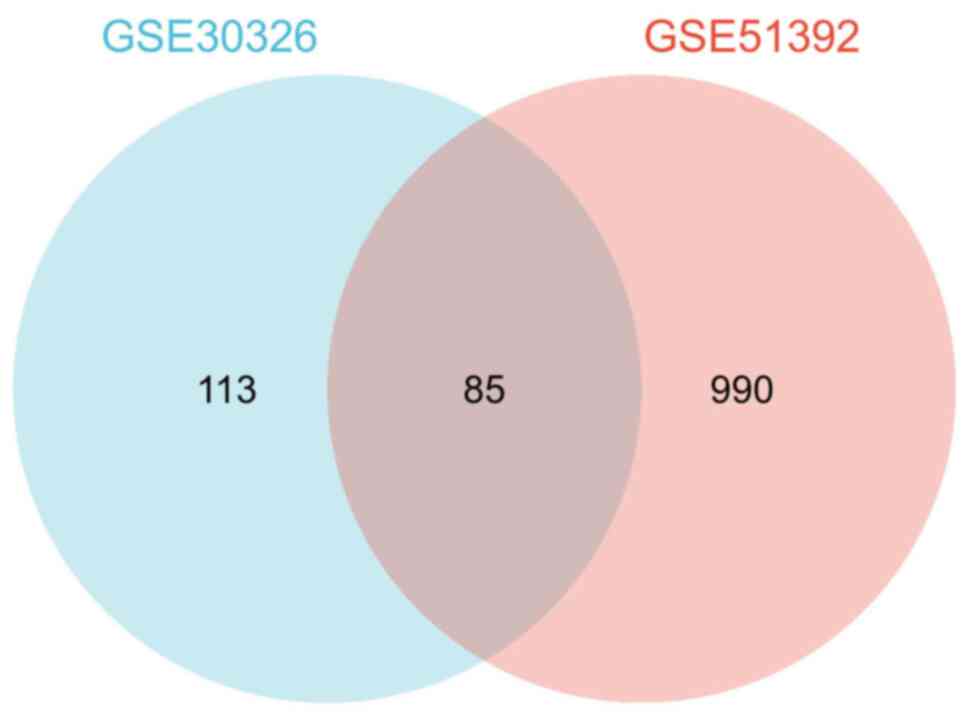
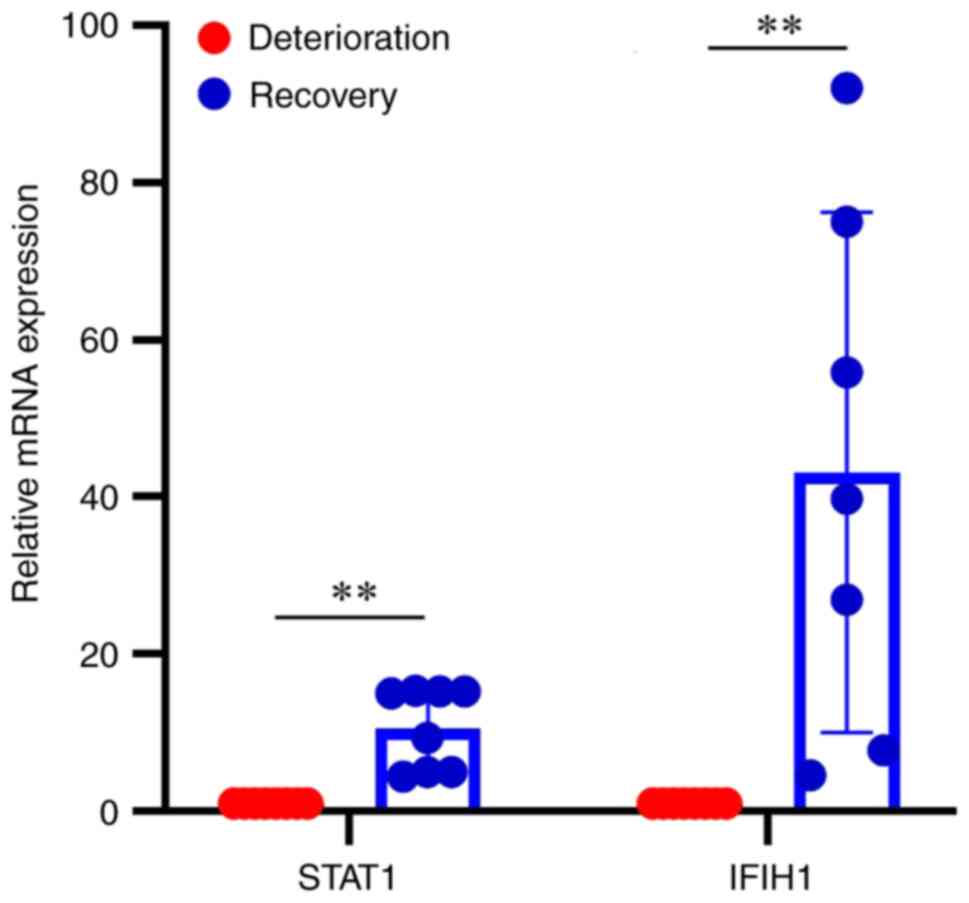
|
1
|
Helou DG, Shafiei-Jahani P, Lo R, Howard
E, Hurrell BP, Galle-Treger L, Painter JD, Lewis G, Soroosh P,
Sharpe AH and Akbari O: PD-1 pathway regulates ILC2 metabolism and
PD-1 agonist treatment ameliorates airway hyperreactivity. Nat
Commun. 11(3998)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cho JL, Ling MF, Adams DC, Faustino L,
Islam SA, Afshar R, Griffith JW, Harris RS, Ng A, Radicioni G, et
al: Allergic asthma is distinguished by sensitivity of
allergen-specific CD4+ T cells and airway structural cells to type
2 inflammation. Sci Transl Med. 8(359ra132)2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhu Z, Lee PH, Chaffin MD, Chung W, Loh
PR, Lu Q, Christiani DC and Liang L: A genome-wide cross-trait
analysis from UK Biobank highlights the shared genetic architecture
of asthma and allergic diseases. Nat Genet. 50:857–864.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Garcia-Marcos L, Asher MI, Pearce N,
Ellwood E, Bissell K, Chiang CY, El Sony A, Ellwood P, Marks GB,
Mortimer K, et al: The burden of asthma, hay fever and eczema in
children in 25 countries: GAN Phase I study. Eur Respir J.
60(2102866)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mortimer K, Reddel HK, Pitrez PM and
Bateman ED: Asthma management in low and middle income countries:
Case for change. Eur Respir J. 60(2103179)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Toussaint M, Jackson DJ, Swieboda D,
Guedan A, Tsourouktsoglou TD, Ching YM, Radermecker C, Makrinioti
H, Aniscenko J, Bartlett NW, et al: Host DNA released by NETosis
promotes rhinovirus-induced type-2 allergic asthma exacerbation.
Nat Med. 23:681–691. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Michi AN, Love ME and Proud D:
Rhinovirus-Induced modulation of epithelial phenotype: Role in
Asthma. Viruses. 12(1328)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Han M, Rajput C, Hinde JL, Wu Q, Lei J,
Ishikawa T, Bentley JK and Hershenson MB: Construction of a
recombinant rhinovirus accommodating fluorescent marker expression.
Influenza Other Respir Viruses. 12:717–727. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Niespodziana K, Stenberg-Hammar K,
Megremis S, Cabauatan CR, Napora-Wijata K, Vacal PC, Gallerano D,
Lupinek C, Ebner D, Schlederer T, et al: PreDicta chip-based high
resolution diagnosis of rhinovirus-induced wheeze. Nat Commun.
9(2382)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bochkov YA, Watters K, Ashraf S, Griggs
TF, Devries MK, Jackson DJ, Palmenberg AC and Gern JE:
Cadherin-related family member 3, a childhood asthma susceptibility
gene product, mediates rhinovirus C binding and replication. P Natl
Acad Sci USA. 112:5485–5490. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mehta AK, Doherty T, Broide D and Croft M:
Tumor necrosis factor family member LIGHT acts with IL-1β and TGF-β
to promote airway remodeling during rhinovirus infection. Allergy.
73:1415–1424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang Q, Nagarkar DR, Bowman ER, Schneider
D, Gosangi B, Lei J, Zhao Y, McHenry CL, Burgens RV, Miller DJ, et
al: Role of double-stranded RNA pattern recognition receptors in
rhinovirus-induced airway epithelial cell responses. J Immunol.
183:6989–6997. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ganjian H, Rajput C, Elzoheiry M and
Sajjan U: Rhinovirus and innate immune function of airway
epithelium. Front Cell Infect Microbiol. 10(277)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bosco A, Ehteshami S, Panyala S and
Martinez FD: Interferon regulatory factor 7 is a major hub
connecting interferon-mediated responses in virus-induced asthma
exacerbations in vivo. J Allergy Clin Immunol. 129:88–94.
2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wagener AH, Zwinderman AH, Luiten S,
Fokkens WJ, Bel EH, Sterk PJ and van Drunen CM: dsRNA-induced
changes in gene expression profiles of primary nasal and bronchial
epithelial cells from patients with asthma, rhinitis and controls.
Respir Res. 15(9)2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhu X, Tang LP, Mao J, Hameed Y, Zhang J,
Li N, Wu D, Huang Y and Li C: Decoding the Mechanism behind the
Pathogenesis of the Focal Segmental Glomerulosclerosis. Comput Math
Method Med. 2022(1941038)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wesolowska-Andersen A, Everman JL,
Davidson R, Rios C, Herrin R, Eng C, Janssen WJ, Liu AH, Oh SS,
Kumar R, et al: Dual RNA-seq reveals viral infections in asthmatic
children without respiratory illness which are associated with
changes in the airway transcriptome. Genome Biol.
18(12)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Thompson EA, Sayers BC, Glista-Baker EE,
Shipkowski KA, Ihrie MD, Duke KS, Taylor AJ and Bonner JC: Role of
signal transducer and activator of transcription 1 in murine
allergen-induced airway remodeling and exacerbation by carbon
nanotubes. Am J Respir Cell Mol Biol. 53:625–636. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kang YH, Biswas A, Field M and Snapper SB:
STAT1 signaling shields T cells from NK cell-mediated cytotoxicity.
Nat Commun. 10(912)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jewell NA, Cline T, Mertz SE, Smirnov SV,
Flano E, Schindler C, Grieves JL, Durbin RK, Kotenko SV and Durbin
JE: Lambda interferon is the predominant interferon induced by
influenza A virus infection in vivo. J Virol. 84:11515–11522.
2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Luu K, Greenhill CJ, Majoros A, Decker T,
Jenkins BJ and Mansell A: STAT1 plays a role in TLR signal
transduction and inflammatory responses. Immunol Cell Biol.
92:761–769. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Matsuyama T, Kubli SP, Yoshinaga SK,
Pfeffer K and Mak TW: An aberrant STAT pathway is central to
COVID-19. Cell Death Differ. 27:3209–3225. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
LeMessurier KS, Rooney R, Ghoneim HE, Liu
BM, Li K, Smallwood HS and Samarainghe AE: Influenza A virus
directly modulates mouse eosinophil responses. J Leukoc Biol.
108:151–168. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mu X and Hur S: Immunogenicity of In
Vitro-Transcribed RNA. Acc Chem Res. 54:4012–4023. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yan K, Zhu W, Yu L, Li N, Zhang X, Liu P,
Chen Q, Chen Y and Han D: Toll-like receptor 3 and RIG-I-like
receptor activation induces innate antiviral responses in mouse
ovarian granulosa cells. Mol Cell Endocrinol. 372:73–85.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Amado-Rodriguez L, Salgado Del Riego E,
Gomez de Ona J, López Alonso I, Gil-Pena H, Lopez-Martinez C,
Martin-Vicente P, Lopez-Vazquez A, Gonzalez Lopez A, Cuesta-Llavona
E, et al: Effects of IFIH1 rs1990760 variants on systemic
inflammation and outcome in critically ill COVID-19 patients in an
observational translational study. Elife. 11(e73012)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dieter C, de Almeida Brondani L, Lemos NE,
Schaeffer AF, Zanotto C, Ramos DT, Girardi E, Pellenz FM, Camargo
JL, Moresco KS, et al: Polymorphisms in ACE1, TMPRSS2, IFIH1,
IFNAR2, and TYK2 genes are associated with worse clinical outcomes
in COVID-19. Genes (Basel). 14(29)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang P, Yang L, Cheng G, Yang G, Xu Z, You
F, Sun Q, Lin RT, Fikrig E and Sutton RE: UBXN1 interferes with
Rig-I-like receptor-mediated antiviral immune response by targeting
MAVS. Cell Rep. 3:1057–1070. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pugh C, Kolaczkowski O, Manny A,
Korithoski B and Kolaczkowski B: Resurrecting ancestral structural
dynamics of an antiviral immune receptor: Adaptive binding pocket
reorganization repeatedly shifts RNA preference. BMC Evol Biol.
16(241)2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Loske J, Rohmel J, Lukassen S, Stricker S,
Magalhães VG, Liebig J, Chua RL, Thürmann L, Messingschlager M,
Seegebarth A, et al: Pre-activated antiviral innate immunity in the
upper airways controls early SARS-CoV-2 infection in children. Nat
Biotechnol. 40:319–324. 2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Selinger C, Tisoncik-Go J, Menachery VD,
Agnihothram S, Law GL, Chang J, Kelly SM, Sova P, Baric RS and
Katze MG: Cytokine systems approach demonstrates differences in
innate and pro-inflammatory host responses between genetically
distinct MERS-CoV isolates. BMC Genomics. 15(1161)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Rachmiel M, Bloch O, Bistritzer T,
Weintrob N, Ofan R, Koren-Morag N and Rapoport MJ: TH1/TH2 cytokine
balance in patients with both type 1 diabetes mellitus and asthma.
Cytokine. 34:170–176. 2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhu J, Message SD, Mallia P, Kebadze T,
Contoli M, Ward CK, Barnathan ES, Mascelli MA, Kon OM, Papi A, et
al: Bronchial mucosal IFN-α/β and pattern recognition receptor
expression in patients with experimental rhinovirus-induced asthma
exacerbations. J Allergy Clin Immunol. 143:114–125 e4.
2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gill MA, Bajwa G, George TA, Dong CC,
Dougherty II, Jiang N, Gan VN and Gruchalla RS: Counterregulation
between the FcepsilonRI pathway and antiviral responses in human
plasmacytoid dendritic cells. J Immunol. 184:5999–6006.
2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu T, Zhou YT, Wang LQ, Li LY, Bao Q,
Tian S, Chen MX, Chen HX, Cui J and Li CW: NOD-like receptor
family, pyrin domain containing 3 (NLRP3) contributes to
inflammation, pyroptosis, and mucin production in human airway
epithelium on rhinovirus infection. J Allergy Clin Immunol.
144:777–787 e9. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Grünvogel O, Esser-Nobis K, Reustle A,
Schult P, Müller B, Metz P, Trippler M, Windisch MP, Frese M,
Binder M, et al: DDX60L is an interferon-stimulated gene product
restricting hepatitis C virus replication in cell culture. J Virol.
89:10548–10568. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wolin SL and Maquat LE: Cellular RNA
surveillance in health and disease. Science. 366:822–827.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kane M, Zang TM, Rihn SJ, Zhang FW, Kueck
T, Alim M, Schoggins J, Rice CM, Wilson SJ and Bieniasz PD:
Identification of interferon-stimulated genes with antiretroviral
activity. Cell Host Microbe. 20:392–405. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cohen JI: GATA2 deficiency and
epstein-barr virus disease. Front Immunol. 8(1869)2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Spinner MA, Sanchez LA, Hsu AP, Shaw PA,
Zerbe CS, Calvo KR, Arthur DC, Gu W, Gould CM, Brewer CC, et al:
GATA2 deficiency: A protean disorder of hematopoiesis, lymphatics,
and immunity. Blood. 123:809–821. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhao X, Fan H, Chen X, Zhao X, Wang X,
Feng YJ, Liu M, Li S and Tang H: Hepatitis B Virus DNA polymerase
restrains viral replication through the CREB1/HOXA distal
transcript antisense RNA Homeobox A13 Axis. Hepatology. 73:503–519.
2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Jenkins DE, Sreenivasan D, Carman F, Samal
B, Eiden LE and Bunn SJ: Interleukin-6-mediated signaling in
adrenal medullary chromaffin cells. J Neurochem. 139:1138–1150.
2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhang Y, Le X, Zheng S, Zhang K, He J, Liu
M, Tu C, Rao W, Du H, Ouyang Y, et al: MicroRNA-146a-5p-modified
human umbilical cord mesenchymal stem cells enhance protection
against diabetic nephropathy in rats through facilitating M2
macrophage polarization. Stem Cell Res Ther. 13(171)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xie LM, Yin X, Bi J, Luo HM, Cao XJ, Ma
YW, Liu YL, Su JW, Lin GL and Guo XG: Identification of potential
biomarkers in dengue via integrated bioinformatic analysis. PLoS
Negl Trop Dis. 15(e0009633)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen Y, Li S, Wei Y, Xu Z and Wu X:
Circ-RNF13, as an oncogene, regulates malignant progression of
HBV-associated hepatocellular carcinoma cells and HBV infection
through ceRNA pathway of circ-RNF13/miR-424-5p/TGIF2. Bosn J Basic
Med Sci. 21:555–568. 2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wei G, Yi C, Ziyun J, Hui L, Hui L and
Yilong X: Network-Based analysis of the genetic effects of
SARS-CoV-2 infection to patients with exacerbation of Virus-Induced
Asthma (VAE). Research Square: https://doi.org/10.21203/rs.3.rs-948407/v1.
|