Introduction
Osteogenesis is the continuous process of bone
formation that occurs throughout an individual's life. This complex
process relies on the coordination between osteoblasts, which are
specialized cells responsible for synthesizing bone tissue, and
osteoclasts, which are involved in the breakdown and remodeling of
existing bone (1,2). Osteogenic differentiation is a
specific cellular process involving the transformation of certain
stem cells or precursor cells into osteoblasts. This complex
process encompasses a series of cellular and molecular events.
Signaling pathways, including the Wnt/β-catenin (3,4) and
bone morphogenetic protein pathways (5,6),
play significant roles in promoting osteoblast differentiation by
activating specific genes. Transcription factors also contribute to
stem cell differentiation toward an osteogenic lineage. Runx2,
known as a master regulator of osteogenesis, controls the
expression of genes critical for osteoblast maturation and bone
mineralization (7,8). Other transcription factors, such as
Osterix and Dlx5, are involved in regulating osteogenic
differentiation (9,10).
Among the mentioned factors affecting osteogenic
differentiation, S100 calcium-binding protein A16 (S100A16), a
novel member of the S100 protein family, had been reported to be
involved in this process based on a mouse model (11). S100A16, which is expressed widely,
including in adipose tissues, has been associated with various
human diseases, including inflammation disorders, prostate cancer
and obesity (12). As a
calcium-binding protein, S100A16 binds one calcium ion per monomer.
In vitro studies suggest that it can promote adipocyte
differentiation. S100A16 overexpression in preadipocytes induces
increased proliferation, enhances adipogenesis, and reduces
insulin-stimulated glucose uptake (11). Gene Ontology (GO) annotations
revealed its involvement in protein homodimerization activity.
Previous studies in mouse models have demonstrated that S100A16
inhibits osteogenesis but stimulates adipogenesis (11). However, the role of S100A16 in the
osteogenic differentiation of bone marrow mesenchymal stem cells
(BMSCs) from rats and underlying mechanisms remain unexplored.
Additionally, extracellular matrix proteins,
including collagen and various growth factors, such as transforming
growth factor-beta (TGF-β), play vital roles in promoting
osteogenic differentiation (13,14).
These factors provide the necessary microenvironment and signaling
cues for stem cells to undergo osteogenic lineage commitment and
subsequent bone formation. As a part of the TGF-β signaling
pathway, the Smad4 gene encodes a protein involved in transmitting
chemical signals from the cell surface to the nucleus, allowing
external factors to influence gene activity and protein production
within the cells (15).
Furthermore, Smad4 serves as both a transcription factor and a
tumor suppressor. However, the specific function of Smad4 in
osteogenic differentiation has not yet been studied.
The mitogen-activated protein kinase (MAPK)/c-Jun
N-terminal kinase (JNK) pathway is another important pathway
implicated in osteogenesis. Smad4 has been reported to have an
impact on this pathway. In fact, in a previous study in human
pancreatic carcinoma it was found that Smad4 downregulated JNK
activity and while also inhibiting thyroid cancer cell growth by
inactivating the MAPK/JNK pathway (16). However, the role of the MAPK/JNK
pathway in osteogenesis remains controversial. A certain study
suggested that bone morphogenetic protein-9 enhances the osteogenic
differentiation of human periodontal ligament stem cells via the
JNK pathway (17), whereas another
found that the MAPK/JNK signaling pathway suppresses the osteogenic
differentiation of MC3T3-E1 osteoblasts under titanium ion exposure
conditions (18).
The current study examined the role of S100A16 and
Smad4 in the osteogenic differentiation of rat BMSCs and
investigated the impact of the novel S100A16-Smad4-MAPK/JNK
signaling axis in this process. Furthermore, MAPK/JNK inhibitors
and small interfering (si)RNAs that repressed the expression of
S100A16 were further applied to determine their effects on the
osteogenic differentiation of rats BMSCs, as well as the expression
of key genes. Additionally, ovariectomized (OVX) rats serve as a
prevalent animal model in osteoporosis research within the medical
field. Rats achieve sexual and endocrine system maturation at 3
months, coinciding with well-formed muscles and skeleton (19). Furthermore, ovariectomy in rats,
which involves the removal of ovaries and subsequent estrogen
deficiency, has been associated with increased differentiation of
BMSCs toward osteogenic pathways (20). Estrogen is a key regulator of bone
homeostasis that inhibits bone resorption and promotes bone
formation. In the absence of estrogen, as observed in OVX rats,
BMSCs are stimulated to undergo osteogenic differentiation. This
response is part of an adaptive mechanism that counteracts the
increased bone resorption, contributing to the accelerated bone
turnover observed in this experimental model. Therefore, OVX female
Sprague-Dawley rats were used in the present study to investigate
the osteogenesis in vivo. Overall, novel regulation factors
that affect osteogenic differentiation were discovered and new
mechanistic evidence for developing targeted and effective
therapeutic strategies for patients with osteoporosis was
ultimately provided.
Materials and methods
Animals and ovariectomy
A total of 30 8-week-old (180-200 g) female
Sprague-Dawley rats were purchased from Charles River Beijing Co.,
Ltd. All rats were housed in a specific pathogen-free facility
under a 12-h light/12-h dark cycle, with a controlled room
temperature of 25˚C, and provided ad libitum access to food
and water. Ovariectomy was performed as previously described
(21). Briefly, rats were injected
intraperitoneally with pentobarbital (30 mg/kg), followed by
exposure and excision of the ovaries from both sides. Rats in the
sham group underwent similar procedures except that their ovaries
were left intact. Following surgery, all animals, including the
sham rats, were sutured and received penicillin injections for
three consecutive days. Sprague-Dawley rats were used for bone
marrow stem cell isolation and establishing the OVX rat model. It
was confirmed that all animals were treated following the IACUC
guidelines. The present study was approved (approval no. 2020018)
by the animal Ethics Committee of Luohe Central Hospital (Luohe,
China).
Isolation of BMSCs and cell
culture
All rats were sacrificed via cervical dislocation
and then the whole rats were immersed in 75% ethanol for 15 min for
sterilization. Bone marrow cells were isolated from Collum femoris
with DMEM (Shanghai BasalMedia Technologies Co., Ltd.) and cultured
at a temperature of 37˚C in the presence of 5% CO2.
After three passages, BMSCs were characterized by hematopoietic
markers (CD11b and CD45) and BMSC markers (CD90 and CD44) using a
CytoFLEX nano flow cytometer (Beckman Coulter, Inc.). The flow
cytometry data were analyzed by Kaluza Analysis Software (version
2.1.1; Beckman Coulter, Inc.). The antibodies used were as follows:
PE-conjugated mouse anti-CD11b antibody (cat. no. sc-53086; Santa
Cruz Biotechnology, Inc.), PE-conjugated mouse anti-CD45 antibody
(cat. no. 202207; BioLegend, Inc.), PE-conjugated mouse anti-CD90
antibody (cat. no. 202523; BioLegend, Inc.), and PE-conjugated
mouse anti-CD44 antibody (cat. no. sc-7297; Santa Cruz
Biotechnology, Inc.). All in vitro experiments were
performed using cells from passages 3-5.
Differentiation of BMSCs
BMSCs (2.0x104) were seeded into HyCyte™
rat BMSC culture medium (Cas9X Biotech Co. Ltd.) and were allowed
to attach for 24 h, after which the medium was replaced with
HyCyte™ osteogenic differentiation medium (Cas9X) and changed every
3 days. After 14 days, cells were fixed with 4% polyformaldehyde
for 15 min and then stained with 0.1% sodium alizarin
sulfonate-Tris-HCL staining solution (pH=8.3) for 30 min at 37˚C.
Stained cells were then washed with PBS and imaged through a light
microscopy. For inhibitor treatments, both U0126 (cat. no. T21332;
TargetMol Chemicals Inc.) and SP600125 (cat. no. T3109; TargetMol
Chemicals Inc.) were used at a concentration of 10 µM.
siRNA interference assay
S100A16 and negative control siRNA were purchased
from Shanghai GenePharma Co., Ltd. with their sequences being as
follows: si-S100A16-1 forward, 5'-GAAUUAGCCUCUUCUCUUC-3' and
reverse, 5'-GAAGAGAAGAGGCUAAUUC-3'; si-S100A16-2 forward,
5'-UAUGUAUCCAAGCACAGCC-3' and reverse, 5'-GGCUGUGCUUGGAUACAUA-3';
si-S100A16-3 forward, 5'- AGAACAAGAUCAGCAAGUC-3' and reverse,
5'-GACUUGCUGAUCUUGUUCU-3'; scrambled negative control forward,
5'-UUCUCCGAACGUGUCACGU-3' and reverse, 5'-ACGUGACACGUUCGGAGAA-3'.
BMSCs were transfected with 50 nM of desired the siRNAs using
Lipofectamine 3000 in Opti-MEM® at 37˚C (Thermo Fisher
Scientific Inc.) on days 0, 3, 5, 7, 9 and 12 of differentiation.
At 6 h after transfection, the medium was replaced with osteogenic
differentiation medium and changed every 3 days. The total RNA of
each sample was isolated and sequenced on day 14 of
differentiation.
Hematoxylin and eosin (H&E)
staining
Rat femurs were dissected, fixed in 4% formaldehyde
at room temperature for 2 days, and subsequently decalcified in
EDTA solution at room temperature over a period of 4 weeks.
Following decalcification, the specimens underwent dehydration and
were processed to form paraffin blocks. Serial transverse and
longitudinal sections, each 5-µm thick, were then prepared from
both the diaphysis and metaphysis of the femurs. These sections
were subjected to H&E and Masson's trichrome staining. All
sections were examined using a light microscope, allowing for a
comprehensive analysis of the bone tissue structure and
composition.
Alizarin red S staining
Cells were initially washed three times with PBS
before fixation in absolute ethanol for 30 min, followed by
complete air drying. Subsequently, Alizarin Red S Solution (0.2%;
pH8.3; Beijing Solarbio Science & Technology Co., Ltd.) was
added to the plates, after which the cells were incubated at room
temperature for 15 min. After incubation, the cells were carefully
rinsed with double-distilled water and allowed to air dry. Stained
cells were examined using light microscopy and photographed.
Western blot analysis
For bone tissue protein extraction, a bone tissue
protein extraction kit (Beijing Solarbio Science & Technology
Co., Ltd.) was used. RIPA buffer (Beyotime Institute of
Biotechnology) was used for cellular protein extraction. The
protein content in all lysates was measured using a BCA protein
quantification kit (Beyotime Institute of Biotechnology) and 10 µg
of protein from each sample was loaded and allowed to run on a 10%
sodium dodecyl-sulfate polyacrylamide gel, connected to an electric
source. The overall protein on the gel was then transferred to a
polyvinylidene difluoride membrane, which was then blocked using 1%
BSA (Beyotime Institute of Biotechnology) for 1 h at room
temperature. Thereafter, the sample was incubated overnight at 4˚C
with primary antibodies that detect and bind to the specific
protein of interest. The membrane was then washed with
Tris-buffered saline with 0.02% Tween 20 (TBST) and then incubated
with horseradish peroxidase (HRP)-linked secondary antibody for 1
h. Afterwards, the membrane was once again washed with TBST. The
chemiluminescent HRP-substrate was then added to the blot, and a
Tanon 5200 automatic chemiluminescence image analysis system
(Shanghai Tianneng Life Science Co., Ltd.) was used to detect the
protein of interest. ImageJ software (v1.50b; National Institutes
of Health) was used for the quantification of the protein bands.
The antibodies used in the present study were as follows: rabbit
anti-S100A16 antibody (1:1,000; cat. no. A16167), rabbit anti-GAPDH
antibody (1:5,000; cat. no. AC001), rabbit anti-RUNX2 antibody
(1:500; cat. no. A11753,), rabbit anti- SMAD4 antibody (1:1,000;
cat. no. A21487), rabbit anti-MAPK antibody (1:1,000; cat. no.
A14401), rabbit anti-phospho-MAPK antibody (1:2,000; cat. no.
AP0526), rabbit anti-JNK1 antibody (1:1,000; cat. no. A2462),
rabbit anti-OSTERIX antibody (1:1,000; cat. no. A18699; all from
ABclonal Biotech Co., Ltd.), rabbit anti-JNK antibody (1:1,000;
cat. no. 80024-1-RR; Proteintech Group, Inc.) and HRP-conjugated
goat anti-rabbit secondary antibody (1:10,000; cat. no. AS014;
ABclonal Biotech Co., Ltd.).
Reverse transcription-quantitative PCR
(RT-qPCR)
The total RNA of each cell lysate sample was
extracted using a SteadyPure RNA extraction kit (Accurate Biology
Co., Ltd.) following the manufacturer's protocol. For bone tissue
RNA extraction, a SPARKeasy bone tissue RNA extraction kit
(Sparkjade biotech Co. Ltd.) was used. The HiFiScript cDNA
Synthesis Kit (CWBio) was used to reverse transcribe cDNA from the
isolated RNAs according to the manufacturer's instructions. The
expression of the genes of interest was determined using qPCR with
SYBR Green Pro Taq HS master mix (Hunan Aikerui Bioengineering Co.,
Ltd.). The thermocycling conditions consisted of an initial
denaturation step at 95˚C for 1 min, followed by denaturation at
95˚C for 10 sec, and annealing and extension at 60˚C for 20 sec.
This cycle was repeated 40 times. The relative expression was
calculated using the 2-ΔΔCq method
as reported previously (22). The
sequences of primers used for the genes of interest are listed in
Table I. The relative mRNA
expression of the desired genes was determined using human GAPDH as
internal control.
 | Table IPrimers used in reverse
transcription-quantitative PCR used in the present study. |
Table I
Primers used in reverse
transcription-quantitative PCR used in the present study.
| Gene name | Primer sequence
(5'-3') |
|---|
| S100A16 | F:
GAGCTGAGGCAGTGAGATGG |
| | R:
ACCAGGCTGTGCTTGGATAC |
| RUNX2 | F:
AGTCTGTCTGGCGACCCTAT |
| | R:
TTGCCAGATCACAACTGGGG |
| OSX | F:
ACCTCTTGAGAGGAGACGGG |
| | R:
CTGTTGAGTCTCGCAGAGGG |
| OCN | F:
GAGGACCCTCTCTCTGCTCA |
| | R:
GGTAGCGCCGGAGTCTATTC |
| ALP | F:
GGCACCATGACTTCCCAGAA |
| | R:
ACCGTCCACCACCTTGTAAC |
| SMAD4 | F:
GCTGGGAACTCAGCCCTCTA |
| | R:
GCAGCTCGTTCAGCAATGAC |
| GAPDH | F:
TGGTGAAGGTCGGTGTGAAC |
| | R:
GATGGTGATGGGTTTCCCGT |
RNA sequencing and bioinformatics
analysis
The total RNA of each cell lysate sample was
extracted from siRNA interference or scramble groups using RNeasy
Mini Kit (cat. no. 74104; Qiagen China Co., Ltd.) following the
manufacturer's instruction. Thereafter, RNA sequencing (RNAseq) was
performed on 2 µg RNA from each sample using Illumina HiSeq™.
Sequencing libraries of nucleotides with length of 300-600 bp
(5'-3') were generated using Unicellular RNAseq Library Prep Kit
(cat. no. AT4205-01; Hangzhou Kaitai Biotech Co., Ltd.). HISAT2
software (version 2.1.0; http://daehwankimlab.github.io/hisat2/) and DESeq2
software (version 1.14.1; https://bioconductor.org/packages/release/bioc/html/DESeq2.html)
were used to identify differentially expressed genes (DEGs) between
each sample using the following filter criteria: P<0.05 and the
absolute value of log2FC >1. DEGs were annotated
using the DAVID database (https://david.ncifcrf.gov/) to examine the enriched GO
terms. Gene set enrichment analysis (GSEA) was performed using GSEA
software (Version 4.3.2; https://www.gsea-msigdb.org/gsea/index.jsp?).
Statistical analysis
Significance differences between the groups were
determined using unpaired Student's t-test or one-way ANOVA
followed by Dunnett's test using GraphPad Prism software (version
9.5.1.733; Dotmatics). Each experiment was performed with at least
three biological repeats and the results were presented as the mean
with standard errors. P<0.05 was considered to indicate a
statistically significant significance.
Results
S100A16 levels in OVX rats and
BMSCs
The OVX rat model was established through bilateral
ovariectomy and utilized to mimic estrogen deficiency-induced bone
loss. H&E and Masson's staining of bone tissues from wild-type
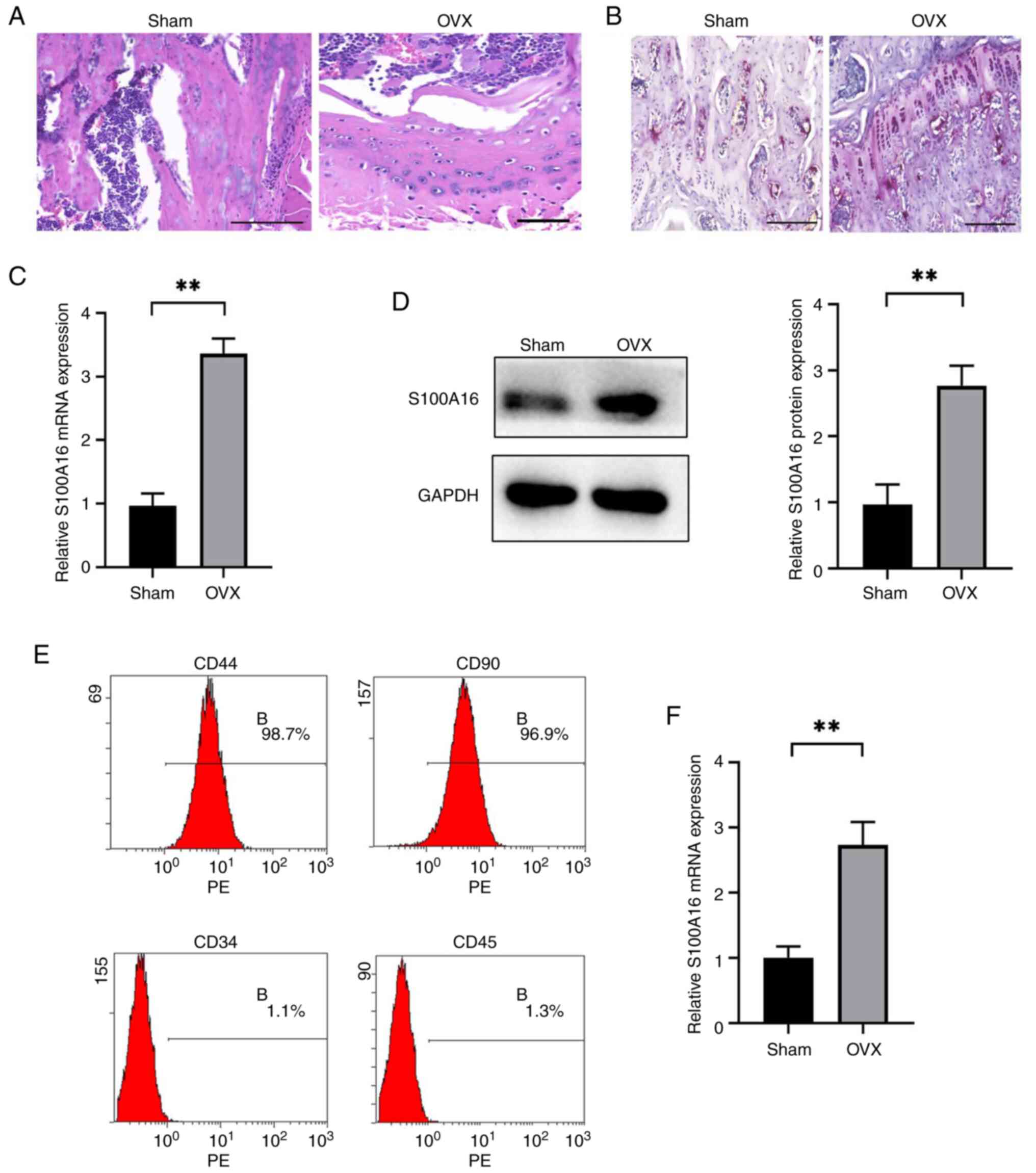
(WT) rats confirmed the occurrence of osteoporosis. Specifically,
H&E staining showed the replacement of trabeculae by adipocytes
in the bone marrow of the femoral shaft of OVX rats (Fig. 1A), whereas Masson's staining
indicated reduced collagen fiber content and decreased levels of
new bone in OVX rats (Fig. 1B). A
comparison of S100A16 expression at the mRNA and protein levels in
bone tissues isolated from OVX and sham-operated rats demonstrated
significantly enhanced expression in the OVX group (Fig. 1C and D). Next, BMSCs were isolated from rat
bone tissues, after which specific markers were characterized using
flow cytometry. As demonstrated in Fig. 1E, BMSCs were positive for
traditional BMSC markers (CD90 and CD44) but were negative for
hematopoietic markers (CD34 and CD45), confirming the successful
isolation of BMSCs suitable for downstream experiment. In addition,
S100A16 mRNA expression levels were significantly higher in BMSCs
from OVX rats than in those from the sham group (Fig. 1F). Collectively, both the in
vivo rat model and in vitro BMSC model demonstrated
enhanced S100A16 levels in the OVX groups, suggesting a potential
pivotal role for this gene in osteoporosis.
Role of S100A16 in osteogenic
differentiation of rat BMSCs
To investigate the role of S100A16 in osteoporosis,
siRNAs were further utilized to knockdown S100A16 in BMSCs and
determine its effects on osteogenic differentiation. First, three
siRNAs targeting S100A16 were tested in terms of their efficiency
in knocking down BMSCs isolated from WT rats. As revealed in
Fig. 2A, siRNAs 1 and 3 exhibited
a knockdown efficiency of over 60% compared with the scramble
control, with siRNA 3 producing the highest efficiency, leading to
its use in subsequent downstream experiments. Next, Alizarin Red S
staining results showed increased mineralized nodule formation in
S100A16-siRNA treated BMSCs than in the scramble control (Fig. 2B and C), indicating that S100A16 knockdown
promoted osteogenic differentiation of BMSCs and in turn revealing
the inhibitory role of S100A16 in this biological process. In
addition, the expression of genes associated with osteogenic
differentiation, including RUNX family transcription factor 2
(RUNX2), Osterix (OSX, also known as Sp7 transcription factor),
Osteocalcin (OCN, also known as bone gamma-carboxyglutamate
protein), and alkaline phosphatase (ALP), was significantly
increased in S100A16-knockdown BMSCs (Fig. 2D), among which the RUNX2 and OSX
were also enhanced at the protein level (Fig. 2E). These results confirmed the
inhibitory role of S100A16 in the osteogenic differentiation of
BMSCs from rats.
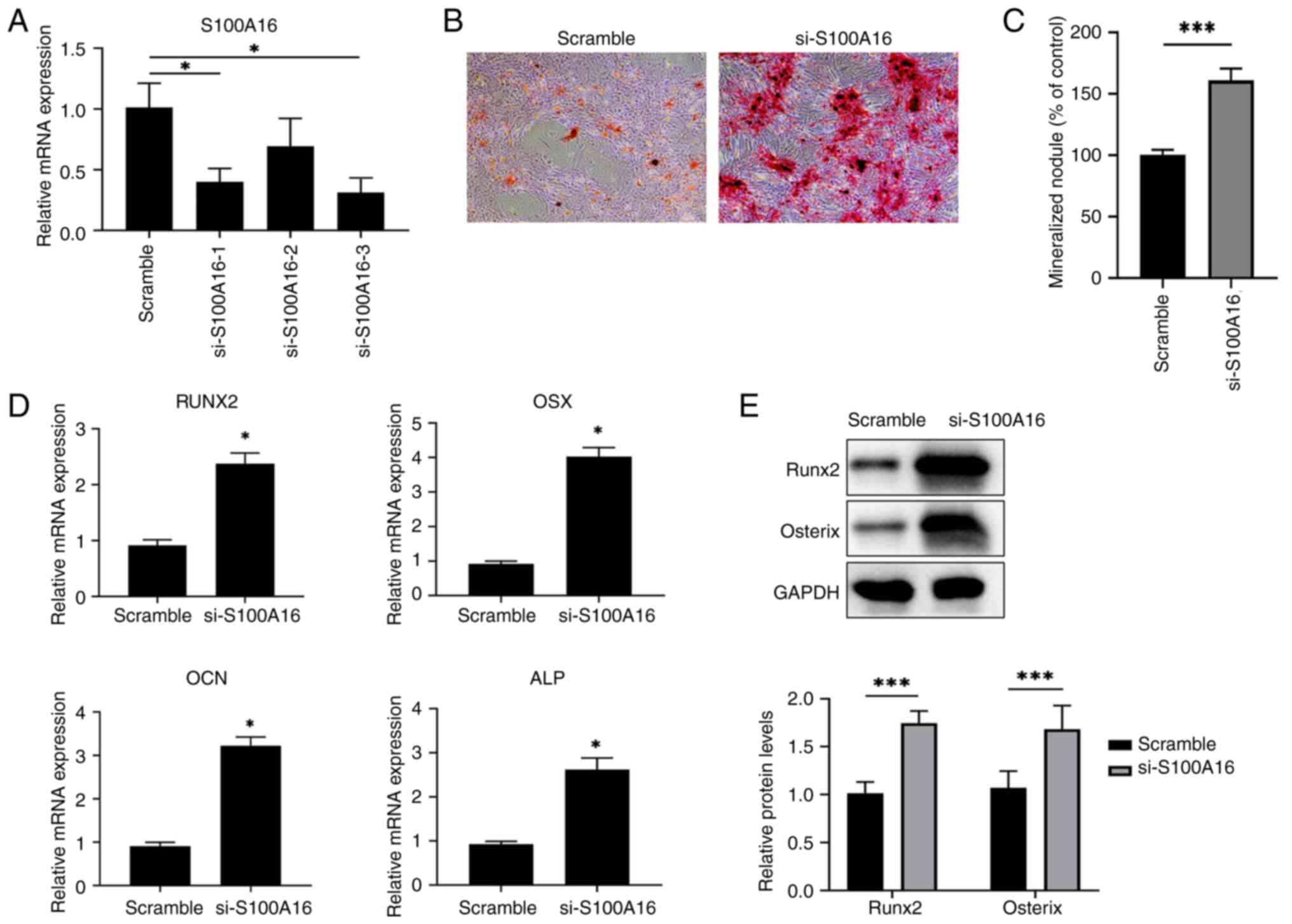 | Figure 2S100A16 silencing promotes osteogenic
differentiation of WT BMSCs in vitro. (A) Validation of
siRNA silencing efficiency in BMSCs isolated from WT rat BMSCs. (B)
Representative images for Alizarin Red S staining showed
differences in osteogenic differentiation between control rat BMSCs
and S100A16-silenced rat BMSCs. (C) Quantification result for
Alizarin Red staining. (D) Transfected BMSCs were cultured in
vitro for 7 days, after which the mRNA expression of Runx2,
OSX, OCN and ALP was analyzed via reverse
transcription-quantitative PCR. (E) The protein expression of Runx2
and Osterix was analyzed via western blotting.
*P<0.05 and ***P<0.001 vs. scramble.
BMSCs, bone marrow mesenchymal stem cells; siRNA, small interfering
RNA; WT, wild-type; Runx2, RUNX family transcription factor 2; OSX,
osterix; OCN, osteocalcin; ALP, alkaline phosphatase. |
Pathways involved in the effects of
S100A16 on osteogenic differentiation of rat BMSCs
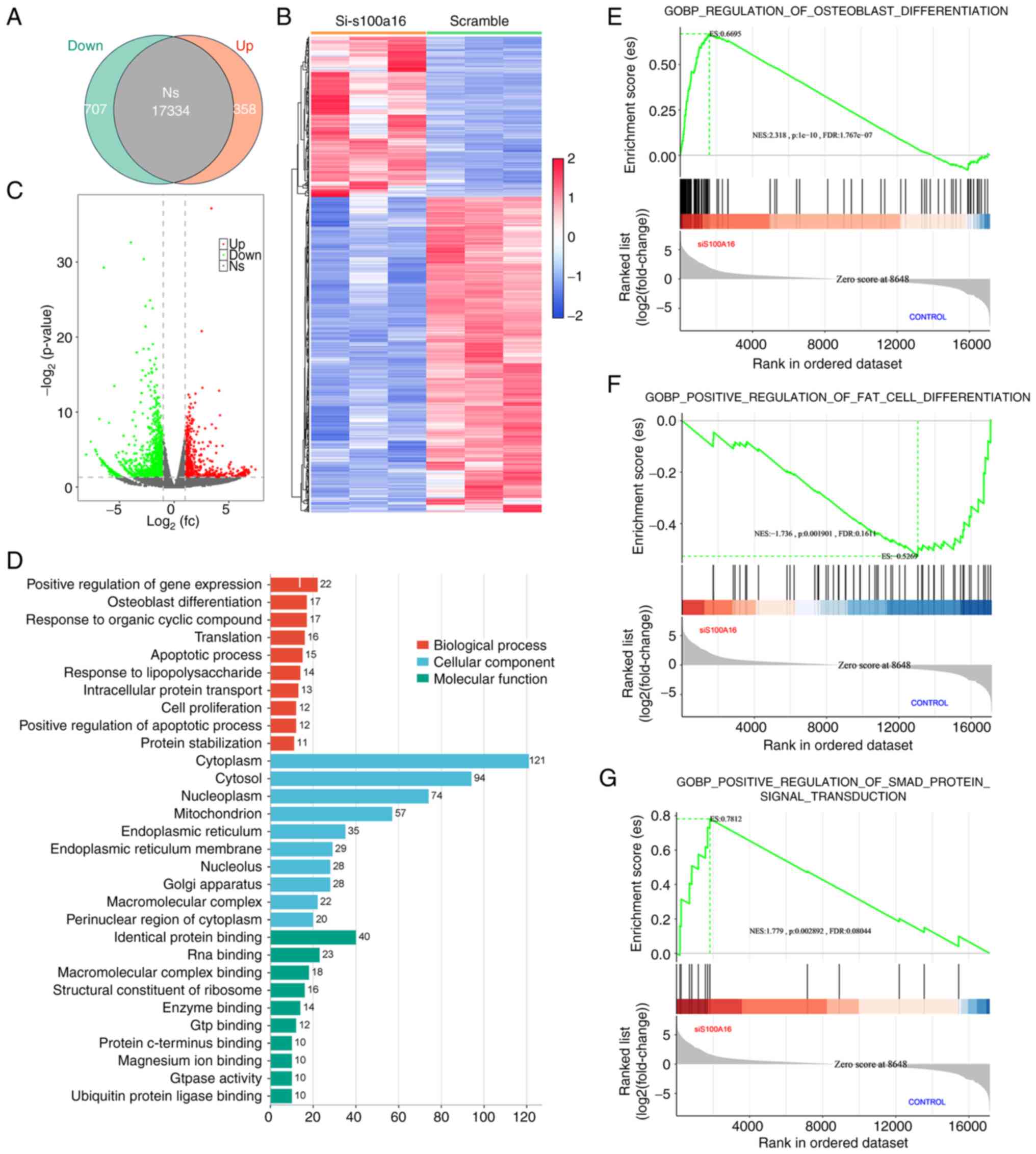
To elucidate the impact of S100A16 on osteogenic
differentiation and identify the underlying biological pathways,
transcriptomic sequencing was performed to identify DEGs between
scramble control and S100A16-knockdown BMSCs. As demonstrated in
Fig. 3A and C, the resulting Venn displayed 358
significantly enriched genes in the S100A16-knockdown groups, with
707 genes downregulated (FDR <0.05, fold change >1.5).
Hierarchical clustering of DEGs identified significantly altered
gene expression pattern in the si-S100A16 group compared with the
control group (Fig. 3B).
Furthermore, these DEGs were analyzed through GO function analysis.
As shown in Fig. 3D, three
ontologies, including biological processes, cellular components and
molecular functions of the transcripts due to S100A16 silencing,
were identified. In addition, GSEA confirmed a significant
upregulation of osteogenic gene set (Fig. 3E) and downregulation of adipogenic
genes in the S100A16-silenced transcriptome (Fig. 3F). Notably, it was further
recognized that S100A16 knockdown profoundly affected the Smad
signaling pathway (Fig. 3G),
indicating the potential functional role of Smad in the effect of
S100A16 on the osteogenic differentiation of BMSCs.
Roles of Smad4 and MAPK/JNK pathways
in the effects of S100A16 on the osteogenic differentiation of rat
BMSCs
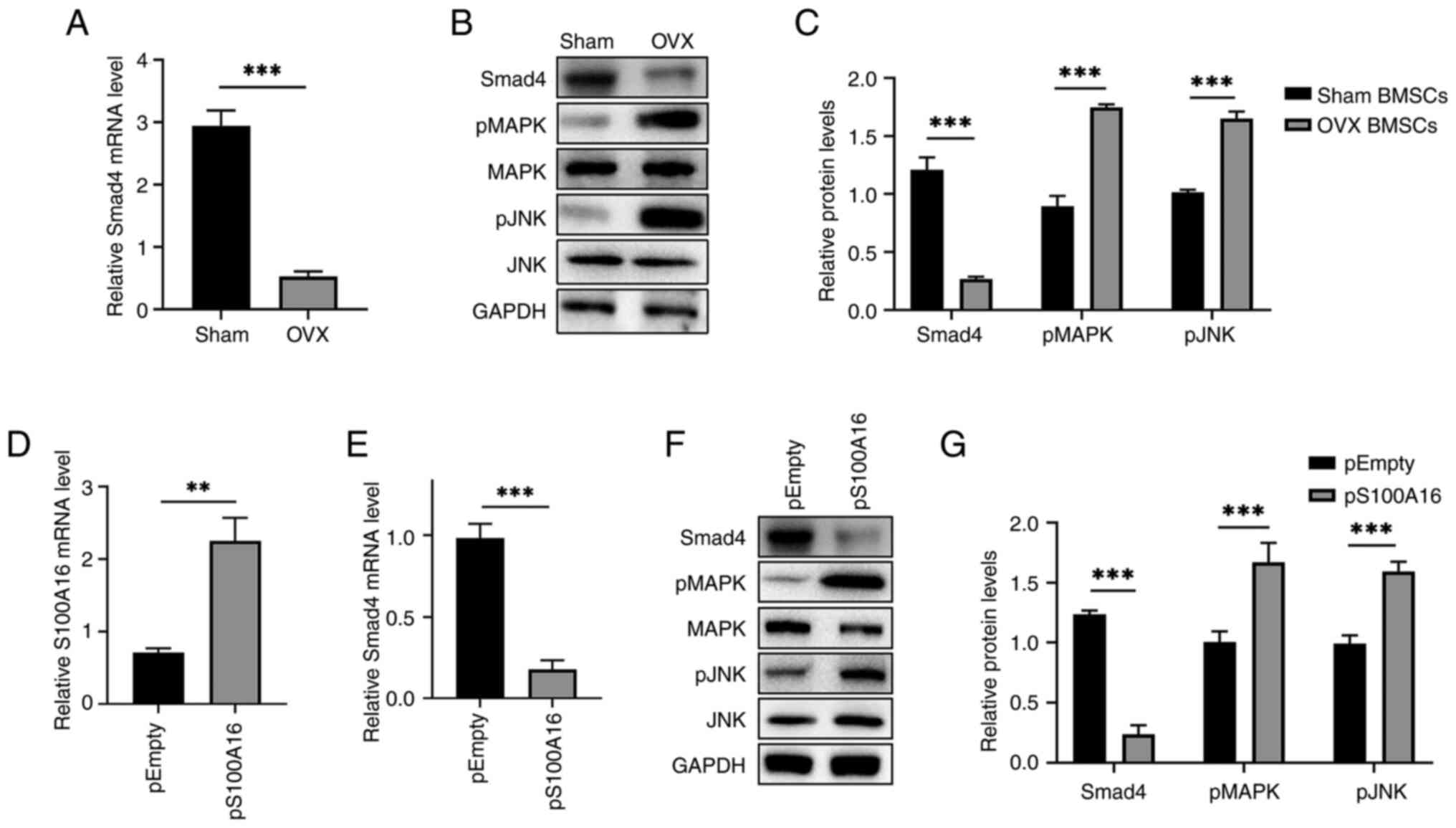
Previous studies have reported that Smad4 plays a
dominant regulatory role in osteogenesis (23-25).
Hence, the interactions between S100A16 and Smad4, as well as the
involved mitogen-activated protein kinase (MAPK)/c-Jun N-terminal
kinase (JNK) signaling pathways were examined. First, Smad4
expression in BMSCs from OVX rats was significantly repressed at
both the mRNA (Fig. 4A) and
protein levels (Fig. 4B).
Meanwhile, the phosphorylation of MAPK and JNK was significantly
upregulated (Fig. 4B and C), indicating that the MAPK/JNK pathways
were involved in osteogenic differentiation. Subsequently, the
effects of S100A16 overexpression on Smad4 and MAPK/JNK pathways
were assessed. Accordingly, the significant overexpression of
S100A16 in BMSCs (Fig. 4D)
significantly downregulated Smad4 (Fig. 4E and F). Furthermore, S100A16 overexpression
significantly enhanced the phosphorylation of MAPK and JNK
(Fig. 4F and G). Overall, these findings revealed
changes in Smad4 and the MAPK/JNK pathways following ovariectomy
and S100A16 overexpression, indicating their roles as mediators in
the inhibitory effects of S100A16 on the osteogenic differentiation
of rat BMSCs.
Suppression of the MAPK/JNK pathways
restores osteogenic differentiation of rat BMSCs
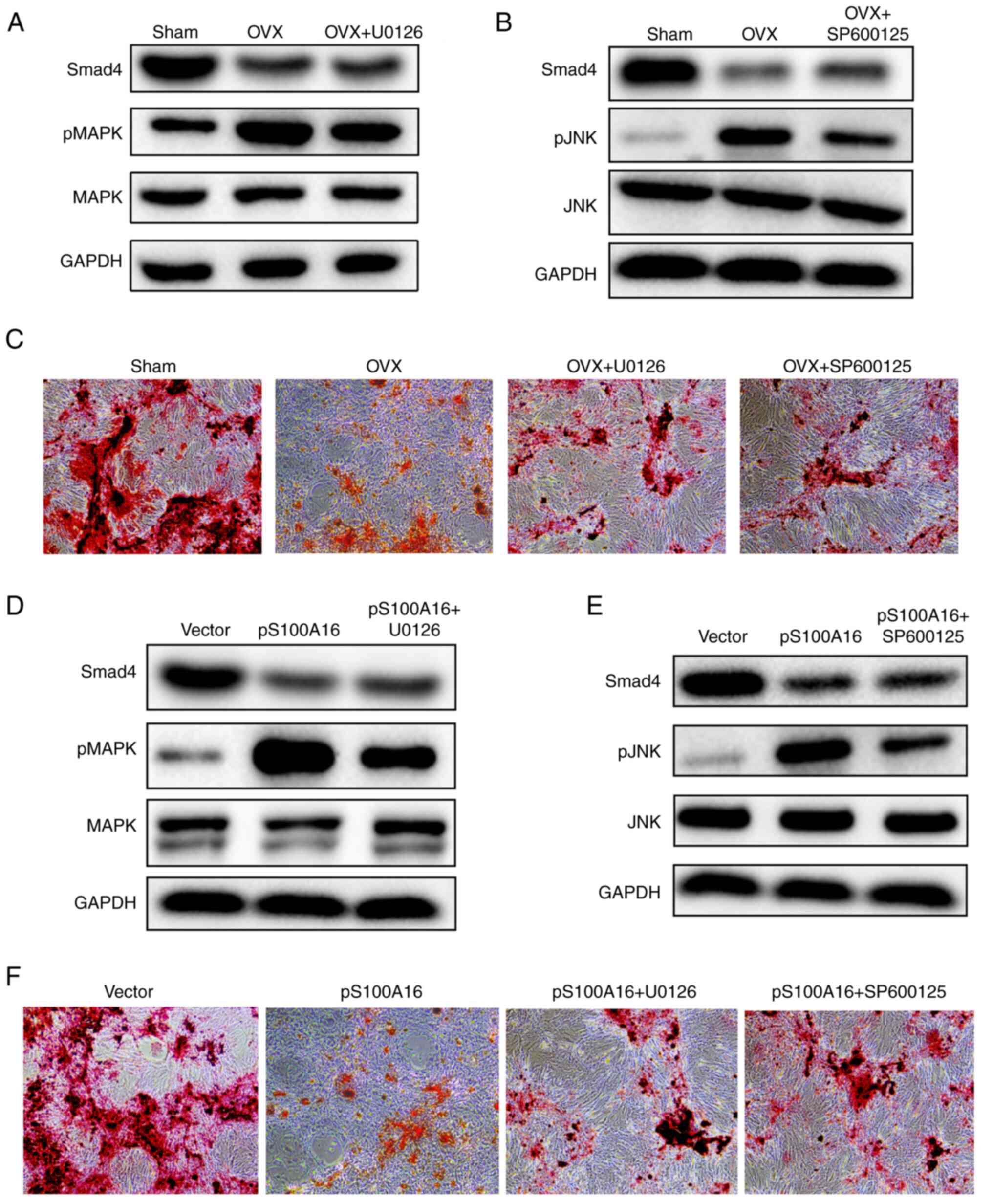
To confirm the regulatory roles of the MAPK/JNK
pathways in the osteogenic differentiation of BMSCs, the inhibitors
U0126 and SP600125 were utilized to suppress MAPK and JNK
signaling, respectively. The application of these inhibitors
efficiently downregulated the phosphorylation of their respective
target enzymes in BMSCs isolated from OVX rats without inducing
changes in the baseline MAPK/JNK levels (Fig. 5A and B). Furthermore, the effects of MAPK/JNK
inhibitors on osteogenic differentiation were compared. As depicted
in Fig. 5C, the Alizarin Red S
staining results clearly indicated that both inhibitors
significantly restored the osteogenic differentiation of BMSCs from
OVX rats. Moreover, the effects of MAPK/JNK pathway inhibitors were
determined in BMSCs isolated from WT rats with S100A16
overexpression. Similarly, Smad4 levels remained stable, whereas
the stimulation of MAPK/JNK phosphorylation was repressed (Fig. 5D and E). More importantly, both inhibitors
alleviated the inhibition of osteogenic differentiation induced by
S100A16 overexpression (Fig. 5F).
In summary, these results demonstrated that blocking the MAPK/JNK
pathways could restore the osteogenic differentiation of BMSCs from
both OVX rats and those with S100A16 overexpression, underscoring
the inhibitory roles of MAPK/JNK phosphorylation in osteogenic
differentiation.
Discussion
The current study investigated the role of S100A16
in the osteogenic differentiation of rat BMSCs and uncovered the
underlying mechanisms and involved pathways from the perspectives
of Smad4 and the MAPK/JNK signaling pathways. Elevated S100A16
levels were first recognized in both bone tissues from OVX rats and
isolated BMSCs. S100A16 knockdown promoted osteogenic
differentiation, confirming the inhibitory role of S100A16.
Furthermore, Smad4 and the MAPK/JNK pathways, identified through
transcriptomic sequencing and pathway analysis, were found to be
involved in the functional effects of S100A16, as further evidenced
by the inhibition of the MAPK/JNK pathways and overexpression of
S100A16.
Overall, the current study unveiled the inhibitory
role of S100A16 in osteogenic differentiation, as well as the roles
of the Smad4 and MAPK/JNK pathways, providing new insights that
could help understand osteogenic differentiation. In fact,
osteogenic differentiation, the process by which undifferentiated
mesenchymal stem cells (MSCs) develop into specialized bone-forming
cells called osteoblasts, has been extensively studied. This
intricate process involves a series of cellular events regulated by
various signaling pathways. Initially, the differentiation of MSCs
into osteoblasts is triggered by specific growth factors, such as
bone morphogenetic proteins, transforming growth factor-beta
(TGF-β) and fibroblast growth factor. These factors activate
intracellular signaling cascades, including the Smad and MAPK
pathways, which initiate the expression of key osteogenic genes,
including Runx2, OSX and ALP. The present study revealed that
changes in S100A16 either via siRNA knockdown or overexpression in
rat BMSCs could lead to changes in the MAPK/JNK pathways and in the
expression of key genes (Figs. 4
and 5), consequently affecting the
osteogenic differentiation. Indeed, other studies using a mouse
model (11) or human in
vitro cell models for osteogenesis (26,27)
have confirmed the inhibitory role of S100A16 in this process. As a
calcium-binding protein, S100A16 has been implicated in the
regulation of various cellular processes, including osteogenic
differentiation and bone metabolism (11). The aforementioned findings along
with the present results highlight the potential significance of
S100A16 as a regulator of osteogenic differentiation and bone
homeostasis.
The present study also provided, to the best of the
authors' knowledge, the first evidence of the link between S100A16
and Smad4 in osteogenic differentiation, where S100A16
overexpression significantly downregulated the expression of Smad4
(Fig. 4). Smad4 is known as a
transcription factor that forms a complex with other Smad proteins
upon TGF-β activation. This complex translocases into the nucleus
and regulates the expression of targeting genes involved in
osteogenic differentiation. In addition, the link between S100A16
and Smad4 has been reported in other diseases, especially cancers
(28-30).
The available knowledge on S100A16 and Smad4 could also provide new
insights into the study of the mechanism governing different
diseases and potential targets for developing therapeutical
strategies.
Furthermore, Smad4 had been reported to downregulate
JNK activity in human pancreatic carcinoma (31). JNK is a member of the MAPK family.
SMAD4 also inhibits thyroid cancer cell growth by inactivating the
MAPK/JNK pathway (16). However,
the role of MAPK/JNK signaling in osteogenesis appears to be
controversial. On the one hand, bone morphogenetic protein-9 had
been reported to enhance the osteogenic differentiation of human
periodontal ligament stem cells via the JNK pathway (17). On the other hand, another study
indicated that the MAPK/JNK signaling pathway suppresses the
osteogenic differentiation of MC3T3-E1 osteoblasts exposed to
titanium ion (18). In the present
study, the inhibition of the MAPK/JNK pathways restored the
osteogenic differentiation of BMSCs (Fig. 5). Together, these results revealed
that the specific role of the MAPK/JNK pathway in osteogenesis
would be context-dependent, with factors including, but not limited
to, species, cell type source and exogenous stimulation.
The current study has several potential limitations.
First, most of it remained at the level of the in vitro cell
models, including the BMSCs isolated from WT and OVX rats. Hence,
the gap between in vitro and in vivo models and
limitations in the conclusions of the present study that could be
extrapolated to in vivo models must be acknowledged. Next,
the interaction between Smad4 and the MAPK/JNK pathways remains to
be explored. Although stable levels of Smad4 were found after
MAPK/JNK pathway inhibition, direct evidence is still required for
studying the potential interaction between these two pathways.
Finally, other pathways involved in the function of S100A16 in
osteogenic differentiation remain to be determined. Together,
future studies addressing these limitations are needed to further
understand the role of S100A16, as well as other inhibitory
factors, in osteogenesis, which would be crucial for developing
strategies that promote and enhance osteogenic differentiation for
potential therapeutic applications in bone tissue engineering and
regenerative medicine.
In summary, the role of S100A16 in the osteogenic
differentiation of rat BMSCs was investigated. The current study
uncovered a novel mechanistic link between S100A16 and Smad4, as
well as the MAPK/JNK signaling pathways, in osteogenic
differentiation. Further studies on these key impact factors may
ultimately contribute to the development of preventive and
therapeutic strategies for osteoporosis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Henan key R&D
and promotion special (scientific and technological research)
support project (grant no. 212102310199) and Key scientific
research projects of colleges and universities in Henan (grant no.
21A320011).
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus under accession number GSE259238 or
at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE259238.
Authors' contributions
JX and YafS conceived the study, designed the
experiments and interpreted the results. JX, YanS, JB and ZW
performed the experiments, and collected and analyzed the data. JX
and JB confirm the authenticity of all the raw data. JX wrote the
manuscript. JB and YafS revised the manuscript. YafS supervised the
study. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal study protocol was approved (approval no.
2020018) by Luohe Central Hospital Animal Ethics Committee (Luohe,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fedarko NS: Osteoblast/osteoclast
development and function in osteogenesis imperfecta. Osteogenesis
Imperfecta. Academic Press, pp45-56, 2014.
|
|
2
|
Kim JM, Lin C, Stavre Z, Greenblatt MB and
Shim JH: Osteoblast-osteoclast communication and bone homeostasis.
Cells. 9(2073)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhang J, Zhang X, Zhang L, Zhou F, van
Dinther M and Ten Dijke P: LRP8 mediates Wnt/β-catenin signaling
and controls osteoblast differentiation. J Bone Miner Res.
27:2065–2074. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Day TF, Guo X, Garrett-Beal L and Yang Y:
Wnt/beta-catenin signaling in mesenchymal progenitors controls
osteoblast and chondrocyte differentiation during vertebrate
skeletogenesis. Dev Cell. 8:739–750. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yamaguchi A, Komori T and Suda T:
Regulation of osteoblast differentiation mediated by bone
morphogenetic proteins, hedgehogs, and Cbfa1. Endocr Rev.
21:393–411. 2000.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Skillington J, Choy L and Derynck R: Bone
morphogenetic protein and retinoic acid signaling cooperate to
induce osteoblast differentiation of preadipocytes. J Cell Biol.
159:135–146. 2002.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gaur T, Lengner CJ, Hovhannisyan H, Bhat
RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS
and Lian JB: Canonical WNT signaling promotes osteogenesis by
directly stimulating Runx2 gene expression. J Biol Chem.
280:33132–33140. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yang W, Li HY, Wu YF, Mi RJ, Liu WZ, Shen
X, Lu YX, Jiang YH, Ma MJ and Shen HY: ac4C acetylation of RUNX2
catalyzed by NAT10 spurs osteogenesis of BMSCs and prevents
ovariectomy-induced bone loss. Mol Ther Nucleic Acids. 26:135–147.
2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang B, Zhang X, Xiao J, Zhou X, Chen Y
and Gao C: Neuropeptide Y upregulates Runx2 and osterix and
enhances osteogenesis in mouse MC3T3-E1 cells via an autocrine
mechanism. Mol Med Rep. 22:4376–4382. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lee KM, Park KH, Hwang JS, Lee M, Yoon DS,
Ryu HA, Jung HS, Park KW, Kim J, Park SW, et al: Inhibition of
STAT5A promotes osteogenesis by DLX5 regulation. Cell Death Dis.
9(1136)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li D, Zhang R, Zhu W, Xue Y, Zhang Y,
Huang Q, Liu M and Liu Y: S100A16 inhibits osteogenesis but
stimulates adipogenesis. Mol Biol Rep. 40:3465–3473.
2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sturchler E, Cox JA, Durussel I, Weibel M
and Heizmann CW: S100A16, a novel calcium-binding protein of the
EF-hand superfamily. J Biol Chem. 281:38905–38917. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu DD, Zhang JC, Zhang Q, Wang SX and
Yang MS: TGF-β/BMP signaling pathway is involved in cerium-promoted
osteogenic differentiation of mesenchymal stem cells. J Cell
Biochem. 114:1105–1114. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang LT, Liu RM, Luo Y, Zhao YJ, Chen DX,
Yu CY and Xiao JH: Hyaluronic acid promotes osteogenic
differentiation of human amniotic mesenchymal stem cells via the
TGF-β/Smad signalling pathway. Life Sci. 232(116669)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhao M, Mishra L and Deng CX: The role of
TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 14:111–123.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cai H, Yang X, Jiang Z, Liang B, Cai Q and
Huang H: Upregulation of SMAD4 inhibits thyroid cancer cell growth
via MAPK/JNK pathway repression. Trop J Pharm Res. 18:2473–2478.
2019.
|
|
17
|
Wang P, Wang Y, Tang W, Wang X, Pang Y,
Yang S, Wei Y, Gao H, Wang D and Cao Z: Bone morphogenetic
protein-9 enhances osteogenic differentiation of human periodontal
ligament stem cells via the JNK pathway. PLoS One.
12(e0169123)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhu WQ, Ming PP, Zhang SM and Qiu J: Role
of MAPK/JNK signaling pathway on the regulation of biological
behaviors of MC3T3-E1 osteoblasts under titanium ion exposure. Mol
Med Rep. 22:4792–4800. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lei Z, Xiaoying Z and Xingguo L:
Ovariectomy-associated changes in bone mineral density and bone
marrow haematopoiesis in rats. Int J Exp Pathol. 90:512–519.
2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Du D, Zhou Z, Zhu L, Hu X, Lu J, Shi C,
Chen F and Chen A: TNF-α suppresses osteogenic differentiation of
MSCs by accelerating P2Y2 receptor in
estrogen-deficiency induced osteoporosis. Bone. 117:161–170.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen W, Chen X, Chen AC, Shi Q, Pan G, Pei
M, Yang H, Liu T and He F: Melatonin restores the
osteoporosis-impaired osteogenic potential of bone marrow
mesenchymal stem cells by preserving SIRT1-mediated intracellular
antioxidant properties. Free Radic Biol Med. 146:92–106.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Song B, Estrada KD and Lyons KM: Smad
signaling in skeletal development and regeneration. Cytokine Growth
Factor Rev. 20:379–388. 2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Park JS, Kim M, Song NJ, Kim JH, Seo D,
Lee JH, Jung SM, Lee JY, Lee J, Lee YS, et al: A reciprocal role of
the Smad4-Taz axis in osteogenesis and adipogenesis of mesenchymal
stem cells. Stem Cells. 37:368–381. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Pakravan K, Razmara E, Mahmud Hussen B,
Sattarikia F, Sadeghizadeh M and Babashah S: SMAD4 contributes to
chondrocyte and osteocyte development. J Cell Mol Med. 26:1–15.
2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gu H, Huang Z, Yin X, Zhang J, Gong L,
Chen J, Rong K, Xu J, Lu L and Cui L: Role of c-Jun N-terminal
kinase in the osteogenic and adipogenic differentiation of human
adipose-derived mesenchymal stem cells. Exp Cell Res. 339:112–121.
2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li Y, Wagner ER, Yan Z, Wang Z, Luther G,
Jiang W, Ye J, Wei Q, Wang J, Zhao L, et al: The calcium-binding
protein S100A6 accelerates human osteosarcoma growth by promoting
cell proliferation and inhibiting osteogenic differentiation. Cell
Physiol Biochem. 37:2375–2392. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Su Y, Qi R, Li L, Wang X, Li S, Zhao X,
Hou R, Ma W, Liu D, Zheng J and Shi M: An immune-related gene
prognostic risk index for pancreatic adenocarcinoma. Front Immunol.
13(945878)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang R, Wu Y, Yu J, Yang G, Yi H and Xu B:
Plasma messenger RNAs identified through bioinformatics analysis
are novel, non-invasive prostate cancer biomarkers. Onco Targets
Ther. 13:541–548. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Leclerc E and Vetter SW: The role of S100
proteins and their receptor RAGE in pancreatic cancer. Biochim
Biophys Acta. 1852:2706–2711. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang X, Cao J, Pei Y, Zhang J and Wang Q:
Smad4 inhibits cell migration via suppression of JNK activity in
human pancreatic carcinoma PANC-1 cells. Oncol Lett.
11:3465–3470. 2016.PubMed/NCBI View Article : Google Scholar
|