Introduction
Calcific aortic valve disease (CAVD) is the most
common cardiovascular disease (1).
As longevity increases and lifestyles adapt, the incidence of CAVD
has risen notably, which could lead to aortic stenosis and heart
failure (2). Currently, early
surgery remains the most effective approach for improving the
prognosis of patients with CAVD. Previous studies have revealed
that the pathological process of CAVD is complex and is associated
with multiple factors, including lipoprotein deposition,
inflammatory response and calcific nodule formation (3,4).
Therefore, it is necessary to further understand the pathological
process of CAVD and discover its unique biomarkers and potential
therapeutic pathways to improve the prognosis of patients with
CAVD. Autophagy, a cellular process that maintains homeostasis via
degrading and recycling proteins and organelles in lysosomes, is
involved in various cellular functions, including metabolic
regulation, immune response, apoptosis and differentiation
(5-8).
This biological process is involved in multiple diseases, including
neurodegenerative diseases (9),
cancer (10), cardiovascular
diseases (11) and metabolic
disorders (12). Autophagy serves
as a ‘regulator’ that maintains cardiovascular redox homeostasis
and preserves cardiovascular functionality via the complex
relationship between oxidant-dependent signaling and autophagic
flux. Peng et al (13) have
reported that atorvastatin bolsters vulnerable atherosclerotic
plaque stability and diminishes aortic lesion sizes by amplifying
autophagy, which in turn mitigates inflammatory cytokine secretion
and lipid accumulation. Similarly, spermidine, an endogenous,
naturally occurring polyamine, significantly reduces necrotic core
areas and lipid accumulation by inducing autophagy to stimulate
cholesterol efflux from vascular smooth muscle cells (VSMCs)
(14). These autophagy-driven
effects underscore the potential of autophagy induction in
cardiovascular disease prevention (15). Additionally, research has
demonstrated that in instances of chronic heart failure following
myocardial infarction, increased levels of matrix metalloproteinase
9 (MMP9) can suppress cardiac autophagic activity, subsequently
exacerbating myocardial ischemia-induced chronic heart failure
(16). Furthermore, according to
Wang et al (17), protein
kinase AMP-activated catalytic subunit α2 (AMPKα2) overexpression
in mouse hearts could blunt the progression of chronic heart
failure by amplifying mitochondrial autophagy and improving
mitochondrial functionality. These studies collectively illustrate
the profound influence of autophagy on cardiovascular health and
disease progression.
Interestingly, previous studies have highlighted the
key role of the RNA-binding protein Sam68 in driving human valve
interstitial cell osteogenic differentiation via the STAT3
signaling pathway and autophagic flux regulation (18). In addition, autophagy upregulation
has been observed in the calcified tissues of patients, showing
that autophagy plays compensatory and pro-survival roles in this
disease (19). Overall, this
compelling evidence emphasizes the promising potential for
identifying autophagy-related targets as therapeutic approaches for
CAVD.
In the present study, a multifaceted approach to
explore the molecular mechanism involved in autophagy in CAVD was
utilized. Microarray data were collected and analyzed to identify
autophagy-related genes, and further enrichment analyses were
conducted to understand their roles and interactions. Key genes
were verified using an independent dataset and quantitative
reverse-transcription quantitative PCR (RT-qPCR) analyses of
patient samples, and their potential as drug targets was examined.
Finally, immune infiltration in response to CAVD was studied to
understand its role in disease progression.
Materials and methods
Microarray data collection
A total of three microarray datasets GSE12644 (10
control and 10 CAVD samples) (20), GSE51472 (5 control and 5 CAVD
samples) (21) and GSE77287 (3
control and 3 CAVD samples) were obtained from the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) (22). Then, mRNA expression profiles
(GSE12644 and GSE51472) were applied to screen for differentially
expressed genes (DEGs) and differentially expressed
autophagy-related genes (DEARGs); GSE77287 was used for validation
of hub genes. When multiple probes corresponded to a gene, the
average expression value was used as the gene expression value
after the probe IDs were converted into gene symbols. The
microarray data from 3 datasets were normalized and log2
transformed.
DEG and DEARG identification
The limma package (version 3.48.1) in R software
(version 4.1; https://www.R-project.org/) was used to identify DEGs
between normal and CAVD samples; thresholds of |log2FC|≥1 and
P<0.05 were adjusted by the Benjamini and Hochberg method. In
addition, autophagy-related genes (ARGs) were extracted from three
widely used autophagy-related databases: The Human Autophagy
Modulator Database (http://hamdb.scbdd.com) (23), the Human Autophagy Database
(http://www.autophagy.lu/index.html)
(24), and the Autophagy Database
(http://www.tanpaku.org/autophagy/index.html) (25). Ultimately, 1167 ARGs were obtained
after the deduplication of genes, and these are listed in Table SI. Overlapping target DEARGs
between the DEGs and ARGs were identified by using the VennDiagram
package (version 1.6.19) in R software (26).
Gene set enrichment analysis
(GSEA)
As a computational method for interpreting gene
expression data based on a molecular signature database, GSEA was
performed using GSEA software (http://software.broadinstitute.org/gsea/index.jsp) to
explore the overall relationship between ARGs and CAVD. The ARG set
contained 1,167 ARGs, and the core genes and enrichment score (ES)
were calculated using the score and rank of genes in GSE12644 and
GSE51472. In addition, |NES|>1.0 and normal P<0.05 were
considered significant in the present study.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of
DEARGs
GO and KEGG analyses were performed with the DEARGs
identified from GSE12644 and GSE51472 by using the Database for
Annotation, Visualization and Integrated Discovery (DAVID)
(http://david.ncifcrf.gov/) to explore
the biological characteristics and signaling pathway related to
DEARGs. A P<0.05 was applied as the cutoff criterion.
Construction of a protein-protein
interaction (PPI) network and identification of hub genes and key
modules
In the present study, the STRING (27) database (http://string-db.org/) and Cytoscape software (version
3.8.2; https://cytoscape.org/) were used to
construct and visualize a PPI network of DEARGs. The hub genes were
screened using Cytohubba, a Cytoscape plugin based on the MCC
algorithm (16). Each DEARG in the
PPI network was assigned a value, and the genes were ranked
accordingly. The top 10 genes were identified as hub genes.
Validation of hub genes in
GSE77287
The independent microarray dataset GSE77287 was
utilized to validate the hub genes. After ID conversion with the
data normalized and log2 transformed, the expression values of 10
hub genes were obtained and compared between normal and CAVD groups
using the Wilcoxon test. Results with P<0.05 were considered
statistically significant.
Immune infiltration analyses
To explore the relationship between the immune
response and CAVD, normalized gene expression data from GSE12644
and GSE51472 were entered in HiPlot software (version 0.1.2;
http://hiplot.com.cn/). The CIBERSORT algorithm
was used to calculate the proportion of infiltrating immune cells,
and a t-test was used to compare between normal and CAVD groups.
Results with P<0.05 were considered statistically
significant.
Potential drug prediction of hub
genes
The DsigDB (28)
database (http://dsigdb.tanlab.org/), which
contains >20,000 gene sets and 17,389 unique compounds covering
19,531 genes, was used to examine potential target drugs associated
with the hub genes by comparing the differences in gene expression
between control and drug-treated groups included in DSigDB database
using the unpaired two-tailed Student's t-test. An FDR<0.05 and
a combined score >2x106 were used as the thresholds
for potential drug selection.
RT-qPCR
In the present study, aortic valves gathered from 3
patients with CAVD were considered the CAVD group, whereas aortic
valves harvested from 3 patients with acute aortic dissection
without calcification were considered the control group. All of the
aortic valves were collected by surgery. The six adult patients (4
men and 2 women, all aged between 41-62 years) included in the
present study had undergone valvular surgery or acute aortic
dissection repair surgery between September 2022 and May 2023 in
the Second Affiliated Hospital of Nanchang University (Nanchang,
China). Patients with any of the following comorbidities were not
be involved in the present study: i) Serious coronary heart
disease, heart tumor, congenital heart disease, or infective
endocarditis; ii) thyroid disease; iii) organ failure, cancer,
infection or autoimmune disease; iv) serious psychiatric or
neurological illness. Detailed information on patients involved in
the present study is listed in Table
SII. Total RNA was extracted from the aortic valve tissues
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed into cDNA using RevertAid
MM (Thermo Fisher Scientific, Inc.) after the RNA quality and yield
were evaluated. The ratio of 260/280 nm, which reflects RNA
quality, was required to be 1.8-2.0. RT was conducted as follows:
42˚C for 1 h, then 72˚C for 10 min. A Light Cycler 480 Real-Time
PCR system (Roche Diagnostics) and TB Green Premix Ex Taq (Takara
Bio, Inc.) were used to identify gene expression levels. The qPCR
process consisted of denaturation at 98˚C for 2 min, accompanied by
thermocycling at 98˚C for 10 sec, 60˚C for 15 sec, and 68˚C for 1
min, repeated for 40 cycles. ACTB was used as an internal control
to normalize relative expressions of mRNA using the
2-ΔΔCq method (29).
The sequences of the primers (Sangon Biotech Co., Ltd.) used are
included in Table SIII. Written
formal consent was acquired from all patients, and the present
study was approved (approval no. SYXK2021-0073) by the Ethics
Committee of the Second Affiliated Hospital of Nanchang University
(Nanchang, China). The experimental protocol was established
according to the ethical guidelines of the Helsinki Declaration and
its revision from October 2013.
Statistical analysis
The statistical analyses involved in the present
study were conducted by the Sangerbox platform or GraphPad Prism
6.0 (Dotmatics) and data are expressed as the mean ± standard
derivation (all experiments were repeated three times). In
addition, comparisons between two groups were made using the
unpaired two-tailed Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
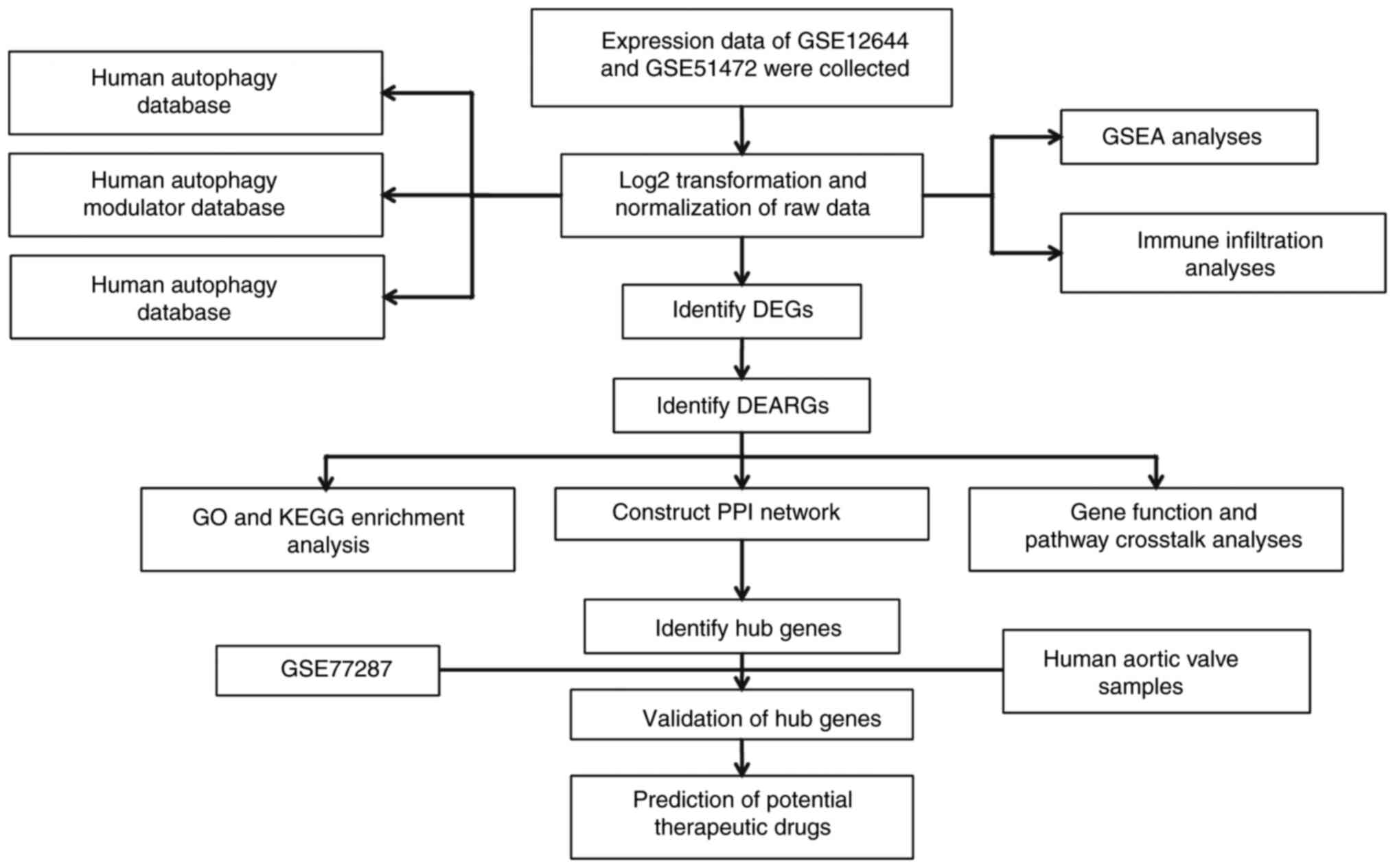
Overall design of the study
The overall flowchart of the study is shown in
Fig. 1.
Relationship between autophagy and
CAVD
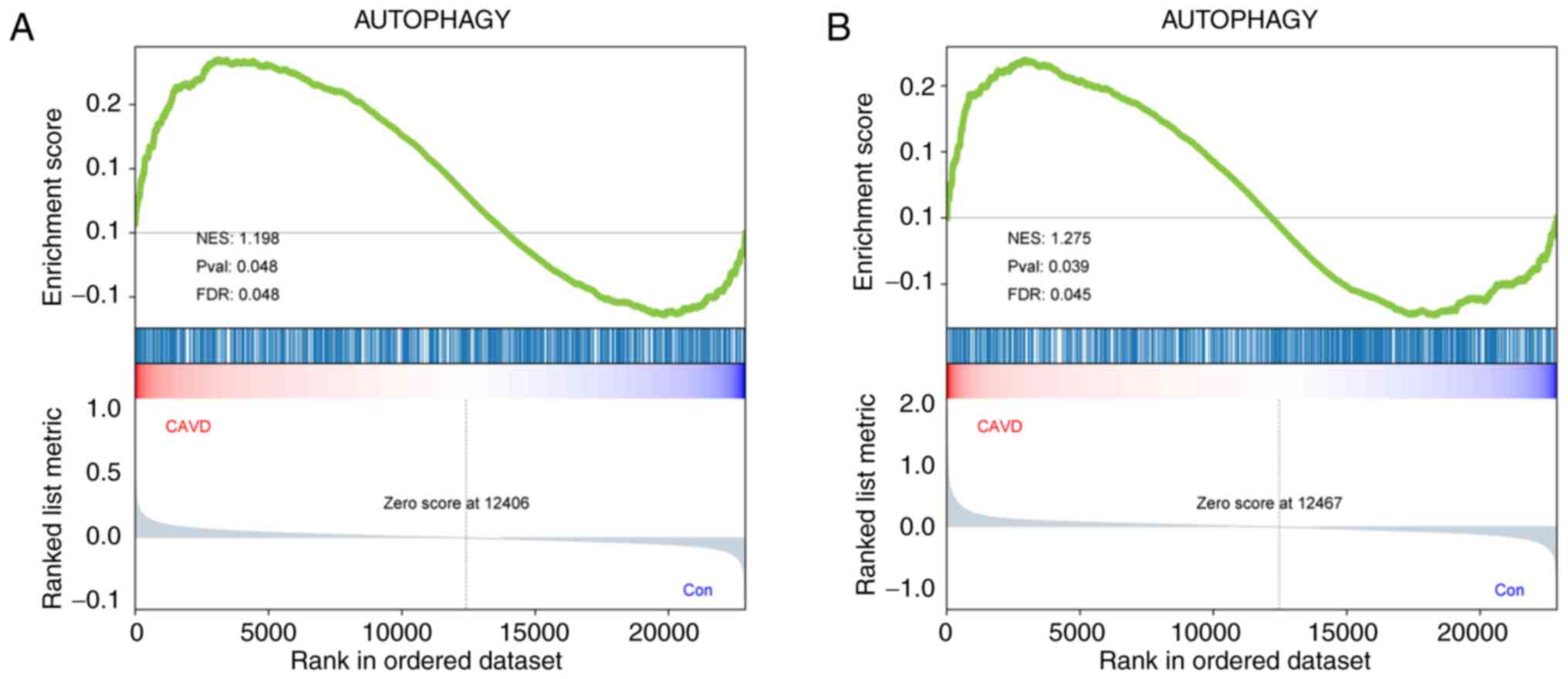
GSEA is a well-known, powerful analytical method for
identifying pathways related to gene expression data. Therefore, a
GSEA was conducted on an ARG dataset to explore the role of the
autophagy pathway in the pathological process of CAVD. The ARG set
was significantly enriched in the CAVD samples of both microarray
datasets, with a nominal P<0.05 and |NES|>1.0; these ARGs
were predominantly upregulated (Fig.
2A and B). This finding
indicated a substantial and significant relationship between
autophagy and CAVD. In addition, all 339 core genes in GSE12644 and
GSE51472 were identified by GSEA.
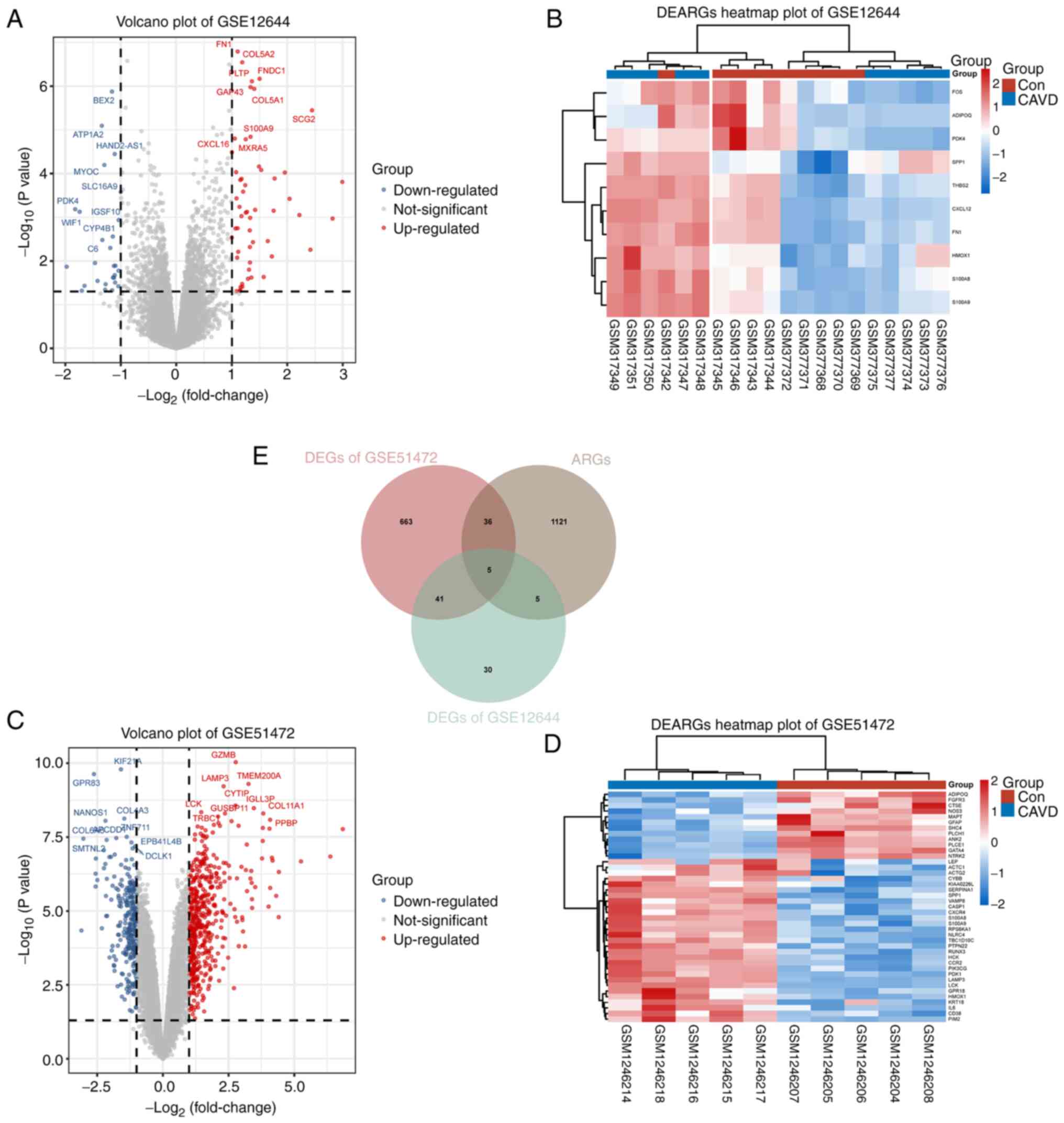
DEG and DEARG identification
A total of 827 DEGs were identified from GSE12644
and GSE51472 using default thresholds (|log2FC|>1.0 and
P<0.05) (Fig. 3A and C). These DEGs included 526 upregulated
genes and 300 downregulated genes. Additionally, after obtaining
1,167 ARGs from multiple databases, 46 DEARGs were identified for
further analysis by overlapping the ARGs with the DEGs from
GSE12644 and GSE51472 (Fig. 3E).
The expression differences of these 46 DEARGs in normal and CAVD
groups were visualized in a clustered heatmap using the Ward.D2
algorithm (Fig. 3B and D). These results indicated that the
DEARGs that were screened out may play a crucial role in CAVD
development by regulating autophagy.
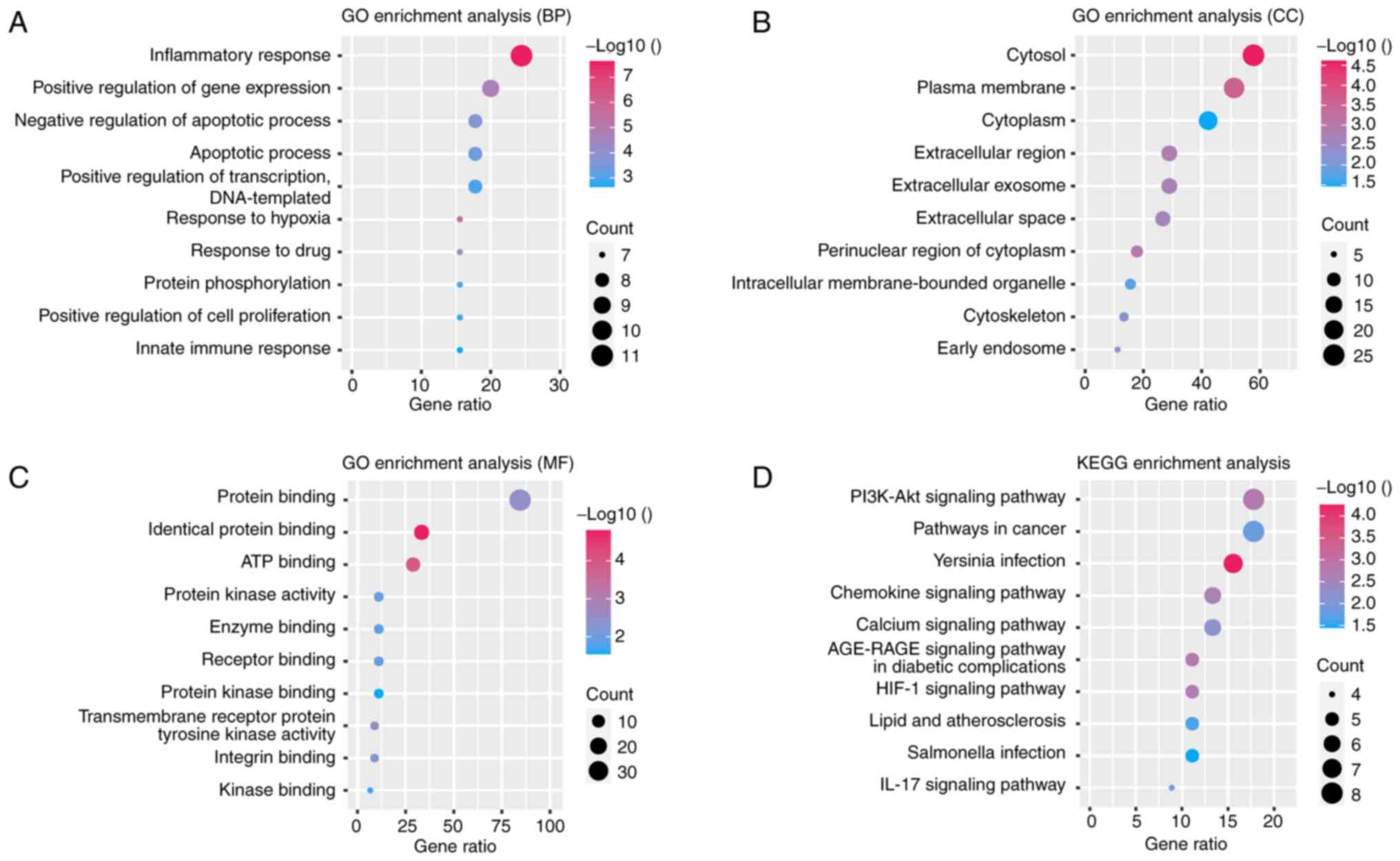
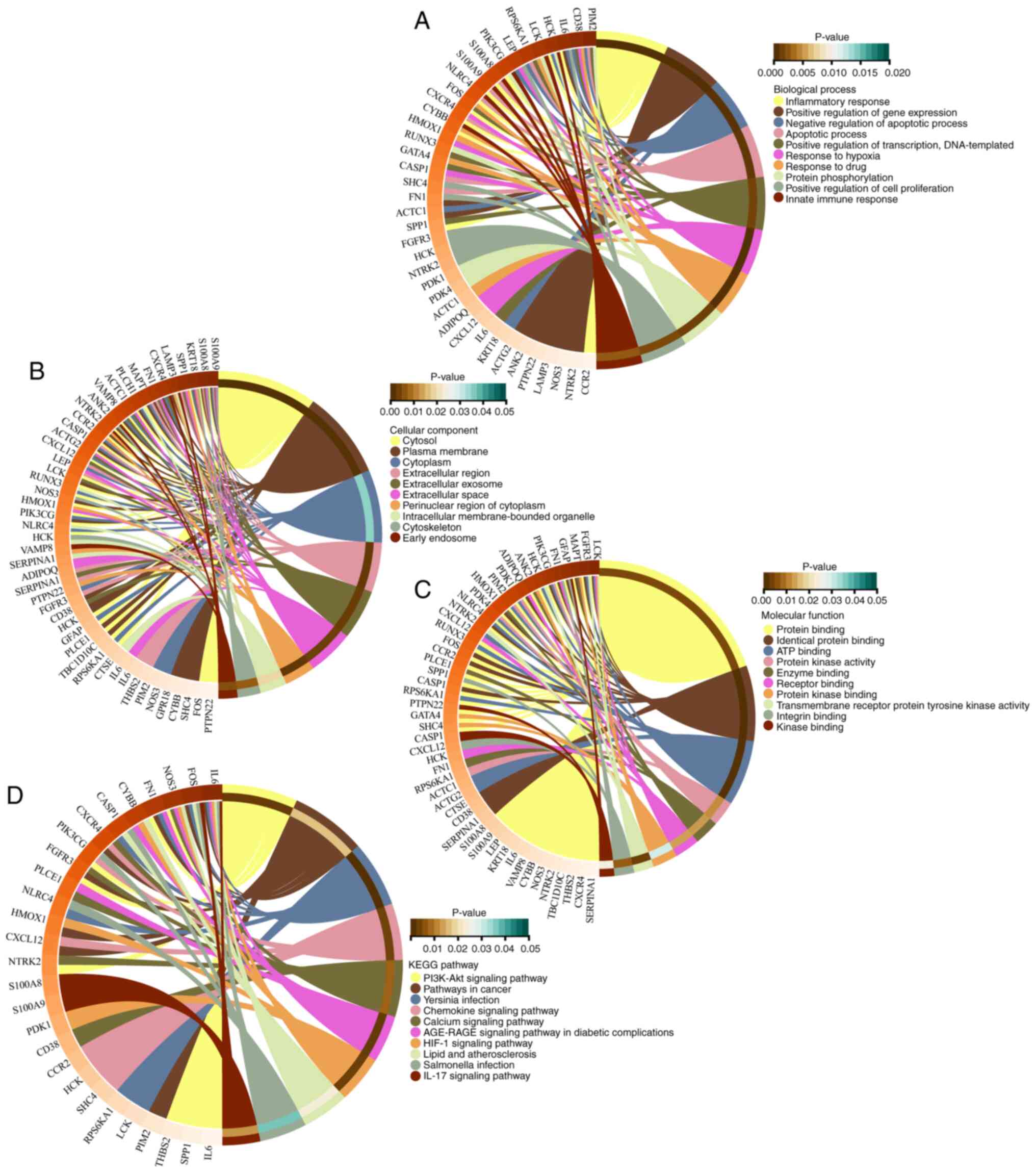
GO and KEGG pathway enrichment
analysis
GO and KEGG pathway enrichment analyses were
conducted to further understand the functions and related pathways
of the DEARGs. As shown in Fig.
4A-D, the GO enrichment analysis indicated that DEARGs in the
biological process category were enriched primarily in the
inflammatory response, positive regulation of gene expression and
negative regulation of apoptotic processes. In the cellular
component category, the DEARGs were enriched mainly in the cytosol,
plasma membrane and cytoplasm. For the molecular function category,
protein binding, identical protein binding and ATP binding were
predominantly enriched. KEGG pathway analysis results revealed that
the DEARGs participated mainly in the PI3K-Akt signaling pathway,
pathways in cancer and Yersinia infection. Furthermore, the
interaction between genes and different functions or pathways was
explored. As demonstrated in Fig.
5A-D, the role of the DEARGs in CAVD regulation is complex and
involves multiple gene functions and pathways. Through both GO and
KEGG pathway enrichment analyses, it is evident that the role of
DEARGs in CAVD regulation is intricate, encompassing various gene
functions and pathways.
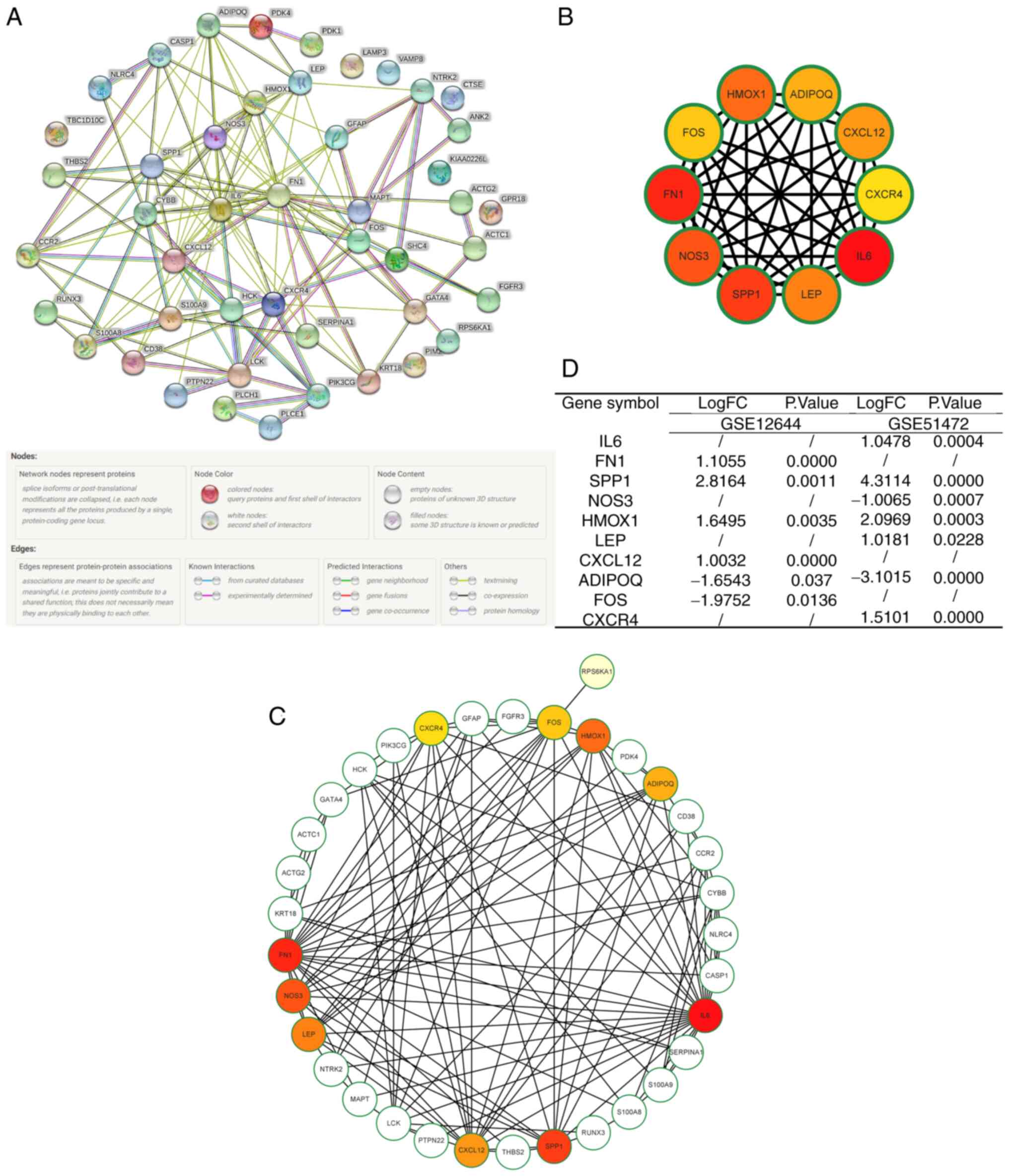
Construction of a PPI network and
identification of hub genes and key modules
A PPI network was constructed to further investigate
the interactions of the DEARGs by the STRING database (Fig. 6A). Additionally, Cytoscape software
was utilized to quantify the data. Based on the MCC algorithm, the
top 10 genes were identified: IL6, FN1, SPP1, NOS3, HMOX1, LEP,
CXCL12, ADIPOQ, FOS and CXCR4 (Fig.
6B and C). Moreover, the
numerous relationships between the hub genes and other DEARGs are
visualized in Fig. 6D.
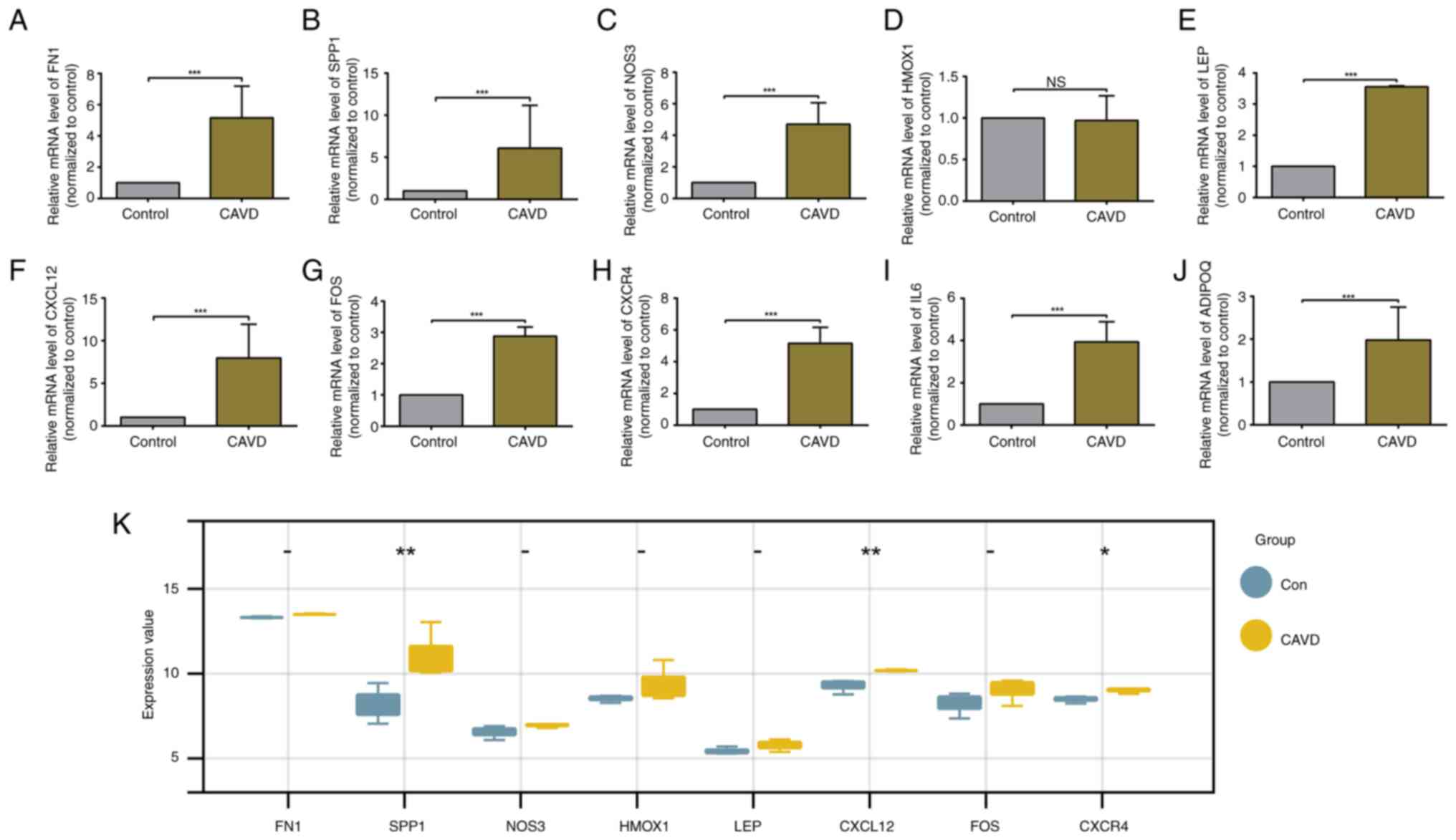
Validation of hub gene expression in
calcified aortic valve samples
In the present study, the expression levels of hub
genes in control and CAVD groups were validated. As revealed in
Fig. 7A-J, the RT-qPCR results
suggested that the hub genes, except for HMOX1, were significantly
upregulated in the calcified aortic valve samples; these particular
genes were thus identified as key genes. The aforementioned results
indicated that the hub genes that were screened out may participate
in the development of CAVD by regulating autophagy.
Validation of hub gene expression
Another independent dataset, GSE77287, was chosen to
validate the key genes. Upon pre-processing the data extracted from
GSE77287, expression data for the 9 key genes were analysed. Three
of the nine selected hub genes (SPP1, CXCL12 and CXCR4) were
remarkably upregulated in CAVD samples, which is consistent with
the GSE12644 and GSE51472 results (Fig. 7K).
Immune infiltration analyses
Recently, an association between the immune response
and CAVD occurrence has been demonstrated (30). An immune infiltration analysis
using CAVD samples to further understand the interaction between
DEARGs and the immune response was consequently performed. As
demonstrated in Fig. 8A and
B, the CIBERSORT algorithm was
used to evaluate the proportions of 22 immune cell types in the
aortic valve samples from GSE12644 and GSE51472. Further
statistical analyses indicated that activated NK cells were
significantly upregulated in the normal aortic valve samples of
both datasets, while M0 macrophages were significantly
downregulated in both datasets (Fig.
8C and D). Additionally, there
were significant differences in the infiltration scores of normal
and calcified aortic valve samples calculated by ssGSEA for both
datasets, GSE51472 (P<0.05) and GSE12644 (P<0.05), suggesting
that immune infiltration plays a significant role in the
pathological process of CAVD. Further research is needed to
investigate the regulatory effects of DEARGs on immune infiltration
in CAVD.
Prediction of potential therapeutic
drugs
The DSigDB database was employed to predict
potential therapeutic drugs related to the hub genes. This
important database includes detailed drug data and comprehensive
information on drug targets and drug actions. A total of 1,599
potential autophagy-related drugs were ultimately identified and
ranked by their combined score. The top 10 predicted drugs are
displayed in Fig. 9A. The top four
drugs, which exhibited strong drug-target associations (adjusted
P<0.0001, combined score >2x106) are as follows:
bisphenol A (Fig. 9B, combined
score=5,183,719.14), resveratrol (Fig.
9C, combined score=4,650,762.31), progesterone (Fig. 9D, combined score=4,246,664.85) and
estradiol (Fig. 9E, combined
score=156640) (Fig. 9F).
Discussion
The present explored the role of autophagy in the
pathological process of CAVD. The main findings are as follows: i)
The GSEA revealed a significant relationship between autophagy and
CAVD, with upregulated ARGs observed in CAVD samples; ii) GO and
KEGG pathway analyses shed light on the functional roles of DEARGs,
including their involvement in the inflammatory response, gene
regulation and apoptotic processes, as well as their participation
in signaling pathways such as PI3K-Akt and pathways in cancer; iii)
the construction of a PPI network identified hub genes, including
IL6, FN1, SPP1, NOS3, HMOX1, LEP, CXCL12, ADIPOQ, FOS and CXCR4,
which play crucial roles in CAVD progression by regulating
autophagy. The expression of these hub genes was validated in
another dataset and calcified aortic valve tissues; iv)
furthermore, immune infiltration analysis highlighted the
involvement of immune cells, such as activated NK cells and
macrophages, in CAVD pathogenesis; and v) the present study also
identified potential therapeutic drugs for CAVD that target
autophagy-related hub genes, including bisphenol A, resveratrol,
progesterone and estradiol.
It is very well established that valve calcification
is a complicated biological process affected by multiple factors,
including lipoprotein deposition, inflammatory responses,
renin-angiotensin-aldosterone system activation, extracellular
matrix remodelling and cytoskeleton changes. This multifactorial
process leads to the destruction of valve-specific cells, such as
endothelial and mesenchymal cells, which causes valve
calcification, valve orifice stenosis and left ventricular outflow
tract obstruction (31-33).
Although numerous of the factors contributing to
valve calcification are understood, the role of autophagy within
this process remains elusive (18,34).
This significant research gap warrants further investigation. In
the present study, in order to find genes related to autophagy in
CAVD, bioinformatics techniques were leveraged. The aim of the
present study was not only to enhance comprehension of the disease
trajectory but also to pinpoint potential therapeutic candidates.
The results of GSEA indicated a substantial and significant
association between autophagy and CAVD. Notably, the DEARGs that
were identified were mostly upregulated and might be crucial in the
pathology of CAVD via autophagy mediation. These genes and their
role in autophagy could be potential therapeutic targets or
biomarkers for CAVD. The functional enrichment analyses, including
GO and KEGG pathway analyses, provided a functional interpretation
of the DEARGs. The significant enrichment of these genes in
multiple biological processes and pathways, such as the
inflammatory response, gene expression regulation, apoptotic
processes, and, notably, the PI3K-Akt signaling pathway,
underscores their roles in these crucial cellular functions.
Autophagy plays an indispensable role in cellular health and
regulates NLRP3 inflammasome activation. Inflammasome
over-activation, due to autophagy impairment, can result in
inflammatory diseases. Hence, balanced interactions between
autophagy and inflammatory responses are paramount in preventing
cellular damage and disease progression (35). Though distinct in nature, autophagy
and apoptosis are intricately intertwined, collaboratively
maintaining cellular and tissue homeostasis. Autophagy is tasked
with the turnover of protein aggregates and damaged organelles,
while apoptosis is responsible for eliminating redundant cells. The
essential nature of caspases in this interplay cannot be
overstated. They not only instigate apoptotic cell death but also
shape autophagic responses by degrading autophagy proteins and
converting pro-autophagic proteins into pro-apoptotic ones; these
functions highlight the sophistication and significance of
autophagy-apoptosis crosstalk (36). Interestingly, research has pointed
to the potential therapeutic use of PI3K/AKT/mTOR signaling pathway
modulation. In fact, previous studies have demonstrated that
inhibiting this pathway can enhance autophagy activity in rat
articular chondrocytes and mitigate inflammatory responses
(37). This suggests a
counterintuitive negative association between the PI3K/AKT
signaling pathway and autophagy, revealing a new layer of
complexity in these biological interactions.
To further identify the mechanism of autophagy
activation in CAVD, a PPI network of DEARGs was established and
several hub genes were identified, including IL6, FN1, SPP1, NOS3,
HMOX1, LEP, CXCL12, ADIPOQ, FOS and CXCR4. The substantial
connectivity of these genes in the PPI network implies their
pivotal roles in the autophagy pathway and their potential
significance in CAVD. Experimental validation was used to
corroborate the upregulated expression of these pivotal genes in
calcified aortic valve tissue, thus highlighting their potential
involvement in CAVD via regulating autophagy. Of these ARGs, SPP1,
CXCL12 and CXCR4 have been affirmed consistently across multiple
datasets. In an insightful study conducted by Lépine et al
(38), depletion of SPP1, an
enzyme within the endoplasmic reticulum (ER) responsible for
dephosphorylating sphingosine-1-phosphate (S1P), triggered
autophagy induction. This event is closely tied to the regulation
of intracellular S1P levels and ER stress. By maintaining
intracellular S1P homeostasis, SPP1 appears to play a critical role
in modulating the unfolded protein response and ER stress-induced
autophagy. Concurrently, heightened SPP1 expression was revealed to
increase matrix mineralization, indicating its integral role in
managing heart valve tissue mineralization (39). These findings suggested that SPP1
may significantly influence CAVD progression through its regulation
of autophagy. Other research also indicated that defective
autophagy in VSMCs results in accelerated cellular senescence and
upregulation of CXCL12, among other factors. These changes
contribute to post-injury neointima formation and atherosclerosis,
suggesting a complex interaction between CXCL12 and autophagy.
Here, CXCL12 appears to be part of a compensatory response
triggered by impaired autophagy, thus contributing to arterial
disease progression (40). In the
context of vascular health and disease, the intricate relationship
between CXCL12 and autophagy unveils potential avenues for
therapeutic intervention. Furthermore, the CXCR4 receptor exerts a
critical effect on SDF-1α-induced autophagy in chondrocytes by
inhibiting mTOR signaling, a key autophagy regulatory pathway.
Interestingly, CXCR4 deficiency in macrophages is associated with
enhanced autophagy and reduced expression of decorin and
inflammatory cytokines (41). This
result suggested that CXCR4 may modulate autophagy in macrophages,
thereby affecting inflammation and angiogenesis under ischemic
conditions. These findings underscore the intricate roles of SPP1,
CXCL12 and CXCR4 in regulating autophagy and their potential
implications in the pathogenesis of CAVD, highlighting new avenues
for therapeutic interventions that could modulate autophagy and the
associated cellular pathways.
The investigation that occurred in the present study
provides substantial insights into the immune infiltration process
in CAVD. Notably, significant differences in the infiltration
levels of activated NK cells and M0 macrophages were observed
between normal and calcified aortic valve tissues. This compelling
outcome suggests a potential association between the immune
response and CAVD. Furthermore, the DSigDB database was used to
identify potential CAVD therapeutic agents. Bisphenol A,
resveratrol and progesterone stood out among the agents identified.
These drugs had strong drug-target associations, highlighting their
potential to intervene in the disease progression by modulating
autophagic flux. Although bisphenol A is a chemical compound
component of polycarbonate plastics and epoxy resins, it exhibits
dual properties as a potent NADPH oxidase promoter (42) and has a substantial combined score
of 5,183,719 for 3 autophagy-related targets. These characteristics
suggest that bisphenol A may significantly affect autophagic flux,
thus potentially influencing CAVD progression. Additionally, it has
been previously demonstrated that resveratrol could improve the
outcomes of patients with CAV through alleviating inflammation
(43). Furthermore, with its
significant binding affinity (combined score: 4,650,762) for 1
autophagy-related target, resveratrol may also modulate autophagic
flux and potentially affect the biological process of CAVD.
Progesterone treatment is commonly used to support and promote
pregnancy, and its use is inversely related to abdominal aortic
aneurysms (44). Moreover,
progesterone exhibits a combined score of 4,246,664 and is
associated with 3 autophagy-related targets, suggesting its
potential to modulate autophagic flux and potentially affect CAVD
progression. These findings provided a solid foundation for these
drugs as potential therapeutic contenders in CAVD management,
thereby underscoring the need for more in-depth studies to validate
their efficacy.
In general, autophagy has been recognized as one of
the key pathological mechanisms of a number of cardiovascular
diseases, including CAVD, and has received widespread attention
(45,46). The innovations of the present study
are mainly reflected in the following aspects: i) The present study
provided new insights into the role of autophagy in CAVD based on
evidence from the human calcific aortic valve transcriptome; ii)
the role of ARGs in the pathological process of CAVD was identified
and verified by aortic valves obtained from patients; iii) the role
of immune response in the process of CAVD was also explored, and
the results showed significant differences between normal and CAVD
samples; iv) moreover, the present study also predicted potential
therapeutic agents for CAVD.
The limitations of the present study have been
acknowledged. Firstly, although the findings offer valuable
insights into the role of DEARGs in CAVD, these results are based
primarily on bioinformatics analyses and experimental validation
based on human samples and need to be validated with comprehensive
in vitro and in vivo experiments and histological
staining of human samples to fully elucidate their underlying
mechanisms in CAVD. Secondly, although human samples for validation
were utilized, due to the limited number of human samples included
in the present study and the heterogeneity among patients, more
human samples are necessary to improve the interpretation and
reliability of the results that were acquired in the present study.
Hence, further functional studies are needed to apply the findings
of the present study to clinical practice.
In conclusion, three dysregulated DEARGs related to
CAVD were identified through bioinformatics analysis and
experimental validation. Additionally, the relationship between
immune infiltration and CAVD was elucidated. Bisphenol A,
resveratrol and progesterone were predicted as therapeutic drugs
for CAVD by mediating autophagy. The present study examined the
role of autophagy in CAVD based on evidence from human CAVD
transcriptome, which helps to elucidate the molecular mechanisms of
CAVD and to identify new diagnostic and therapeutic targets for
CAVD.
Supplementary Material
Merged autophagy-related gene set
Information of patients involved in
this study.
Primer sequences
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 82070303 and 81860054).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JCL and LJW conceptualized and designed the present
study. YY and FJH collected and assembled the data. TH, YJ and JSY
analysed and interpreted the data. TH performed the experiments.
All authors wrote the manuscript. JCL and LJW confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The experimental protocol was established according
to the ethical guidelines of the Declaration of Helsinki and its
revision from October 2013 and was approved by the Human Ethics
Committee of The Second Affiliated Hospital of Nanchang University
(Nanchang, China; approval no. SYXK2021-0073). Written informed
consent was obtained from the patients prior to enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen Y, Xiao F and Wang R: Calcified
aortic valve disease complicated with and without diabetes
mellitus: The underlying pathogenesis. Rev Cardiovasc Med.
23(7)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lindman BR, Clavel MA, Mathieu P, Iung B,
Lancellotti P, Otto CM and Pibarot P: Calcific aortic stenosis. Nat
Rev Dis Primers. 2(16006)2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kraler S, Blaser MC, Aikawa E, Camici GG
and Lüscher TF: Calcific aortic valve disease: From molecular and
cellular mechanisms to medical therapy. Eur Heart J. 43:683–697.
2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jiang T, Hasan SM, Faluk M and Patel J:
Evolution of transcatheter aortic valve replacement | review of
literature. Curr Probl Cardiol. 46(100600)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kim KH and Lee MS: Autophagy-a key player
in cellular and body metabolism. Nat Rev Endocrinol. 10:322–337.
2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Levine B, Mizushima N and Virgin HW:
Autophagy in immunity and inflammation. Nature. 469:323–335.
2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Maiuri MC, Zalckvar E, Kimchi A and
Kroemer G: Self-eating and self-killing: Crosstalk between
autophagy and apoptosis. Nat Rev Mol Cell Biol. 8:741–752.
2007.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Clarke AJ and Simon AK: Autophagy in the
renewal, differentiation and homeostasis of immune cells. Nat Rev
Immunol. 19:170–183. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nixon RA: The role of autophagy in
neurodegenerative disease. Nat Med. 19:983–997. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Marsh T and Debnath J: Autophagy
suppresses breast cancer metastasis by degrading NBR1. Autophagy.
16:1164–1165. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bravo-San Pedro JM, Kroemer G and Galluzzi
L: Autophagy and mitophagy in cardiovascular disease. Circ Res.
120:1812–1824. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Luciani A, Schumann A, Berquez M, Chen Z,
Nieri D, Failli M, Debaix H, Festa BP, Tokonami N, Raimondi A, et
al: Impaired mitophagy links mitochondrial disease to epithelial
stress in methylmalonyl-CoA mutase deficiency. Nat Commun.
11(970)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Peng S, Xu LW, Che XY, Xiao QQ, Pu J, Shao
Q and He B: Atorvastatin inhibits inflammatory response, attenuates
lipid deposition, and improves the stability of vulnerable
atherosclerotic plaques by modulating autophagy. Front Pharmacol.
9(438)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Michiels CF, Kurdi A, Timmermans JP, De
Meyer GRY and Martinet W: Spermidine reduces lipid accumulation and
necrotic core formation in atherosclerotic plaques via induction of
autophagy. Atherosclerosis. 251:319–327. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chen HY, Xiao ZZ, Ling X, Xu RN, Zhu P and
Zheng SY: ELAVL1 is transcriptionally activated by FOXC1 and
promotes ferroptosis in myocardial ischemia/reperfusion injury by
regulating autophagy. Mol Med. 27(14)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Nandi SS, Katsurada K, Sharma NM, Anderson
DR, Mahata SK and Patel KP: MMP9 inhibition increases autophagic
flux in chronic heart failure. Am J Physiol Heart Circ Physiol.
319:H1414–H1437. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang B, Nie J, Wu L, Hu Y, Wen Z, Dong L,
Zou MH, Chen C and Wang DW: AMPKα2 protects against the development
of heart failure by enhancing mitophagy via PINK1 phosphorylation.
Circ Res. 122:712–729. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu X, Zheng Q, Wang K, Luo J, Wang Z, Li
H, Liu Z, Dong N and Shi J: Sam68 promotes osteogenic
differentiation of aortic valvular interstitial cells by
TNF-α/STAT3/autophagy axis. J Cell Commun Signal. 17:863–879.
2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Carracedo M, Persson O, Saliba-Gustafsson
P, Artiach G, Ehrenborg E, Eriksson P, Franco-Cereceda A and Bäck
M: Upregulated autophagy in calcific aortic valve stenosis confers
protection of valvular interstitial cells. Int J Mol Sci.
20(1486)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bossé Y, Miqdad A, Fournier D, Pépin A,
Pibarot P and Mathieu P: Refining molecular pathways leading to
calcific aortic valve stenosis by studying gene expression profile
of normal and calcified stenotic human aortic valves. Circulation.
Circ Cardiovasc Genet. 2:489–498. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Rysä J: Gene expression profiling of human
calcific aortic valve disease. Genom Data. 7:107–108.
2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang NN, Dong J, Zhang L, Ouyang D, Cheng
Y, Chen AF, Lu AP and Cao DS: HAMdb: A database of human autophagy
modulators with specific pathway and disease information. J
Cheminform. 10(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Moussay E, Kaoma T, Baginska J, Muller A,
Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G and
Janji B: The acquisition of resistance to TNFα in breast cancer
cells is associated with constitutive activation of autophagy as
revealed by a transcriptome analysis using a custom microarray.
Autophagy. 7:760–770. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Homma K, Suzuki K and Sugawara H: The
autophagy database: An all-inclusive information resource on
autophagy that provides nourishment for research. Nucleic Acids
Res. 39:D986–D990. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lin G, Chai J, Yuan S, Mai C, Cai L,
Murphy RW, Zhou W and Luo J: VennPainter: A tool for the comparison
and identification of candidate genes based on venn diagrams. PLoS
One. 11(e0154315)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee
S, Jeon M, Kang J and Tan AC: DSigDB: Drug signatures database for
gene set analysis. Bioinformatics. 31:3069–3071. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Broeders W, Bekkering S, El Messaoudi S,
Joosten LAB, van Royen N and Riksen NP: . Innate immune cells in
the pathophysiology of calcific aortic valve disease: Lessons to be
learned from atherosclerotic cardiovascular disease? Basic Res
Cardiol. 117(28)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mathieu P and Boulanger MC: Basic
mechanisms of calcific aortic valve disease. Can J Cardiol.
30:982–993. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kostyunin AE, Yuzhalin AE, Ovcharenko EA
and Kutikhin AG: Development of calcific aortic valve disease: Do
we know enough for new clinical trials? J Mol Cell Cardiol.
132:189–209. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gould ST, Srigunapalan S, Simmons CA and
Anseth KS: Hemodynamic and cellular response feedback in calcific
aortic valve disease. Circ Res. 113:186–197. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Xie W, Shan Y, Wu Z, Liu N, Yang J, Zhang
H, Sun S, Chi J, Feng W, Lin H and Guo H: Herpud1 deficiency
alleviates homocysteine-induced aortic valve calcification. Cell
Biol Toxicol. 39:2665–2684. 2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Biasizzo M and Kopitar-Jerala N: Interplay
between NLRP3 inflammasome and autophagy. Front Immunol.
11(591803)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wu H, Che X, Zheng Q, Wu A, Pan K, Shao A,
Wu Q, Zhang J and Hong Y: Caspases: A molecular switch node in the
crosstalk between autophagy and apoptosis. Int J Biol Sci.
10:1072–1083. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xue JF, Shi ZM, Zou J and Li XL:
Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of
articular chondrocytes and attenuates inflammatory response in rats
with osteoarthritis. Biomed Pharmacother. 89:1252–1261.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lépine S, Allegood JC, Park M, Dent P,
Milstien S and Spiegel S: Sphingosine-1-phosphate
phosphohydrolase-1 regulates ER stress-induced autophagy. Cell
Death Differ. 18:350–361. 2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Peacock JD, Huk DJ, Ediriweera HN and
Lincoln J: Sox9 transcriptionally represses Spp1 to prevent matrix
mineralization in maturing heart valves and chondrocytes. PLoS One.
6(e26769)2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Grootaert MO, da Costa Martins PA, Bitsch
N, Pintelon I, De Meyer GR, Martinet W and Schrijvers DM: Defective
autophagy in vascular smooth muscle cells accelerates senescence
and promotes neointima formation and atherogenesis. Autophagy.
11:2014–2032. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ma Q, Zhang N, You Y, Zhu J, Yu Z, Chen H,
Xie X and Yu H: CXCR4 blockade in macrophage promotes angiogenesis
in ischemic hindlimb by modulating autophagy. J Mol Cell Cardiol.
169:57–70. 2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Priego AR, Parra EG, Mas S,
Morgado-Pascual JL, Ruiz-Ortega M and Rayego-Mateos S: Bisphenol A
modulates autophagy and exacerbates chronic kidney damage in mice.
Int J Mol Sci. 22(7189)2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Samiei N, Hosseini S, Maleki M, Moradi L,
Joghataei MT and Arabian M: Modulatory role of SIRT1 and resistin
as therapeutic targets in patients with aortic valve stenosis. Arch
Med Res. 50:333–341. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ohlsson C, Langenskiöld M, Smidfelt K,
Poutanen M, Ryberg H, Norlén AK, Nordanstig J, Bergström G and
Tivesten Å: Low progesterone and low estradiol levels associate
with abdominal aortic aneurysms in men. J Clin Endocrinol Metab.
107:e1413–e1425. 2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Feng Y, Chen Y, Wu X, Chen J, Zhou Q, Liu
B, Zhang L and Yi C: Interplay of energy metabolism and autophagy.
Autophagy. 20:4–14. 2024.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Fang J, Qian Y, Chen J, Xu D, Cao N, Zhu
G, Hu W, Hu H, Qian N, Yang S, et al: Human antigen R regulates
autophagic flux by stabilizing autophagy-associated mRNA in
calcific aortic valve disease. Cardiovasc Res. 119:2117–2129.
2023.PubMed/NCBI View Article : Google Scholar
|