmental disorder or abnormality of the heart and large blood
vessels that occurs during the embryonic stage (63). Based on the clinical presentation,
CHD is classified as cyanotic heart disease, septal defects and
left-sided obstructive defects (64,65).
At present, diagnostic methods, such as ultrasound-guided
measurement of nuchal translucency, and biomarkers, such as β-human
chorionic gonadotropin and pregnancy-associated plasma protein-A,
are used to screen for fetal CHD. However, false-positive results
are common owing to the non-specificity of these methods (66).
Although numerous microRNAs play an important role
in regulating cardiac function (64,67-74),
microRNA-1 is most commonly associated with CHD (75). In addition to regulating embryonic
heart development, microRNA-1 targets the cardiac transcription
factor heart- and neural crest derivatives-expressed protein 2,
which is involved in cardiovascular development during the
embryonic stage (76). In a
previous study, mice with microRNA-1 deficiency were revealed to
have excessive proliferation of cardiomyocytes and defects in
cardiac conduction, indicating that dysregulation of microRNA-1 may
contribute to CHD (77).
Additionally, microRNAs serve as potential biomarkers in clinical
settings owing to their stability in blood, urine and other
biological fluids and their ability to resist degradation by
RNA-degrading enzymes (78). Yu
et al (79) indicated that
microRNAs in maternal serum can be used to detect fetal CHD. Zhu
et al (80) used reverse
transcription-quantitative PCR followed by sequencing by
oligonucleotide ligation and detection to demonstrate that
microRNA-19b, microRNA-22, microRNA-29c and microRNA-375 are
upregulated in fetal CHD.
Liver diseases in children are more complex and
varied compared with those in adults because it is common for liver
diseases to be underrecognized or diagnosed late in children;
under-diagnosis of liver disease in children is largely due to the
absence of symptoms in most cases, especially in the early stages
(81). As the majority of liver
diseases cannot be easily detected because they do not present
obvious symptoms until the late stage of the disease, most patients
develop severe liver fibrosis or cirrhosis before diagnosis, which
seriously affects their health (82). Biopsy is considered the
gold-standard method for diagnosing liver disease and fibrosis;
however, it can lead to severe bleeding and pain owing to its
invasiveness (83). In addition,
biopsy is limited to a small area of the liver and results in
sampling errors, thereby leading to a potentially inaccurate
diagnosis of heterogeneously distributed liver diseases (84). Therefore, a non-invasive diagnostic
method is required for accurate and safe assessment of the extent
of liver disease and fibrosis. MicroRNAs are involved in regulating
several biological and pathological processes in hepatocytes, and
their aberrant expression is associated with different liver
pathologies. For example, the serum levels of miR-138 and miR-143
are characteristic of liver fibrosis in its later stages (85). In addition, the expression profile
of microRNAs is specific to different etiologies of liver disease
in both adults and children (86).
Therefore, microRNAs may be used as potential biomarkers for the
diagnosis of liver diseases in children.
Biliary atresia (BA) refers to the complete fibrous
obstruction of a part of or entire extrahepatic bile ducts, and it
is the most common cause of cholestasis in newborns (87,88).
Owing to a poor prognosis, its diagnosis and treatment are
challenging, with 70% of children with BA requiring a liver
transplant for long-term survival (89). Mice with biliary obstruction
exhibit upregulated expression of microRNA-let7a-5p (a 4-fold
increase in expression compared with control mice), which is
associated with the expression of adenosine triphosphate-binding
cassette subfamily C member 2 (ABCC2). ABCC2 is needed for the
biliary excretion of numerous endogenous and heterogeneous
compounds and promotes bile flow independently of bile acids
(90). Upregulation of
microRNA-155 enhances pro-inflammatory activity via activating
JAK2/STAT3 and suppressing cytokine signaling 1, whereas its
downregulation reduces the incidence of BA (91). A study on mouse models of rhesus
rotavirus infection-induced BA demonstrated that microRNA-222
regulates fibrosis by targeting protein phosphatase 2 regulatory
subunit B-α (PPP2R2A); notably, microRNA-222 inhibits PPP2R2A
dephosphorylation and promotes Akt activation (25). In addition, clinical trials have
demonstrated that aberrant expression levels of microRNA-214,
microRNA-19b, microRNA-222 and microRNA-21 are strongly associated
with liver fibrosis in patients with BA (92-94).
Single-nucleotide polymorphisms in microRNAs have been reported to
affect the development and prognosis of various diseases. In
particular, polymorphisms in microRNA-100 (rs1834306 A>G),
microRNA-499 (rs3746444 A>G), microRNA-492 (rs2289030 G>C)
and microRNA-938 (rs2505901 T>C) may contribute to BA
susceptibility (95-97).
A clinical trial has demonstrated that circulating
levels of microRNAs are higher in patients with cystic fibrosis
without liver disease (CFLD; n=30) compared with in those with
liver disease (n=52). In addition, reverse
transcription-quantitative PCR has been used to test healthy
children and children with CFLD (n=20). The results revealed that
the combination of serum microRNA-122, microRNA-21 and microRNA-25
is clinically relevant for the early diagnosis of CFLD, whereas the
combination of serum microRNA-210, microRNA-148a and microRNA-19a
facilitates the early diagnosis of liver fibrosis non-invasively
(98).
MicroRNAs play an important role in the pathogenesis
of asthma by regulating inflammation. For example, childhood asthma
has been associated with the downregulation of let-7 microRNA
family members and upregulation of microRNA-155, microRNA-21,
microRNA-146a/b, microRNA-142-3p, microRNA-223 and microRNA-142-5p
(107,108). Let-7 is a highly conserved
microRNA family that is most abundantly expressed in the lungs.
Reduced levels of let-7 microRNA family members have been reported
in the ovalbumin-sensitized mouse model (109). Let-7 microRNA family members play
a pro-inflammatory role in asthma by inhibiting the secretion of
interleukin-13(110).
MicroRNAs have been demonstrated to regulate airway
remodeling in mouse models. Ras homolog family member A (RhoA)
participates in airway remodeling by regulating the differentiation
of mesenchymal stem cells. MicroRNA-133a decreases the expression
of RhoA, leading to the shrinkage of bronchial smooth muscle cells
(111).
Epilepsy is a chronic condition characterized by
abnormal neural discharge that leads to transient malfunctions in
the brain (114). It is
associated with sudden, spontaneously terminating, recurrent
motor-sensory, mental and consciousness disorders (115). Epilepsy is the most common
neurological disorder in children characterized by a persistent
predisposition to developing seizures (116). Medical advancements (such as
genetic testing, electroencephalography and neuroimaging) have
improved the diagnosis, treatment and quality of life of children
with epilepsy (114). However,
delayed diagnosis and treatment may affect their health
adversely.
A study on hippocampal sections revealed that
temporal lobe epilepsy is associated with the increased expression
of microRNA-135a-5p. Similarly, the expression of microRNA-135a-5p
is upregulated in epileptiform discharges of neonatal rat
hippocampal neurons. In addition, inhibition of caspase activity
and apoptosis inhibitor 1 expression demonstrates that
microRNA-135a-5p promotes apoptosis in the epileptic temporal lobe,
thereby reducing cell survival (116).
In another study including 63 patients with temporal
lobe epilepsy (mean age, 9.81±2.79 years), serum analysis revealed
significantly reduced expression of microRNA-15a-5p. In addition, a
primary hippocampal cell culture (without magnesium) from newborn
rats was used to mimic temporal lobe epilepsy in children, and the
results showed that overexpression of microRNA-15a-5p could
attenuate temporal lobe epilepsy-induced reductions in cell
viability, and could reversed the cell apoptosis induced by
temporal lobe epilepsy. This finding indicates that microRNA-15a-5p
may serve as a highly specific and sensitive biomarker for the
diagnosis of temporal lobe epilepsy in children (43).
Previous studies have reported that microRNAs are
involved in the pathophysiology of epilepsy and represent an
advanced tool for developing diagnostic and therapeutic strategies
that are more effective and less invasive compared with traditional
clinical strategies (drug treatment and ketogenic diet) (117-119).
However, to the best of our knowledge, studies using microRNAs as
diagnostic biomarkers for epilepsy, especially in children, are
limited. An in-depth understanding of the role of microRNAs in
early-stage epilepsy may guide the development of more rapid and
accurate diagnostic strategies, as well as more effective
prevention and therapeutic strategies for improving the quality of
life of children with epilepsy.
The primary goal of pediatricians is to ensure the
healthy and safe growth of children; however, children often
develop chronic diseases, which are difficult to prevent and treat
(120). MicroRNAs play a notable
role in chronic diseases in children (121-124).
To develop and classify microRNAs as effective biomarkers, further
research is warranted to gain an in-depth understanding of the
mechanisms and functional significance of various microRNAs in
chronic diseases in children. Identification of disease-specific
microRNAs and their target genes may guide the development of novel
therapeutic strategies. Altogether, microRNAs serve as promising
biomarkers for the diagnosis and treatment of chronic diseases in
children (Fig. 1).
Not applicable.
Funding: No funding was received.
Not applicable.
MZ and YH reviewed the literature and wrote the
manuscript. Both authors have read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Perrin JM, Bloom SR and Gortmaker SL: The
increase of childhood chronic conditions in the United States.
JAMA. 297:2755–2759. 2007.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Janse AJ, Uiterwaal CSPM, Gemke RJBJ,
Kimpen JLL and Sinnema G: A difference in perception of quality of
life in chronically ill children was found between parents and
pediatricians. J Clin Epidemiol. 58:495–502. 2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Omran A, Elimam D and Yin F: MicroRNAs:
New insights into chronic childhood diseases. Biomed Res Int.
2013(291826)2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Dong H, Lei J, Ding L, Wen Y, Ju H and
Zhang X: MicroRNA: Function, detection, and bioanalysis. Chem Rev.
113:6207–6233. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mohr AM and Mott JL: Overview of MicroRNA
biology. Semin Liver Dis. 35:3–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wu Y, Li Q, Zhang R, Dai X, Chen W and
Xing D: Circulating microRNAs: Biomarkers of disease. Clin Chim
Acta. 516:46–54. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Calderari S, Diawara MR, Garaud A and
Gauguier D: Biological roles of microRNAs in the control of insulin
secretion and action. Physiol Genomics. 49:1–10. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Romakina VV, Zhirov IV, Nasonova SN,
Zaseeva AV, Kochetov AG, Liang OV and Tereshchenko SN: MicroRNAs as
biomarkers of cardiovascular diseases. Kardiologiya. 58:66–71.
2018.PubMed/NCBI View Article : Google Scholar : (In Russian).
|
|
10
|
Ward BP, Tsongalis GJ and Kaur P:
MicroRNAs in chronic lymphocytic leukemia. Exp Mol Pathol.
90:173–178. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cao RY, Li Q, Miao Y, Zhang Y, Yuan W, Fan
L, Liu G, Mi Q and Yang J: The emerging role of MicroRNA-155 in
cardiovascular diseases. Biomed Res Int.
2016(9869208)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lukiw WJ: microRNA-146a signaling in
Alzheimer's disease (AD) and prion disease (PrD). Front Neurol.
11(462)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fernandez-Valverde SL, Taft RJ and Mattick
JS: MicroRNAs in β-cell biology, insulin resistance, diabetes and
its complications. Diabetes. 60:1825–1831. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang K: The ubiquitous existence of
MicroRNA in body fluids. Clin Chem. 63:784–785. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sims EK, Lakhter AJ, Anderson-Baucum E,
Kono T, Tong X and Evans-Molina C: MicroRNA 21 targets BCL2 mRNA to
increase apoptosis in rat and human beta cells. Diabetologia.
60:1057–1065. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Nielsen LB, Wang C, Sørensen K,
Bang-Berthelsen CH, Hansen L, Andersen ML, Hougaard P, Juul A,
Zhang CY, Pociot F and Mortensen HB: Circulating levels of microRNA
from children with newly diagnosed type 1 diabetes and healthy
controls: Evidence that miR-25 associates to residual beta-cell
function and glycaemic control during disease progression. Exp
Diabetes Res. 2012:896362. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Erener S, Mojibian M, Fox JK, Denroche HC
and Kieffer TJ: Circulating miR-375 as a biomarker of β-cell death
and diabetes in mice. Endocrinology. 154:603–608. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Osipova J, Fischer DC, Dangwal S, Volkmann
I, Widera C, Schwarz K, Lorenzen JM, Schreiver C, Jacoby U,
Heimhalt M, et al: Diabetes-associated microRNAs in pediatric
patients with type 1 diabetes mellitus: a cross-sectional cohort
study. J Clin Endocrinol Metab. 99:E1661–E1665. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu X, Wang Y, Sun Z, Ren S, Yang W, Deng
Y, Tian C, Yu Y and Gao B: Molecular expression and functional
analysis of genes in children with temporal lobe epilepsy. J Integr
Neurosci. 18:71–77. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Han S, Wang WJ, Duan L, Hou ZL, Zeng JY,
Li L, Meng MY, Zhang YY, Wang Y, Xie YH, et al: MicroRNA profiling
of patients with sporadic atrial septal defect. Biotechnol
Biotechnol Equip. 33:510–519. 2019.
|
|
21
|
Sánchez-Gómez MC, García-Mejía KA,
Pérez-Díaz Conti M, Díaz-Rosas G, Palma-Lara I, Sánchez-Urbina R,
Klünder-Klünder M, Botello-Flores JA, Balderrábano-Saucedo NA and
Contreras-Ramos A: MicroRNAs association in the cardiac hypertrophy
secondary to complex congenital heart disease in children. Pediatr
Cardiol. 38:991–1003. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Xiao Y, Wang J, Chen Y, Zhou K, Wen J,
Wang Y, Zhou Y, Pan W and Cai W: Up-regulation of miR-200b in
biliary atresia patients accelerates proliferation and migration of
hepatic stallate cells by activating PI3K/Akt signaling. Cell
Signal. 26:925–932. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shen W, Chen G, Dong R, Zhao R and Zheng
S: MicroRNA-21/PTEN/Akt axis in the fibrogenesis of biliary
atresia. J Pediatr Surg. 49:1738–1741. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hand NJ, Horner AM, Master ZR, Boateng LA,
LeGuen C, Uvaydova M and Friedman JR: MicroRNA profiling identifies
miR-29 as a regulator of disease-associated pathways in
experimental biliary atresia. J Pediatr Gastroenterol Nutr.
54:186–192. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shen WJ, Dong R, Chen G and Zheng S:
microRNA-222 modulates liver fibrosis in a murine model of biliary
atresia. Biochem Biophys Res Commun. 446:155–159. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xiao Y, Wang J, Yan W, Zhou Y, Chen Y,
Zhou K, Wen J, Wang Y and Cai W: Dysregulated miR-124 and miR-200
expression contribute to cholangiocyte proliferation in the
cholestatic liver by targeting IL-6/STAT3 signalling. J Hepatol.
62:889–896. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yu DS, An FM, Gong BD, Xiang XG, Lin LY,
Wang H and Xie Q: The regulatory role of microRNA-1187 in
TNF-α-mediated hepatocyte apoptosis in acute liver failure. Int J
Mol Med. 29:663–668. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
An F, Gong B, Wang H, Yu D, Zhao G, Lin L,
Tang W, Yu H, Bao S and Xie Q: miR-15b and miR-16 regulate TNF
mediated hepatocyte apoptosis via BCL2 in acute liver failure.
Apoptosis. 17:702–716. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Salehi S, Brereton HC, Arno MJ, Darling D,
Quaglia A, O'Grady J, Heaton N and Aluvihare VR: Human liver
regeneration is characterized by the coordinated expression of
distinct microRNA governing cell cycle fate. Am J Transplant.
13:1282–1295. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dattaroy D, Pourhoseini S, Das S, Alhasson
F, Seth RK, Nagarkatti M, Michelotti GA, Diehl AM and Chatterjee S:
Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via
leptin-mediated NADPH oxidase in experimental and human
nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver
Physiol. 308:G298–G312. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Israelow B, Mullokandov G, Agudo J,
Sourisseau M, Bashir A, Maldonado AY, Dar AC, Brown BD and Evans
MJ: Hepatitis C virus genetics affects miR-122 requirements and
response to miR-122 inhibitors. Nat Commun. 5(5408)2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Karam RA and Abd Elrahman DM: Differential
expression of miR-155 and Let-7a in the plasma of childhood asthma:
Potential biomarkers for diagnosis and severity. Clin Biochem.
68:30–36. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang Y, Sun E, Li X, Zhang M, Tang Z, He
L and Lv K: miR-155 contributes to Df1-induced asthma by increasing
the proliferative response of Th cells via CTLA-4 downregulation.
Cell Immunol. 314:1–9. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou Y, Yang Q, Xu H, Zhang J, Deng H, Gao
H, Yang J, Zhao D and Liu F: miRNA-221-3p enhances the secretion of
interleukin-4 in mast cells through the phosphatase and tensin
homolog/p38/nuclear factor-kappaB pathway. PLoS One.
11(e0148821)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tsitsiou E, Williams AE, Moschos SA, Patel
K, Rossios C, Jiang X, Adams OD, Macedo P, Booton R, Gibeon D, et
al: Transcriptome analysis shows activation of circulating CD8+ T
cells in patients with severe asthma. J Allergy Clin Immunol.
129:95–103. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kärner J, Wawrzyniak M, Tankov S, Runnel
T, Aints A, Kisand K, Altraja A, Kingo K, Akdis CA, Akdis M and
Rebane A: Increased microRNA-323-3p in IL-22/IL-17-producing T
cells and asthma: A role in the regulation of the TGF-β pathway and
IL-22 production. Allergy. 72:55–65. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu F, Qin HB, Xu B, Zhou H and Zhao DY:
Profiling of miRNAs in pediatric asthma: Upregulation of miRNA-221
and miRNA-485-3p. Mol Med Rep. 6:1178–1182. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tian M, Zhou Y, Jia H, Zhu X and Cui Y:
The clinical significance of changes in the expression levels of
MicroRNA-1 and inflammatory factors in the peripheral blood of
children with acute-stage asthma. Biomed Res Int.
2018(7632487)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liang Y, Feng Y, Wu W, Chang C, Chen D,
Chen S and Zhen G: microRNA-218-5p plays a protective role in
eosinophilic airway inflammation via targeting δ-catenin, a novel
catenin in asthma. Clin Exp Allergy. 50:29–40. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ren L, Zhu R and Li X: Silencing miR-181a
produces neuroprotection against hippocampus neuron cell apoptosis
post-status epilepticus in a rat model and in children with
temporal lobe epilepsy. Genet Mol Res. 15:2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Peng J, Omran A, Ashhab MU, Kong H, Gan N,
He F and Yin F: Expression patterns of miR-124, miR-134, miR-132,
and miR-21 in an immature rat model and children with mesial
temporal lobe epilepsy. J Mol Neurosci. 50:291–297. 2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Risbud RM, Lee C and Porter BE:
Neurotrophin-3 mRNA a putative target of miR21 following status
epilepticus. Brain Res. 1424:53–59. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li N, Pan J, Liu W, Li Y, Li F and Liu M:
MicroRNA-15a-5p serves as a potential biomarker and regulates the
viability and apoptosis of hippocampus neuron in children with
temporal lobe epilepsy. Diagn Pathol. 15(46)2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Achkar NP, Cambiagno DA and Manavella PA:
miRNA biogenesis: A dynamic pathway. Trends Plant Sci.
21:1034–1044. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kandhavelu M and Kandhavelu J: Pre-piRNA
biogenesis mimics the pathway of miRNA. Biochem Syst Ecol.
43:200–204. 2012.
|
|
46
|
Murchison EP and Hannon GJ: miRNAs on the
move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol.
16:223–229. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Saj A and Lai EC: Control of microRNA
biogenesis and transcription by cell signaling pathways. Curr Opin
Genet Dev. 21:504–510. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Suzuki HI and Miyazono K: Dynamics of
microRNA biogenesis: Crosstalk between p53 network and microRNA
processing pathway. J Mol Med (Berl). 88:1085–1094. 2010.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009.PubMed/NCBI View Article : Google Scholar
|
|
50
|
American Diabetes Association. 2.
Classification and diagnosis of diabetes. Diabetes Care. 38 (Suppl
1):S8–S16. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
American Diabetes Association. Diagnosis
and classification of diabetes mellitus. Diabetes Care. 37 (Suppl
1):S81–S90. 2014.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Tuomi T, Santoro N, Caprio S, Cai M, Weng
J and Groop L: The many faces of diabetes: A disease with
increasing heterogeneity. Lancet. 383:1084–1094. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ma RCW: Epidemiology of diabetes and
diabetic complications in China. Diabetologia. 61:1249–1260.
2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Moheet A, Mangia S and Seaquist ER: Impact
of diabetes on cognitive function and brain structure. In: Year in
Diabetes and Obesity. Powers AC and Ahima RS (eds). pp60-71,
2015.
|
|
55
|
Baroukh N, Ravier MA, Loder MK, Hill EV,
Bounacer A, Scharfmann R, Rutter GA and Van Obberghen E:
MicroRNA-124a regulates Foxa2 expression and intracellular
signaling in pancreatic beta-cell lines. J Biol Chem.
282:19575–19588. 2007.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Keller DM, Clark EA and Goodman RH:
Regulation of microRNA-375 by cAMP in pancreatic β-cells. Mol
Endocrinol. 26:989–999. 2012.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Poy MN, Hausser J, Trajkovski M, Braun M,
Collins S, Rorsman P, Zavolan M and Stoffel M: miR-375 maintains
normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci
USA. 106:5813–5818. 2009.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Regazzi R: MicroRNAs as therapeutic
targets for the treatment of diabetes mellitus and its
complications. Expert Opin Ther Targets. 22:153–160.
2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Sun LL, Jiang BG, Li WT, Zou JJ, Shi YQ
and Liu ZM: MicroRNA-15a positively regulates insulin synthesis by
inhibiting uncoupling protein-2 expression. Diabetes Res Clin
Pract. 91:94–100. 2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Shen Z, Yu Y, Yang Y, Xiao X, Sun T, Chang
X, Tang W, Zhu Y and Han X: miR-25 and miR-92b regulate insulin
biosynthesis and pancreatic β-cell apoptosis. Endocrine.
76:526–535. 2022.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Corno AF, Koerner TS and Salazar JD:
Innovative treatments for congenital heart defects. World J
Pediatr. 19:1–6. 2023.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wu Y, Ma XJ, Wang HJ, Li WC, Chen L, Ma D
and Huang GY: Expression of Cx43-related microRNAs in patients with
tetralogy of Fallot. World J Pediatr. 10:138–144. 2014.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Dueñas A, Exposit A, Aranega A and Franco
D: The role of non-coding RNA in congenital heart diseases. J
Cardiovasc Dev Dis. 6(15)2019.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Gu H, Chen L, Xue J, Huang T, Wei X, Liu
D, Ma W, Cao S and Yuan Z: Expression profile of maternal
circulating microRNAs as non-invasive biomarkers for prenatal
diagnosis of congenital heart defects. Biomed Pharmacother.
109:823–830. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Hoelscher SC, Doppler SA, Dreßen M, Lahm
H, Lange R and Krane M: MicroRNAs: Pleiotropic players in
congenital heart disease and regeneration. J Thorac Dis. 9 (Suppl
1):S64–S81. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Kalayinia S, Arjmand F, Maleki M,
Malakootian M and Singh CP: MicroRNAs: Roles in cardiovascular
development and disease. Cardiovasc Pathol.
50(107296)2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Arabian M, Mirzadeh Azad F, Maleki M and
Malakootian M: Insights into role of microRNAs in cardiac
development, cardiac diseases, and developing novel therapies. Iran
J Basic Med Sci. 23:961–969. 2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Paul S, Ruiz-Manriquez LM, Ledesma-Pacheco
SJ, Benavides-Aguilar JA, Torres-Copado A, Morales-Rodríguez JI, De
Donato M and Srivastava A: Roles of microRNAs in chronic pediatric
diseases and their use as potential biomarkers: A review. Arch
Biochem Biophys. 699(108763)2021.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Smith T, Rajakaruna C, Caputo M and
Emanueli C: MicroRNAs in congenital heart disease. Ann Transl Med.
3(333)2015.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Tian J, An X and Niu L: Role of microRNAs
in cardiac development and disease. Exp Ther Med. 13:3–8.
2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Xie WQ, Zhou L, Chen Y and Ni B:
Circulating microRNAs as potential biomarkers for diagnosis of
congenital heart defects. World J Emerg Med. 7:85–89.
2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Yan HL and Hua YM: Research advances on
role of microRNAs in congenital heart diseases. Zhongguo Dang Dai
Er Ke Za Zhi. 16:1070–1074. 2014.PubMed/NCBI(In Chinese).
|
|
74
|
Zajkowska A and Małecki M: microRNAs role
in heart development. Postepy Biol Komorki. 42:107–126. 2015.
|
|
75
|
Wei Y, Peng S, Wu M, Sachidanandam R, Tu
Z, Zhang S, Falce C, Sobie EA, Lebeche D and Zhao Y: Multifaceted
roles of miR-1s in repressing the fetal gene program in the heart.
Cell Res. 24:278–292. 2014.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Xin M, Olson EN and Bassel-Duby R: Mending
broken hearts: Cardiac development as a basis for adult heart
regeneration and repair. Nat Rev Mol Cell Biol. 14:529–541.
2013.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Bruneau BG: The developmental genetics of
congenital heart disease. Nature. 451:943–948. 2008.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Ho PTB, Clark IM and Le LTT:
MicroRNA-based diagnosis and therapy. Int J Mol Sci.
23(7167)2022.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Yu Z, Han S, Hu P, Zhu C, Wang X, Qian L
and Guo X: Potential role of maternal serum microRNAs as a
biomarker for fetal congenital heart defects. Med Hypotheses.
76:424–426. 2011.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Zhu S, Cao L, Zhu J, Kong L, Jin J, Qian
L, Zhu C, Hu X, Li M, Guo X, et al: Identification of maternal
serum microRNAs as novel non-invasive biomarkers for prenatal
detection of fetal congenital heart defects. Clin Chim Acta.
424:66–72. 2013.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Della Corte C, Mosca A, Vania A, Alterio
A, Alisi A and Nobili V: Pediatric liver diseases: Current
challenges and future perspectives. Expert Rev Gastroenterol
Hepatol. 10:255–265. 2016.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Calvopina DA, Coleman MA, Lewindon PJ and
Ramm GA: Function and regulation of MicroRNAs and their potential
as biomarkers in paediatric liver disease. Int J Mol Sci.
17(1795)2016.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Makri E, Goulas A and Polyzos SA:
Epidemiology, pathogenesis, diagnosis and emerging treatment of
nonalcoholic fatty liver disease. Arch Med Res. 52:25–37.
2021.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Gheorghe G, Bungău S, Ceobanu G, Ilie M,
Bacalbaşa N, Bratu OG, Vesa CM, Găman MA and Diaconu CC: The
non-invasive assessment of hepatic fibrosis. J Formos Med Assoc.
120:794–803. 2021.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Tadokoro T, Morishita A and Masaki T:
Diagnosis and therapeutic management of liver fibrosis by MicroRNA.
Int J Mol Sci. 22(8139)2021.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wang X, He Y, Mackowiak B and Gao B:
MicroRNAs as regulators, biomarkers and therapeutic targets in
liver diseases. Gut. 70:784–795. 2021.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Hartley JL, Davenport M and Kelly DA:
Biliary atresia. Lancet. 374:1704–1713. 2009.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Bezerra JA, Wells RG, Mack CL, Karpen SJ,
Hoofnagle JH, Doo E and Sokol RJ: Biliary atresia: Clinical and
research challenges for the twenty-first century. Hepatology.
68:1163–1173. 2018.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Sokol RJ, Shepherd RW, Superina R, Bezerra
JA, Robuck P and Hoofnagle JH: Screening and outcomes in biliary
atresia: Summary of a national institutes of health workshop.
Hepatology. 46:566–581. 2007.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Balasubramaniyan N, Devereaux MW, Orlicky
DJ, Sokol RJ and Suchy FJ: Up-regulation of miR-let7a-5p leads to
decreased expression of ABCC2 in obstructive cholestasis. Hepatol
Commun. 3:1674–1686. 2019.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Zhao R, Dong R, Yang Y, Wang Y, Ma J, Wang
J, Li H and Zheng S: MicroRNA-155 modulates bile duct inflammation
by targeting the suppressor of cytokine signaling 1 in biliary
atresia. Pediatr Res. 82:1007–1016. 2017.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Dong R, Zheng Y, Chen G, Zhao R, Zhou Z
and Zheng S: miR-222 overexpression may contribute to liver
fibrosis in biliary atresia by targeting PPP2R2A. J Pediatr
Gastroenterol Nutr. 60:84–90. 2015.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Zhao D, Luo Y, Xia Y, Zhang JJ and Xia Q:
MicroRNA-19b expression in human biliary atresia specimens and its
role in BA-related fibrosis. Dig Dis Sci. 62:689–698.
2017.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Yoneyama T, Ueno T, Masahata K, Toyama C,
Maeda A, Tazuke Y, Oue T, Miyagawa S and Okuyama H: Elevation of
microRNA-214 is associated with progression of liver fibrosis in
patients with biliary atresia. Pediatr Surg Int. 38:115–122.
2022.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Chang J, Liang J, Chai C, Liu F, Tao B,
Wang H, Tong Y, Wang Z and Xia H: MiR-100 rs1834306 A>G
increases biliary atresia risk in Southern Han Chinese children.
Biomed Res Int. 2023(4835839)2023.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Su L, Tian Y, Fu M, Zhang RZ, Ou XF, Xia
HM and Li RZ: Association of miRNA-492 rs2289030 G>C and
miRNA-938 rs2505901 T>C gene polymorphisms with biliary atresia
susceptibility. Biomed Environ Sci. 34:577–580. 2021.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Shan Y, Shen N, Han L, Chen Q, Zhang J,
Long X and Xia Q: MicroRNA-499 Rs3746444 polymorphism and biliary
atresia. Dig Liver Dis. 48:423–428. 2016.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Cook NL, Pereira TN, Lewindon PJ, Shepherd
RW and Ramm GA: Circulating microRNAs as noninvasive diagnostic
biomarkers of liver disease in children with cystic fibrosis. J
Pediatr Gastroenterol Nutr. 60:247–254. 2015.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Boonpiyathad T, Sözener ZC, Satitsuksanoa
P and Akdis CA: Immunologic mechanisms in asthma. Semin Immunol.
46(101333)2019.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Papi A, Brightling C, Pedersen SE and
Reddel HK: Asthma. Lancet. 391:783–800. 2018.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Liang J, Liu XH, Chen XM, Song XL, Li W
and Huang Y: Emerging roles of non-coding RNAs in childhood asthma.
Front Pharmacol. 13(856104)2022.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Kai W, Qian XU and Qun WUZ: MicroRNAs and
asthma regulation. Iran J Allergy Asthma Immunol. 14:120–125.
2015.PubMed/NCBI
|
|
103
|
Midyat L, Gulen F, Karaca E, Ozkinay F,
Tanac R, Demir E, Cogulu O, Aslan A, Ozkinay C, Onay H and Atasever
M: MicroRNA expression profiling in children with different asthma
phenotypes. Pediatr Pulmonol. 51:582–587. 2016.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Mousavi SR, Ahmadi A, Jamalkandi SA and
Salimian J: Involvement of microRNAs in physiological and
pathological processes in asthma. J Cell Physiol. 234:21547–21559.
2019.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Jat KR and Kabra SK: Awareness about
childhood asthma. Indian J Med Res. 145:581–583. 2017.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Foster PS, Maltby S, Rosenberg HF, Tay HL,
Hogan SP, Collison AM, Yang M, Kaiko GE, Hansbro PM, Kumar RK and
Mattes J: Modeling TH 2 responses and airway
inflammation to understand fundamental mechanisms regulating the
pathogenesis of asthma. Immunol Rev. 278:20–40. 2017.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Specjalski K and Niedoszytko M: MicroRNAs:
Future biomarkers and targets of therapy in asthma? Curr Opin Pulm
Med. 26:285–292. 2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
van den Berge M and Tasena H: Role of
microRNAs and exosomes in asthma. Curr Opin Pulm Med. 25:87–93.
2019.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Cañas JA, Rodrigo-Muñoz JM, Sastre B,
Gil-Martinez M, Redondo N and Del Pozo V: MicroRNAs as potential
regulators of immune response networks in asthma and chronic
obstructive pulmonary disease. Front Immunol.
11(608666)2021.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Xu L, Yi M, Tan Y, Yi Z and Zhang Y: A
comprehensive analysis of microRNAs as diagnostic biomarkers for
asthma. Ther Adv Respir Dis. 14(1753466620981863)2020.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Chiba Y: Non-coding RNAs and bronchial
smooth muscle hyperresponsiveness in allergic bronchial asthma.
Nihon Yakurigaku Zasshi. 155:364–368. 2020.PubMed/NCBI View Article : Google Scholar : (In Japanese).
|
|
112
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4(307)2013.PubMed/NCBI View Article : Google Scholar
|
|
113
|
He X, Jing Z and Cheng G: MicroRNAs: New
regulators of Toll-like receptor signalling pathways. Biomed Res
Int. 2014(945169)2014.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Katyayan A and Diaz-Medina G: Epilepsy:
Epileptic syndromes and treatment. Neurol Clin. 39:779–795.
2021.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Paul S, Reyes PR, Garza BS and Sharma A:
MicroRNAs and child neuropsychiatric disorders: A brief review.
Neurochem Res. 45:232–240. 2020.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Ma Y: The challenge of microRNA as a
biomarker of epilepsy. Curr Neuropharmacol. 16:37–42.
2018.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Brennan GP and Henshall DC: microRNAs in
the pathophysiology of epilepsy. Neurosci Lett. 667:47–52.
2018.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Cava C, Manna I, Gambardella A, Bertoli G
and Castiglioni I: Potential role of miRNAs as theranostic
biomarkers of epilepsy. Mol Ther Nucleic Acids. 13:275–290.
2018.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Ghafouri-Fard S, Hussen BM, Abak A, Taheri
M and Jalili Khoshnoud R: Aberrant expression of miRNAs in
epilepsy. Mol Biol Rep. 49:5057–5074. 2022.PubMed/NCBI View Article : Google Scholar
|
|
120
|
van der Lee JH, Mokkink LB, Grootenhuis
MA, Heymans HS and Offringa M: Definitions and measurement of
chronic health conditions in childhood: A systematic review. JAMA.
297:2741–2751. 2007.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Groot M and Lee H: Sorting mechanisms for
MicroRNAs into extracellular vesicles and their associated
diseases. Cells. 9(1044)2020.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Laggerbauer B and Engelhardt S: MicroRNAs
as therapeutic targets in cardiovascular disease. J Clin Invest.
132(e159179)2022.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Vegter EL, van der Meer P, de Windt LJ,
Pinto YM and Voors AA: MicroRNAs in heart failure: From biomarker
to target for therapy. Eur J Heart Fail. 18:457–468.
2016.PubMed/NCBI View Article : Google Scholar
|