Introduction
Gastric cancer is a malignant tumor of the digestive
system, and is the fifth most common type of cancer and the fourth
leading cause of cancer mortality worldwide (1). In East Asia, China is one of the
countries with the highest incidence of gastric cancer (1). Notably, gastric cancer displays
heterogeneity with regard to phenotypes and genotypes (2). Stomach adenocarcinoma (STAD), the
main pathological subtype of gastric cancer, arises from the
malignant transformation of somatic cells in the gastric glands,
accounting for ~95% of all cases (3). The primary treatment approach for
gastric cancer is surgical resection. Despite advancements in
endoscopic, surgical and systemic therapies, as well as an
increased focus on multidisciplinary evaluation and treatment, the
5-year survival rates remain unsatisfactory (4). Therefore, it is necessary to urgently
explore new and specific molecular biomarkers for the diagnosis and
prognosis of STAD. These biomarkers may facilitate the development
of targeted diagnostic and therapeutic strategies.
Fas-activated serine/threonine kinase domain 1
(FASTKD1) belongs to the FASTK family, which comprises six members,
including FASTK and its homologs, FASTKD1-5. These proteins have
exclusive expression in the mitochondrial matrix and extensively
regulate diverse mitochondrial functions (5-7).
Each member has distinct functions in governing various aspects of
mitochondrial RNA biology, such as mRNA processing, maturation,
ribosome assembly and translation (5-7).
All six proteins are found solely in vertebrates and share a
conserved arrangement of three homology domains: FAST_1, FAST_2 and
RAP (8). FASTKD1 specifically
regulates the mitochondrial ND3 domain and is thought to serve a
role in RNA stability (8). It has
previously been suggested that FASTKD1 may function as a sensitive
RNA biomarker for endometrial carcinoma when detected in uterine
aspirates (9). Additionally,
upregulation of FASTKD1 has been associated with negative prognoses
in pediatric and adult patients with acute lymphoblastic leukemia
(10). However, to the best of our
knowledge, there is currently no available evidence supporting the
involvement of FASTKD1 in STAD.
The present study aimed to assess the possible
importance of FASTKD1 in STAD by examining The Cancer Genome Atlas
(TCGA) STAD dataset and Gene Expression Omnibus (GEO) datasets.
Using R software and GENT2 online database (http://gent2.appex.kr), the variation in FASTKD1
expression was examined across different tumor types. Additionally,
immunohistochemical staining was performed to verify the
differential expression of FASTKD1 in STAD tumors in comparison to
adjacent tissues. Furthermore, a comprehensive analysis of the
enriched pathways of FASTKD1 was performed, exploring their
biological functions and mechanisms of signal transduction.
Additionally, the correlation between FASTKD1 expression levels,
and immune cell infiltration and m6A modification we investigated.
The present study emphasizes the significant involvement of FASTKD1
in STAD, and indicates its potential as a biomarker for the
diagnosis and prognosis of patients with STAD.
Materials and methods
Expression of FASTKD1 in STAD
The RNA-sequencing (RNA-seq) data utilized in the
present study were sourced from the XENA-TCGA database (https://xena.ucsc.edu) and consist of transcript per
million values. The dataset comprises 10,534 samples, which were
uniformly processed by UCSC XENA using the Toil process (11). To analyze the differences in
FASTKD1 expression across various tumors, the GPL570 platform data
from the GENT2 database was used, which includes microarray
expression data from 72 tumor tissues or cell lines from the GEO
database (12). Furthermore, the
RNA-Seq data from 407 patients diagnosed with STAD were obtained
from TCGA website (https://portal.gdc.cancer.gov); the dataset used
consists of 32 normal adjacent samples and 375 tumor samples from
patients with STAD. The dataset also provides relevant clinical
characteristics for further analysis (13). To explore the differences in
FASTKD1 expression between STAD and normal tissue, GSE27342,
GSE29272, GSE33335 and GSE63089 datasets were collected from the
GEO database (www.ncbi.nlm.nih.gov/geo) (14-17).
The datasets chosen for the present study were obtained from
patients diagnosed with STAD and the number of tumor samples and
normal adjacent samples analyzed in the study were matched
accordingly.
Receiver operating characteristic (ROC) and Kaplan
Meier (KM) curves served as tools to objectively evaluate the
diagnostic and prognostic significance of FASTKD1 in patients with
STAD. The TCGA STAD dataset was utilized to evaluate the ROC
predictive efficacy of FASTKD1. To assess the prognostic
significance of FASTKD1 mRNA expression on overall survival (OS),
the Kaplan Meier Plotter (https://kmplot.com/analysis/) was used, using data
from various resources within the GEO, including GSE14210,
GSE15459, GSE22377, GSE29272 and GSE51105 (18-22).
GSE62254 was excluded from the analysis because it exhibited
significantly distinct clinical and genomic data (longer survival
and shifted expression) upon examination with the Kaplan Meier
Plotter. The resulting KM plots displayed hazard ratios (HR), 95%
confidence intervals (CI) and log-rank P-values. P<0.05 was
considered to indicate a statistically significant difference in
the prognostic outcomes. Furthermore, TCGA STAD dataset was
utilized to investigate the correlation between the expression
levels of FASTKD1 and the clinicopathological characteristics of
patients diagnosed with STAD.
STAD tissue samples
Tumor tissues were collected from 48 patients (the
subjects were between the ages of 28-78, with a median age of 56.5
years and a male to female ratio of 1.82:1) with STAD who underwent
surgical resection at the Taihe Hospital (Shiyan, China) between
January 2019 and April 2022. The research protocol was authorized
by the Ethics Committee of Taihe Hospital affiliated with Hubei
University of Medicine (approval no. 2022KS010). The research was
conducted following the principles stated in the Declaration of
Helsinki and its succeeding amendments.
IHC staining
To prepare for IHC staining, the collected tumor
tissues were fixed in 10% neutral buffered formalin for 24 h at
room temperature, then the tissues were dehydrated using disposable
tissue embedding cassettes and then embedded after being dipped in
wax. The 5-micron pathology sections underwent deparaffinization
with xylene, followed by dehydration with alcohol. Endogenous
peroxidase activity was blocked by applying a 3%
H2O2 solution for 5 min at room temperature.
Antigen retrieval was achieved by incubating the samples in a
pressure cooker for 3 min with sodium citrate buffer (10 mM Sodium
Citrate; 0.05% tween-20; pH 6.0). After blocking with 5% goat serum
(Guangzhou Dingguo Biotechnology Co., Ltd.) at room temperature for
30 min, the sections were incubated overnight at 4˚C with a rabbit
monoclonal FASTKD1 antibody (1:50; cat. no. D122349-0100; Sangon
Biotech). Subsequently, a goat anti-mouse IgG-HRP secondary
antibody (1:200; cat. no. ab6802; Abcam) was applied to the
sections for 1 h at room temperature. Finally, the sections were
stained using DAB (A:B; 1:50) reagent for 3-5 min at room
temperature, followed by counterstaining with hematoxylin for 4
min. After sealing the slices with neutral gum, the results were
analyzed using a light microscope.
Enrichment analysis of the gene
co-expression network for FASTKD1 in STAD
LinkedOmics (https://linkedomics.org/) was used to investigate the
co-expressed genes of FASTKD1 in TCGA STAD. The results were
visually represented by volcano plots and heatmaps (23). In order to further illuminate the
functional characteristics of the co-expressed genes, Gene Ontology
(GO) functional analysis and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis were conducted using the
clusterProfiler package (version 3.14.3) (24) in R software (version 3.6.3)
(25). The resulting bubble chart
displays the top five significant findings based on count scores,
meeting the criteria of the corrected p-value obtained by the
p-value correction method (P.adj) <0.05 and q-value <0.2. The
resulting data were then graphically visualized using the ggplot2
software package (version 3.1.0; https://ggplot2.tidyverse.org).
Gene set enrichment analysis
(GSEA)
To gain deeper insights into the underlying
mechanisms of FASTKD1, TCGA STAD dataset was stratified into high
and low expression groups based on the median expression levels of
FASTKD1. Subsequently, single-gene differential analysis was
conducted using the DESeq2 package (version 1.26.0) (26) to assess variations between the two
groups. Furthermore, all genes displaying differential expression
were subjected to GSEA using the clusterProfiler package (version
3.14.3) (24) to explore
associated biological enrichment processes. The dataset utilized
for GSEA originated from the MSigDB database, accessible at
https://www.gsea-msigdb.org/gsea/msigdb/index.jsp
(27). For GSEA,
c2.cp.all.v2022.1.Hs.symbols.gmt [ALL Canonical Pathways] was
employed as the reference gene set. Enriched pathways meeting the
criteria of FDR (q-value) <0.25 and P<0.05 were visualized
using the ggplot2 package (version 3.1.0).
Tumor-infiltrating immune cells
To investigate the possible role of FASTKD1 in the
regulation of tumor-infiltrating immune cells, the Tumor Immune
Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/) (28) was used. Using this tool, the
correlation between FASTKD1 expression and the presence of
immune-infiltrating cells in TCGA STAD samples was evaluated. The
study focused on B cells, neutrophils, CD4+ T cells,
macrophages, CD8+ T cells and dendritic cells (DCs).
Furthermore, the association between FASTKD1 copy number variation
(CNV) and the infiltration of immune cells was explored utilizing
the SCNA module presented in the TIMER database. The present study
employed the survival module in the TIMER database to illustrate
the correlation between clinical outcomes, immune cell
infiltration, and FASTKD1 gene expression. The GSVA software
package (version 1.34.0) (29) was
used to analyze the expression differences of 24 immune cells in
STAD samples between the groups with high and low expression of
FASTKD1.
FASTKD1 expression and m6A
modification in STAD
The present study utilized the R software (version
3.6.3) to investigate the correlation between FASTKD1 expression
and the expression of 21 m6A-related genes in TCGA STAD dataset.
These genes include eight writers (METTL3, METTL14, RBM15, RBM15B,
WTAP, VIRMA/KIAA1429, CBLL1 and ZC3H13), two erasers (ALKBH5 and
FTO) and 11 readers (YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3,
IGF2BP1, HNRNPA2B1, HNRNPC, FMR1, LRPPRC and ELAVL1) (30). The data obtained from this analysis
were subsequently visualized and assessed using the ggplot2
software package (version 3.1.0). Furthermore, the Kaplan-Meier
Plotter (31) was utilized to
evaluate the prognostic value of these 21 m6A-associated genes in
STAD samples.
Statistical analysis
All statistical analyses in the present study were
performed using R software (version 3.6.3). P<0.05 was
considered to indicate a statistically significant difference in
all analyses. When comparing categorical variables between groups,
either the χ2 test, the χ2 test with Yates
correction, or the Fisher's exact test was used. The statistical
methods used to analyze differences between subgroups of clinical
variables were the independent samples t-test, and one-way ANOVA or
Welch one-way ANOVA followed by the Tukey HSD or Games Howell
post-hoc tests. A comparison was made between changes in 24 immune
cell subtypes in STAD tumor samples using the Wilcoxon rank-sum
test.
Results
Pan-cancer analysis of FASTKD1
expression
FASTKD1 mRNA expression was analyzed across diverse
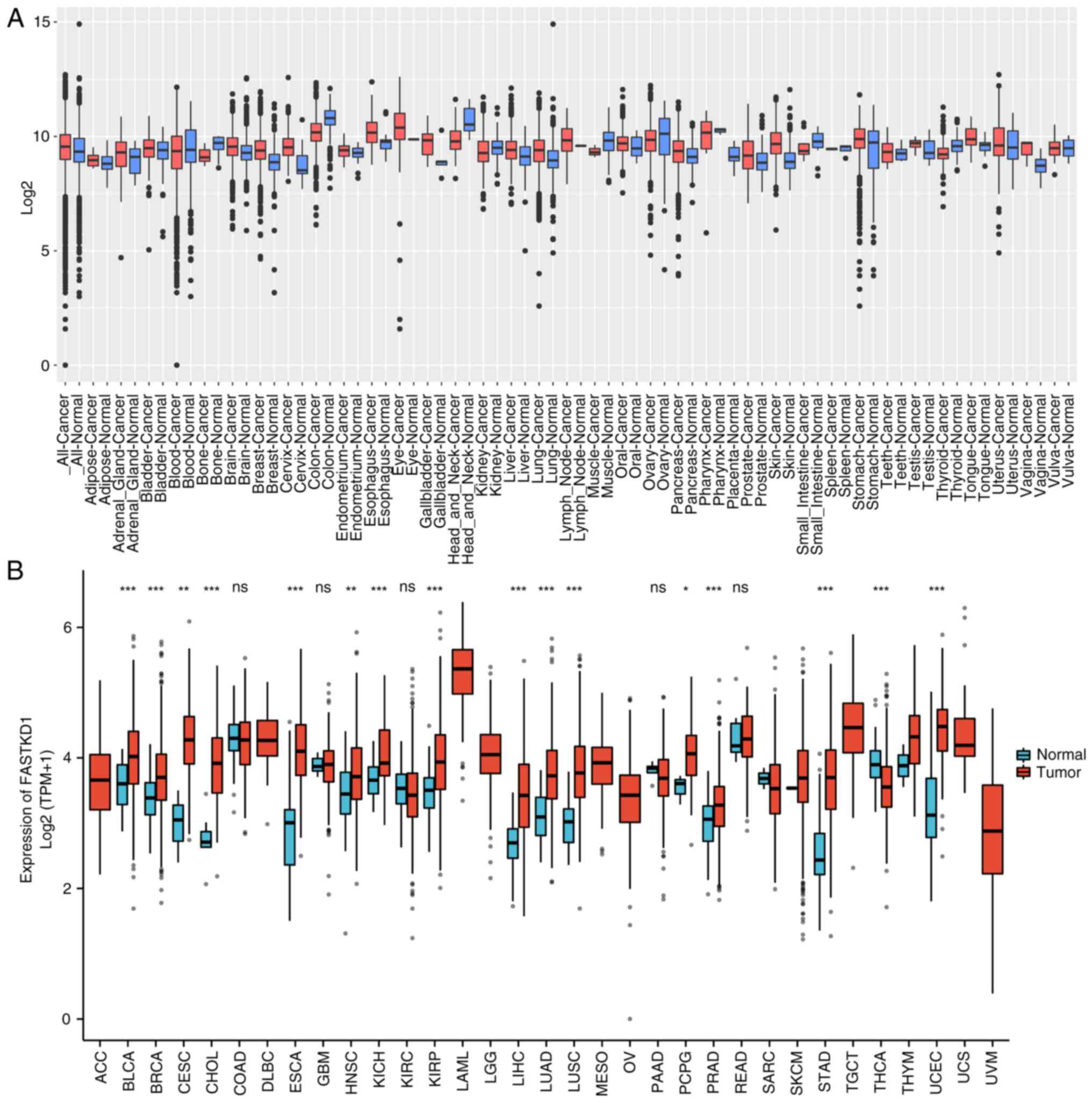
tumor types using the GENT2 and XENA-TCGA datasets. Fig. 1 displays the observed differences
in FASTKD1 expression levels between various tumor types and some
normal adjacent tissues. The findings revealed that FASTKD1
expression was elevated in several cancer types compared with in
normal tissues (Fig. 1A). Further
analysis revealed a significant increase in the mRNA expression
levels of FASTKD1 in various types of tumors, such as bladder
cancer, breast cancer, cervical cancer, bile duct cancer,
esophageal cancer, head and neck cancer, kidney chromophobe, liver
cancer, lung adenocarcinoma, lung squamous cell carcinoma,
pheochromocytoma and paraganglioma, prostate cancer, STAD and
endometrioid cancer (Fig. 1B).
Expression levels of FASTKD1 in
patients with STAD
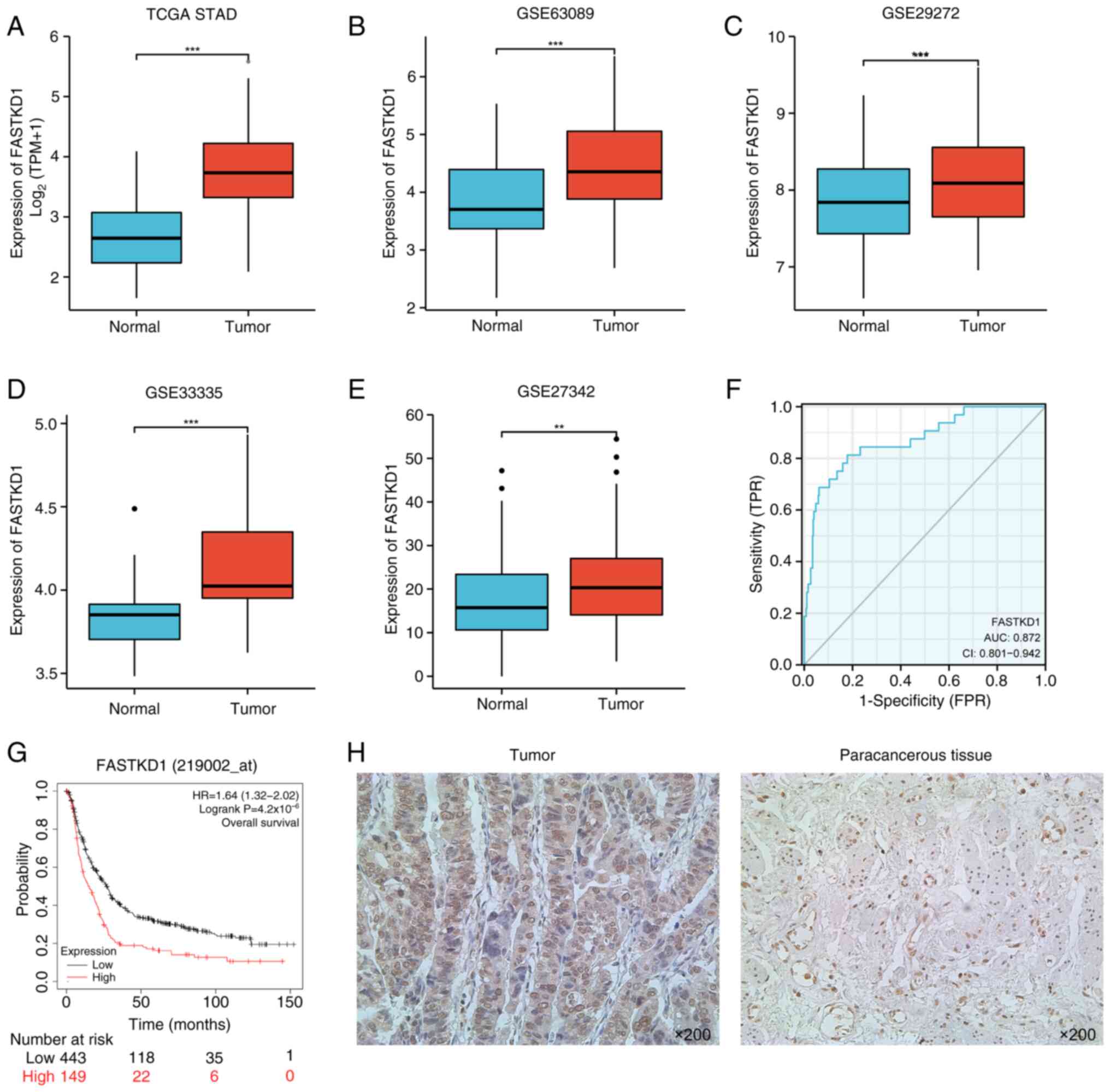
A comparison of FASTKD1 expression between STAD and
some normal adjacent tissues was conducted by analyzing the STAD
datasets from TCGA and GEO. In TCGA dataset, there was a
significant increase in FASTKD1 mRNA expression levels observed in
STAD samples compared with those in control normal samples
(Fig. 2A). Moreover, analysis of
the GSE63089, GSE29272, GSE33335 and GSE27342 datasets demonstrated
a marked increase in the expression levels of FASTKD1 in STAD
samples compared with in control samples (Fig. 2B-E). The ROC analysis revealed an
area under the ROC curve of 0.87 (95% CI, 0.801-0.942; Fig. 2F). This finding indicated that
FASTKD1 expression may accurately differentiate between STAD and
normal samples. Additionally, the survival analysis showed that
high expression of FASTKD1 in STAD could significantly predict poor
survival (HR, 1.64; 95% CI, 1.32-2.02; P=4.2x10-6;
Fig. 2G). In addition, IHC
analysis indicated a marked increase in FASTKD1 protein levels in
STAD tumor tissue compared with in adjacent normal tissue (Fig. 2H). Taken together, the increased
expression levels of FASTKD1 mRNA and protein in STAD tissues
suggest its potential as a diagnostic marker.
Further analysis was conducted on clinical data
obtained from TCGA STAD cohort to investigate the relationship
between FASTKD1 expression and various clinical characteristics. A
total of 407 clinical samples were included in the investigation.
The data revealed that the expression levels of FASTKD1 were higher
in patients aged >65 years in comparison to those aged ≤65 years
(Fig. 3B). However, there were no
significant differences in FASTKD1 expression based on sex or
ethnicity (Fig. 3A and C). In addition, the results indicated
that FASTKD1 expression levels were higher in patients with T1
stage STAD compared with T2 and T3. In addition, higher expression
levels of FASTKD1 were observed in patients with T4 stage STAD
compared with T2 (Fig. 3E).
However, no significant differences were identified among the
analyzed clinical samples in terms of lymph node involvement,
metastasis and pathological stage (Fig. 3D, F and G).
Subsequently, it was revealed that clinical patients who did not
undergo anti-reflux therapy exhibited considerably higher FASTKD1
expression levels compared with the treated individuals (Fig. 3I). However, no notable distinction
was observed in the presence or absence of Helicobacter
pylori infection (Fig. 3H).
The FASTKD1 expression levels were also not significantly different
between deceased and surviving patients in relation to OS,
disease-specific survival and progression-free interval events
(Fig. 3J-L). The clinical features
of FASTKD1 in STAD are summarized in detail in Table I.
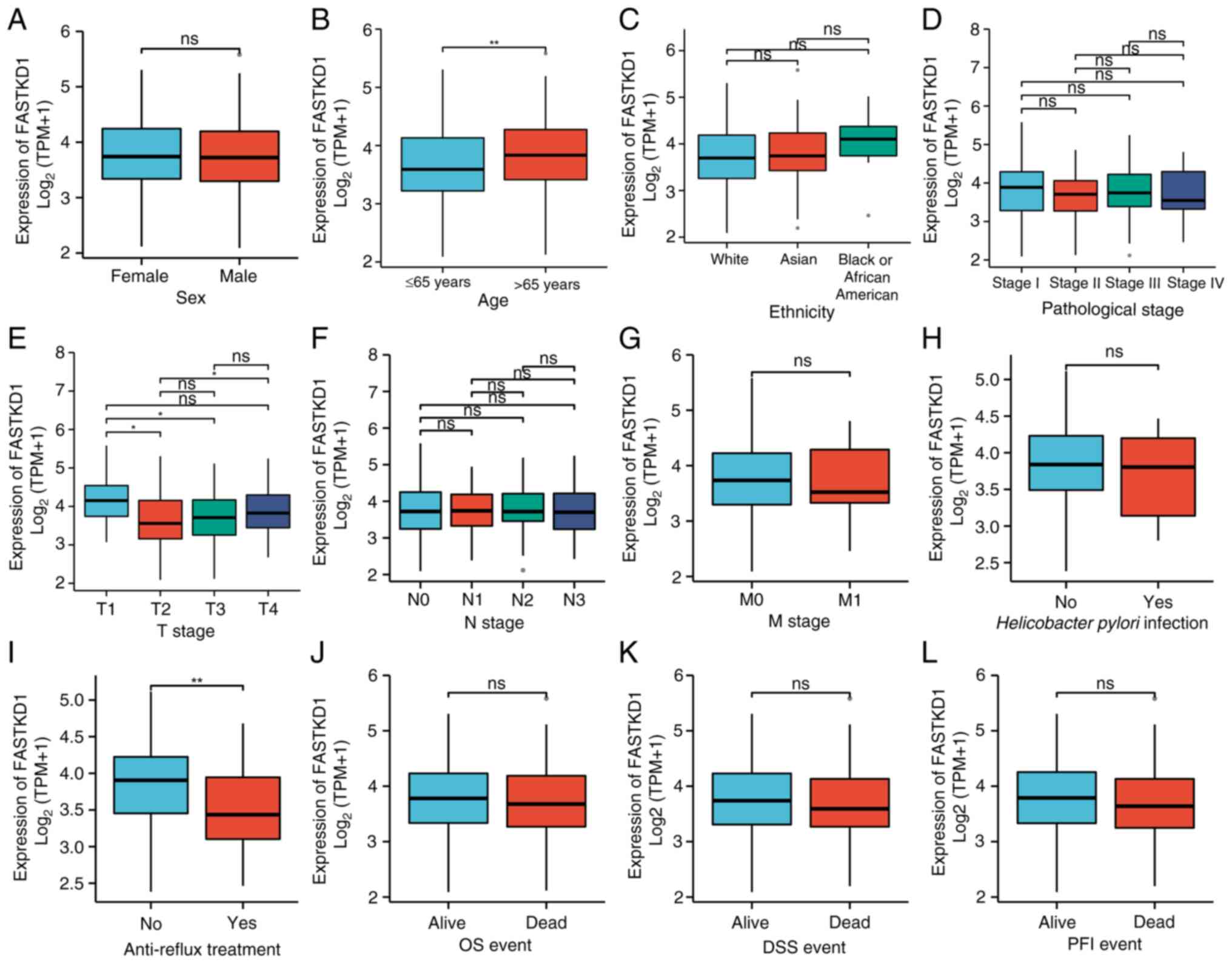 | Figure 3Association of FASTKD1 mRNA
expression levels with clinicopathological characteristics in
patients with stomach adenocarcinoma. (A) Sex, (B) age, (C)
ethnicity, (D) pathological stage, (E) T stage, (F) N stage, (G) M
stage, (H) Helicobacter pylori infection, (I) anti-reflux
treatment, (J) OS event, (K) DSS event, (L) PFI event.
*P<0.05, **P<0.01, ns not significant;
FASTKD1, Fas-activated serine/threonine kinase domain 1; OS,
overall survival; DSS, disease-specific survival; PFI,
progression-free interval. |
 | Table IClinical characteristics of FASTKD1
in stomach adenocarcinoma. |
Table I
Clinical characteristics of FASTKD1
in stomach adenocarcinoma.
|
Characteristics | Low expression of
FASTKD1 (n=41) (%) | High expression of
FASTKD1 (n=54) (%) | P-value |
|---|
| Sex, n (%) | | | 0.639 |
|
Female | 14 (14.7) | 16 (16.8) | |
|
Male | 27 (28.4) | 38 (40.0) | |
| Age, n (%) | | | 0.361 |
|
≤65 | 19 (20.0) | 20 (21.1) | |
|
>65 | 22 (23.2) | 34 (35.8) | |
| Ethnicity, n
(%) | | | 0.247 |
|
Asian | 3 (3.2) | 5 (5.3) | |
|
Black or
African American | 1 (1.1) | 6 (6.3) | |
|
White | 37 (38.9) | 43 (45.3) | |
| Pathological T
stage, n (%) | | | 0.020 |
|
T1 | 1 (1.1) | 0 (0.0) | |
|
T2 | 14 (14.7) | 6 (6.3) | |
|
T3 | 18 (18.9) | 33 (34.7) | |
|
T4 | 8 (8.4) | 15 (15.8) | |
| Pathological N
stage, n (%) | | | 0.923 |
|
N0 | 9 (9.5) | 11 (11.6) | |
|
N1 | 9 (9.5) | 14 (14.7) | |
|
N2 | 11 (11.6) | 16 (16.8) | |
|
N3 | 12 (12.6) | 13 (13.7) | |
| Pathological M
stage, n (%) | | | >0.999 |
|
M0 | 38 (40.0) | 51 (53.7) | |
|
M1 | 3 (3.2) | 3 (3.2) | |
| Pathological stage,
n (%) | | | 0.067 |
|
Stage I | 8 (8.4) | 3 (3.2) | |
|
Stage
II | 5 (5.3) | 13 (13.7) | |
|
Stage
III | 23 (24.2) | 35 (36.8) | |
|
Stage
IV | 5 (5.3) | 3 (3.2) | |
| Helicobacter
pylori infection, n (%) | | | 0.901 |
|
No | 36 (37.9) | 49 (51.6) | |
|
Yes | 5 (5.3) | 5 (5.3) | |
| Reflux history, n
(%) | | | 0.075 |
|
No | 33 (34.7) | 51 (53.7) | |
|
Yes | 8 (8.4) | 3 (3.2) | |
| Anti-reflux
treatment, n (%) | | | 0.023 |
|
No | 30 (31.6) | 49 (51.6) | |
|
Yes | 11 (11.6) | 5 (5.3) | |
| OS event, n
(%) | | | 0.854 |
|
Alive | 22 (23.2) | 30 (31.6) | |
|
Dead | 19(20) | 24 (25.3) | |
| DSS event, n
(%) | | | 0.964 |
|
No | 26 (27.4) | 34 (35.8) | |
|
Yes | 15 (15.8) | 20 (21.1) | |
| PFI event, n
(%) | | | 0.309 |
|
No | 20 (21.1) | 32 (33.7) | |
|
Yes | 21 (22.1) | 22 (23.2) | |
STAD co-expression network analysis of
FASTKD1
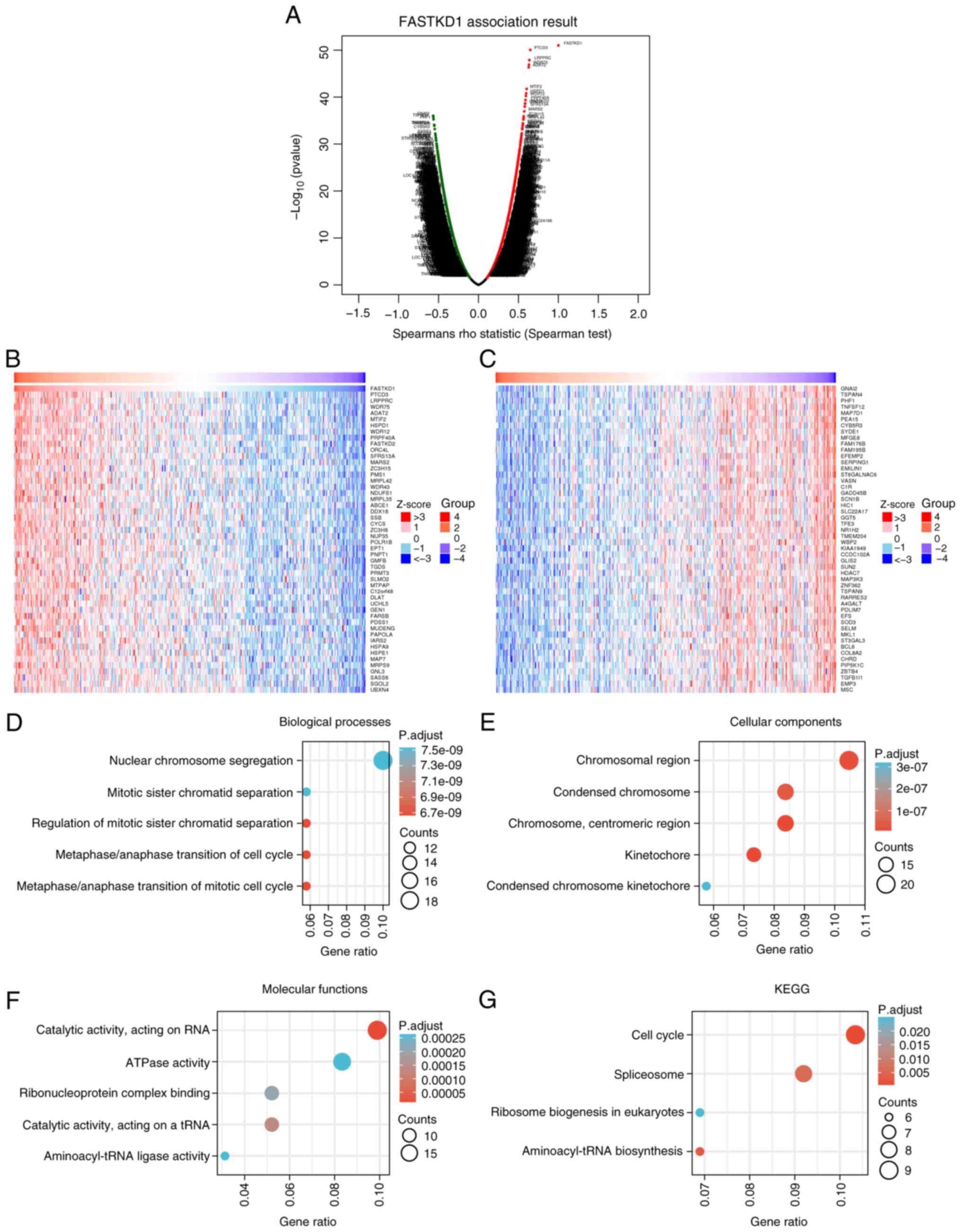
To analyze the co-expressed genes of FASTKD1 in
STAD, LinkedOmics was used. The results showed a positive
correlation between FASTKD1 and 10,229 genes, whereas 9,994 genes
were negatively correlated with FASTKD1 (Fig. 4A). Heatmaps were generated to
visually represent the top 50 genes that exhibited a positive or
negative correlation with FASTKD1 expression. Fig. 4B displays the heatmap for genes
with a positive correlation to FASTKD1 expression, whereas Fig. 4C presents the heatmap for genes
with a negative correlation to FASTKD1 expression. The top 200
co-expressed genes that showed a positive correlation with FASTKD1
expression were subjected to GO/KEGG functional enrichment
analysis. When P.adj <0.05 and q-value <0.2, there were 183
biological process (BP) terms, 46 cellular component (CC) terms, 31
molecular function (MF) terms and four KEGG pathways identified.
The bubble charts present the five most significantly enriched GO
BP, CC and MF terms, and KEGG pathways. The GO bubble map analysis
revealed that FASTKD1 co-expression was mainly linked to ‘nuclear
chromosome segregation’, ‘chromosomal region’, ‘catalytic activity,
acting on RNA’ and ‘ATPase activity’ (Fig. 4D-F). The KEGG pathway bubble map
analysis demonstrated that FASTKD1 co-expression was predominantly
related to ‘Cell cycle’ and ‘Spliceosome’ (Fig. 4G).
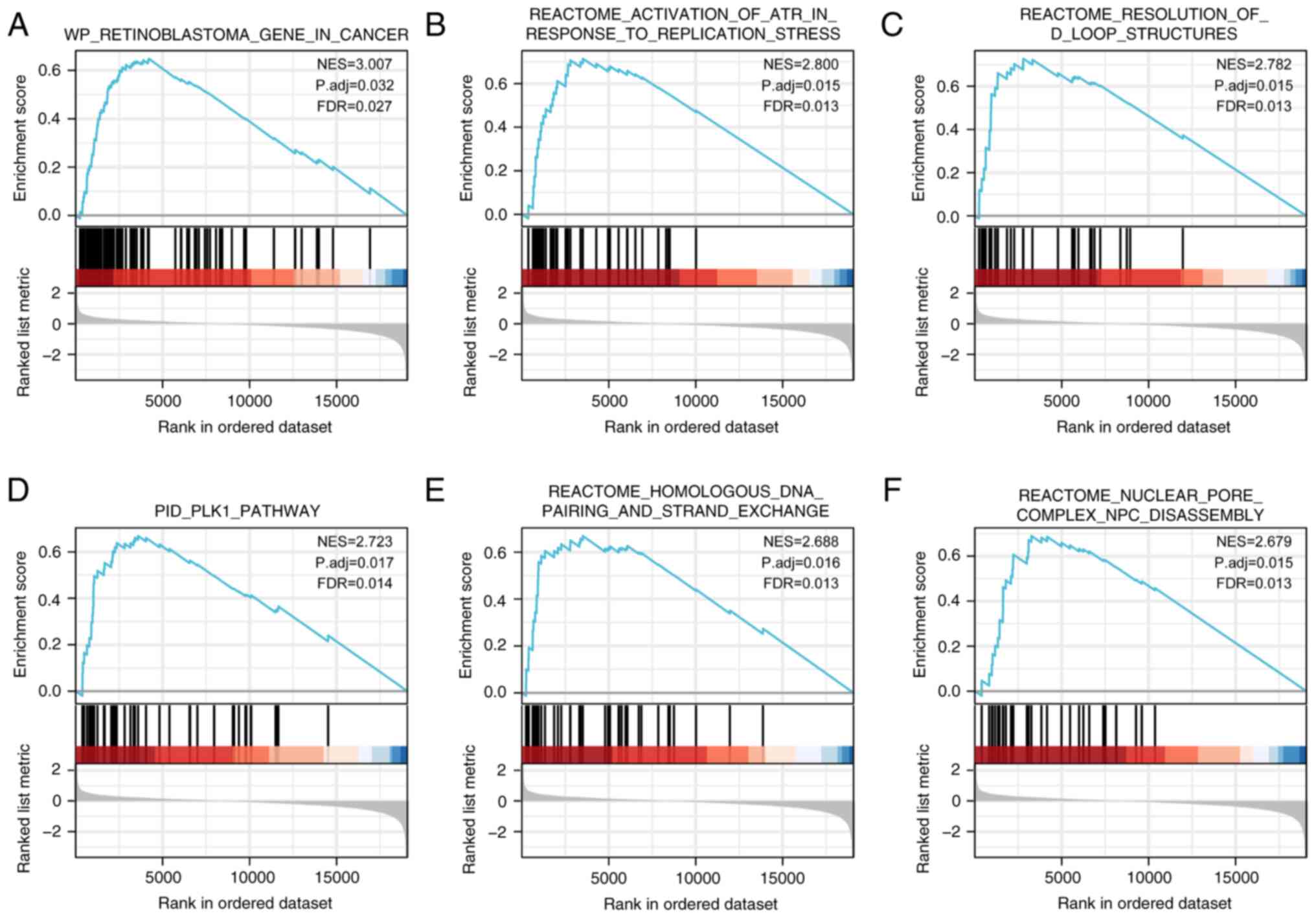
GSEA
To examine the potential role of FASTKD1 in STAD,
GSEA was performed using the single gene differential analysis
associated with FASTKD1. Out of a total of 249 genesets, the top
six results were obtained according to the normalized enrichment
score. These results included ‘WP RETINOBLASTOMA GENE IN CANCER’
(FDR=0.027, P.adj=0.032), ‘REACTOME ACTIVATION OF ATR IN RESPONSE
TO REPLICATION STRESS’ (FDR=0.013, P.adj=0.015), ‘REACTOME
RESOLUTION OF D LOOP STRUCTURES’ (FDR=0.013, P.adj=0.015), ‘PID
PLK1 PATHWAY’ (FDR=0.014, P.adj=0.017), ‘REACTOME HOMOLOGOUS DNA
PAIRING AND STRAND EXCHANGE’ (FDR=0.013, P.adj=0.016) and ‘REACTOME
NUCLEAR PORE COMPLEX NPC DISASSEMBLY’ (FDR=0.013, P.adj=0.015)
(Fig. 5).
Associations between FASTKD1 and
tumor-infiltrating immune cells
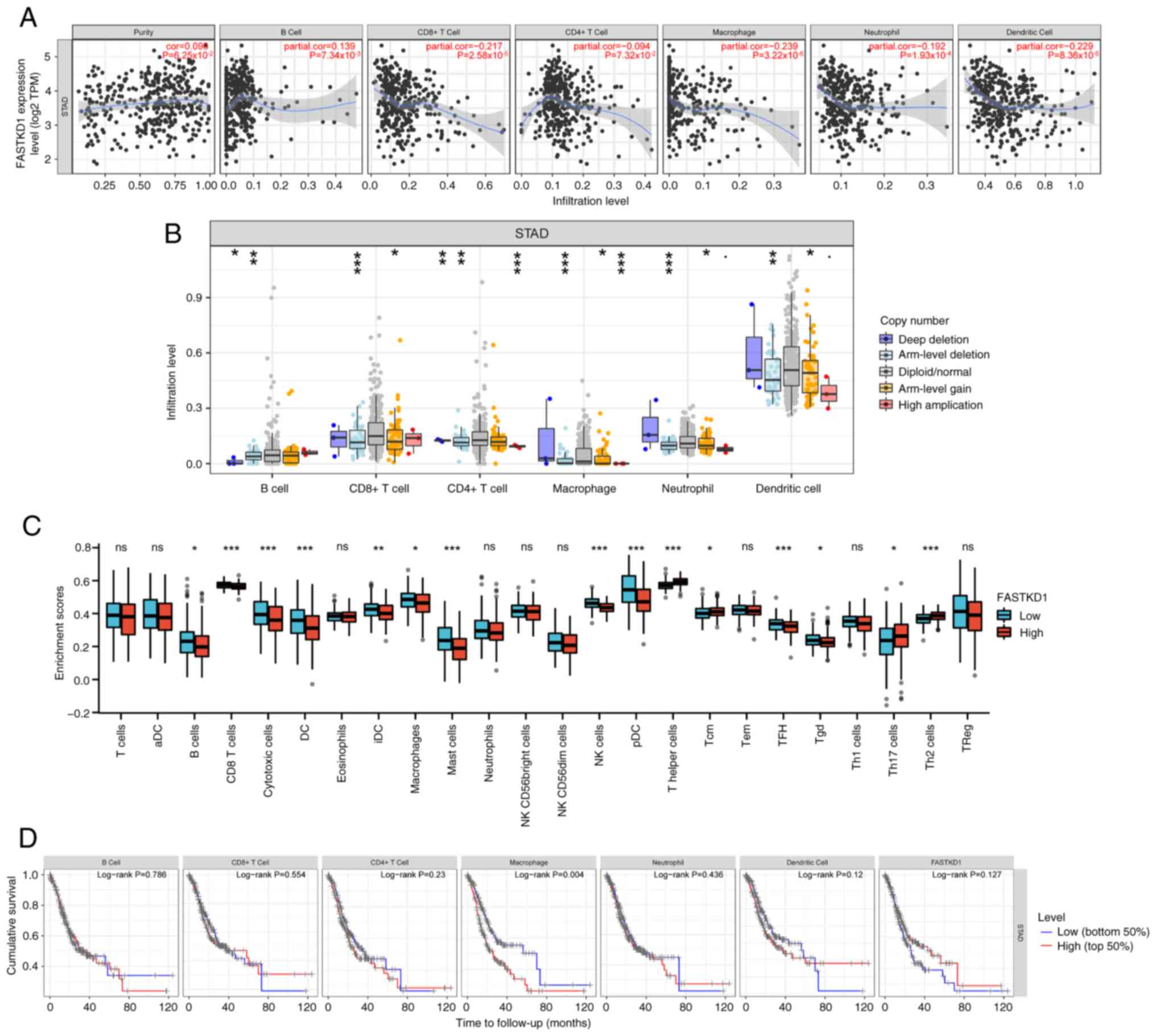
To investigate the possible role of FASTKD1 in tumor
immunity, the present study examined the relationship between
FASTKD1 expression and the presence of immune-infiltrating cells in
STAD. The analysis used the TIMER database and TCGA STAD dataset to
evaluate correlations between FASTKD1 expression and immune cell
infiltration (Fig. 6A) illustrates
a positive correlation between the expression of FASTKD1 and B
cells (ρ=0.139; P=7.34x10-3), and a negative correlation
between the expression of FASTKD1 and CD8+ T cells
(ρ=-0.217; P=2.58x10-5), CD4+ T cells
(ρ=-0.094; P=7.32x10-2), macrophages (ρ=-0.239;
P=3.22x10-6), neutrophils (ρ=-0.192;
P=1.93x10-4) and dendritic cells (ρ=-0.229;
P=8.36x10-6). (Fig. 6B)
illustrates that varying copy numbers of FASTKD1 may affect the
degree of immune infiltration in STAD. The results demonstrated
that the generalized change in FASTKD1 copy number significantly
influenced the level of immune infiltration in STAD, mainly
including deep deletion, arm-level deletion, diploid/normal,
arm-level gain. Based on R package (GSVA package, version 1.34.0)
analysis, it was observed that the expression levels of FASTKD1
were associated with tumor-infiltrating immune cells, such as B
cells (P<0.05), CD8 T cells (P<0.001), cytotoxic cells
(P<0.001), DCs (P<0.001), immature DCs (P<0.01),
macrophages (P<0.05), mast cells (P<0.001), natural killer
(NK) cells (P<0.001), plasmacytoid DCs (P<0.001), T helper
(Th) cells (P<0.001), central memory T cells (P<0.05), T
follicular helper cells (P<0.001), γδ T cells (P<0.05), Th17
cells (P<0.05) and Th2 cells (P<0.001) (Fig. 6C). Kaplan-Meier plots of immune
infiltration and the FASTKD1 gene were generated using the TIMER
online database to visualize survival differences, the use of KM
curves demonstrated that the prognosis of STAD was associated with
macrophages (P=0.004; Fig.
6D).
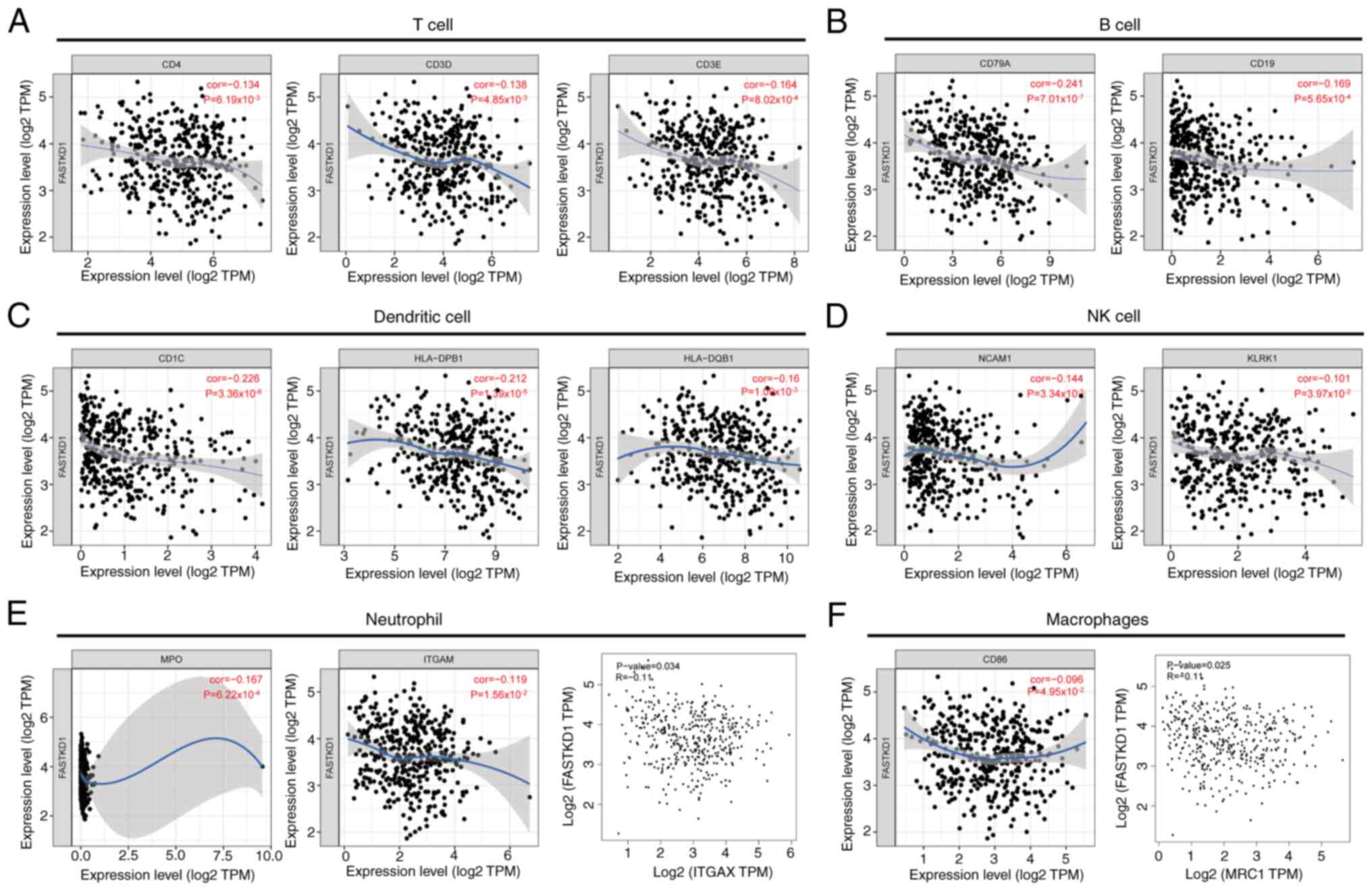
Further exploration is required to investigate the
role of FASTKD1 in tumor immunity. Therefore, the present study
analyzed the correlation between the expression levels of FASTKD1
in STAD and various immune infiltration markers, using the TIMER
and GEPIA databases. The expression of FASTKD1 was weakly
negatively correlated with T-cell biomarkers (CD4, CD3D and CD3E),
B-cell biomarkers (CD79A and CD19), DC biomarkers (CD1C, HLA-DPB1
and HLA-DQB1), NK cell biomarkers (NCAM1 and KLRK1), neutrophil
biomarkers (MPO, ITGAM and ITGAX) and macrophage biomarkers (CD86
and MRC1) (Fig. 7).
Correlations between the expression
levels of FASTKD1 and m6A modification in STAD
Accumulating evidence has supported the crucial
involvement of m6A modifications in processes such as inflammation,
innate immunity and antitumor responses. These processes are
mediated by interactions with various m6A regulatory factors
(32-34).
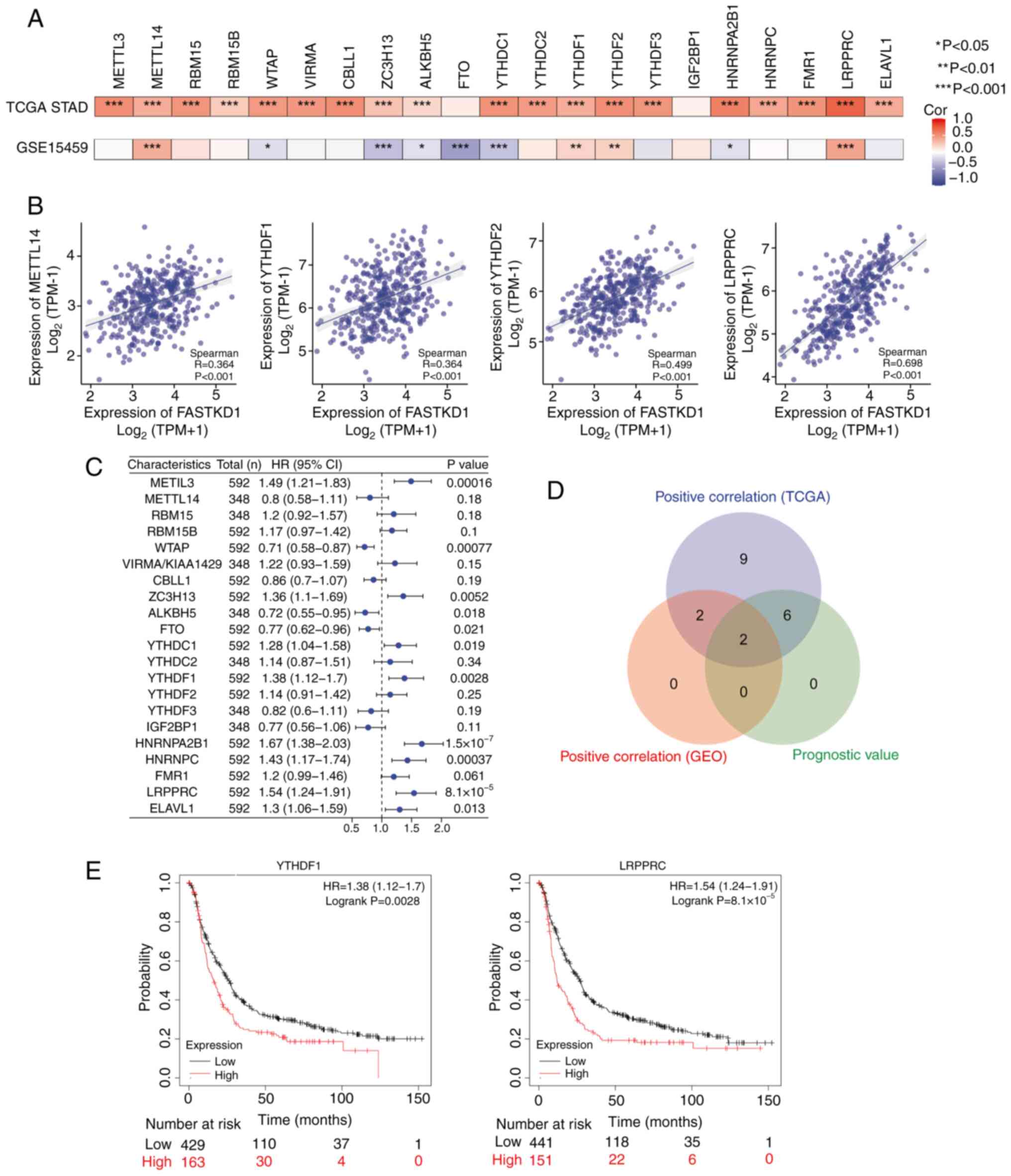
The present study analyzed both TCGA STAD cohort and the GSE15459
cohort to investigate the correlation between the expression levels
of FASTKD1 and 21 m6A-related genes in STAD (Fig. 8A) shows TCGA STAD and GSE15459
datasets. In TCGA STAD dataset, significant positive correlations
were detected between FASTKD1 and the expression levels of METTL3,
METTL14, RBM15, RBM15B, WTAP, VIRMA/KIAA1429, CBLL1, ZC3H13,
ALKBH5, YTHDC1, YTHDC2, YTHDF3, HNRNPA2B1, HNRNPC, FMR1, LRPPRC and
ELAVL1. In the GSE15459 dataset, significant positive correlations
were detected between FASTKD1 and the expression levels of METTL14,
YTHDF1, YTHDF2 and LRPPRC. In the GSE15459 dataset, there were also
negative correlations detected between FASTKD1 and the expression
levels of WTAP, ZC3H13, ALKBH5, FTO, YTDHDC1 and HNRNPA2B1. Scatter
plots (Fig. 8B) were created to
illustrate the correlation between FASTKD1 and m6A
modification-related genes. Furthermore, a forest plot (Fig. 8C) was employed to present the
prognostic importance of the 21 m6A-related genes in STAD. A Venn
plot (Fig. 8D) was also generated
to display the intersection of m6A expression-related genes and
prognostic genes. For the overlapping genes, KM survival analysis
was performed. The KM-plotter indicated that high expression levels
of YTHDF1 (HR=1.38; log-rank P=0.0028) and LRPPRC (HR=1.54;
log-rank P=8.1x10-5) were associated with a worse
prognosis in STAD (Fig. 8E). The
present study identified an association between FASTKD1 and m6A
modifications in STAD, especially through its interaction with the
LRPPRC and YTHDF1 genes. This interaction may affect the
progression and prognosis of STAD.
Discussion
STAD is a type of cancer resulting from the
malignant transformation of somatic cells in the gastric glands. It
is one of the most prevalent types of gastrointestinal cancer, with
an estimated annual incidence of >1 million cases worldwide
(2). Due to its frequently
advanced stage upon diagnosis, the mortality rates associated with
gastric cancer remain high. In addition, although the cancer
incidence rate is decreasing in most countries, clinicians predict
an increase in cancer cases in the future due to the aging
population. Therefore, researching new molecular targets and
pathways is crucial for providing innovative insights into the
diagnosis, treatment and prognosis of STAD.
FASTKD1 is a member of the FASTK family, which
includes six members: FASTK, the original member, and its homologs,
FASTKD1-5. These FASTK family proteins are found exclusively in
vertebrates and have widespread expression in mitochondria across
multiple body tissues. They serve a vital role in preserving and
stabilizing mitochondria, emerging as critical regulators of
post-transcriptional gene expression within these organelles
(6). FASTKD1 is situated on
chromosome 2q31.1 and is expressed in the mitochondrial matrix. It
features an amino-terminal mitochondrial targeting signal and a
C-terminal region with three conserved domains, FAST1, FAST2 and
RAP (35). FASTKD1 controls the
ND3 domain within mitochondria and may participate in processes
associated with RNA stability. Furthermore, FASTKD1 operates as a
protective factor against reactive oxygen species-induced oxidative
stress and cell death, although the exact mechanism of this
protection is currently unknown (5). Additionally, FASTKD1 alters
mitochondrial dynamics in a CypD-independent manner and impacts
processes, such as autophagy/mitophagy and caspase-3 activation
(35). However, limited research
has examined the role of FASTKD1 in carcinogenesis (36).
The present study analyzed XENA-TCGA and GEO
datasets to observe the expression levels of FASTKD1 in various
tumors, including STAD. The elevated expression of FASTKD1 in STAD
tissue was subsequently confirmed through IHC staining. In
addition, the present study revealed that increased levels of
FASTKD1 expression may aid the diagnosis of STAD, and could be
strongly linked to unfavorable prognosis and clinical features in
patients with STAD. For example, administering anti-reflux therapy
was shown to significantly reduce the expression levels of FASTKD1
in patients with STAD.
The GO/KEGG enrichment analysis revealed that the
co-expressed genes of FASTKD1 were active in a variety of terms and
pathways, such as ‘nuclear chromosome segregation’, ‘chromosomal
region’, ‘catalytic activity, acting on RNA’, ‘ATPase activity’,
‘Cell cycle’ and ‘Spliceosome’. Several studies have demonstrated
the significant involvement of the aforementioned biological
functions in the initiation and progression of tumors (37-40).
Proper segregation of nuclear chromosomes is crucial to maintaining
genomic stability during cell division and errors in this process
can lead to aneuploidy, a condition in which cells have an abnormal
number of chromosomes, which is often observed in cancer cells.
Aneuploidy contributes to genomic instability, promotes
tumorigenesis and enables the acquisition of genetic alterations
that propel tumor progression (41). Structural variations,
amplifications, deletions and rearrangements in particular
chromosomal regions are associated with different types of cancer,
and these changes bring about alterations in gene dosage,
disruption of regulatory elements, and activation or inactivation
of oncogenes and tumor suppressor genes found within these regions.
These alterations can lead to uncontrolled growth and survival of
cells, and promote the development and progression of tumors
(42). Catalytic activity,
specifically acting on RNA, includes RNAs involved in processing,
modification and degradation, and dysregulation of these RNA
activities can affect critical cellular processes, including the
stability, translation and splicing of mRNA. Aberrant RNA
metabolism is frequently observed in cancer cells and can lead to
altered gene expression patterns, which promote tumor cell
proliferation and survival (43).
ATPase activity is crucial to multiple cellular processes, such as
DNA repair, chromatin remodeling and protein folding, whereas
dysregulated ATPase activity in cancer is associated with defects
in DNA damage response and repair mechanisms, compromised chromatin
structure and altered protein homeostasis; these disruptions can
contribute to genomic instability, a hallmark of cancer, and
promote tumor progression (44).
The cell cycle is regulated to guarantee precise DNA replication
and cell division, whereas excessive cell proliferation and
accumulation of genetic abnormalities can be caused by the
dysregulation of checkpoints. Notably, dysregulation of the cell
cycle is a hallmark of cancer, contributing to uncontrolled cell
proliferation, genomic instability and tumor progression (45). The spliceosome is a sophisticated
molecular apparatus responsible for RNA splicing, which is a key
process in the regulation of gene expression; notably,
dysregulation of splicing events can lead to the production of
abnormal isoforms with oncogenic properties. In addition,
disruptions in spliceosome components or splicing factors have been
reported in numerous types of cancer, and markedly affect essential
cellular processes, including cell proliferation, survival and
metastasis, thereby contributing to tumor progression (46).
Single gene differential GSEA analysis revealed that
the differentially expressed genes associated with FASTKD1 were
enriched in the following pathways: ‘Wp Retinoblastoma Gene in
Cancer’, ‘Reactome Activation Of Atr in Response to Replication
Stress’, ‘Reactome Resolution of D Loop Structures’, ‘Pid Plk1
Pathway’, ‘Reactome Homologous Dna Pairing and Strand Exchange’ and
‘Reactome Nuclear Pore Complex Npc Disassembly’. These cellular
processes and pathways serve a critical role in the initiation and
progression of cancer. A complete understanding of their
dysregulation and functional implications may provide valuable
insights into the underlying mechanisms of STAD. The retinoblastoma
pathway in cancer involves the central role of the RB1 gene in
regulating cell cycle progression and inhibiting tumorigenesis;
notably, RB1 disruptions, caused by mutations or inactivation, can
significantly contribute to the development of various types of
cancer (47). Activation of ATR in
response to replication stress is a pathway related to the role of
ATR proteins in response to DNA replication stress; ATR serves a
critical role in maintaining genomic integrity by initiating
signaling pathways involved in the DNA damage response, leading to
cell cycle arrest and DNA repair (48). The association between D-loop
resolution and homologous recombination suggests a DNA repair
mechanism responsible for the accurate repair of DNA double-strand
breaks; accurate D-loop resolution has a critical role in
maintaining genomic stability and preventing the accumulation of
genetic abnormalities (49). PLK1
is a critical regulator of cell division and exerts its influence
at multiple stages of mitosis, including mitotic entry, spindle
assembly, chromosome segregation and cytokinesis; by contrast,
disruption of PLK1 activity has been reported in several cancer
types, and is associated with aberrant cell division, chromosomal
instability and tumor progression (50). Homologous DNA pairing and strand
exchange is a pathway associated with the critical steps in DNA
repair by homologous recombination; specifically, this pathway
highlights the importance of homologous DNA pairing and subsequent
strand exchange events, since dysregulation of these complex
processes can lead to genomic instability and increase the risk of
tumorigenesis (51). The NPC
disassembly pathway refers to the process of NPC disassembly during
mitosis to facilitate accurate chromosome segregation;
dysregulation of NPC disassembly disrupts normal cell division and
contributes to chromosomal instability, a common feature observed
in cancer cells (52). The
intricate relationship between these processes and pathways
suggests their potential involvement in the biological functions
and mechanisms associated with FASTKD1 in STAD. Further
investigation of these pathways may provide valuable insights into
the role of FASTKD1 in the development and progression of STAD.
The assessment of immune infiltration of tumor cells
has become increasingly important in cancer research and clinical
practice. Understanding the composition and functional properties
of the immune infiltrate can assist in identifying potential
therapeutic targets and developing immunotherapies. The present
study provided valuable insights into the interplay between FASTKD1
and the immune microenvironment in the context of STAD. The results
indicated a negative correlation between the expression levels of
FASTKD1 in STAD and the presence of several immune cell types.
Specifically, a negative correlation was observed between FASTKD1
expression and the infiltration of CD8+ T cells,
CD4+ T cells, macrophages, neutrophils and DCs. These
results suggested that FASTKD1 may serve a role in modulating the
tumor immune microenvironment in STAD by potentially affecting the
abundance or activity of these immune cell populations (53).
The present study also revealed that CNVs in FASTKD1
may have an impact on the levels of infiltrating immune cells in
STAD. Changes in FASTKD1 CNV were shown to be associated with
changes in the abundance of B cells, CD4+ T cells,
macrophages, neutrophils and DCs within the tumor microenvironment.
In the present study, it was demonstrated that the prevalent copy
number alterations associated with FASTKD1 in STAD were
characterized by two patterns: Arm-level deletion and arm-level
gain. Arm-level deletion is a genomic alteration commonly observed
in various tumor types, including STAD. It is characterized by the
loss of genetic material spanning an entire chromosomal arm. The
loss of genetic material from an entire chromosomal arm can result
in the inactivation or loss of multiple genes located within that
region. These genes may include tumor suppressor genes, which
normally regulate cell proliferation and prevent tumor formation
(54). By contrast, arm-level gain
is often associated with amplification of oncogenes, which are
genes that promote tumor growth when abnormally activated or
amplified. The increased copy number of these oncogenes leads to
increased expression or functional activity, which drives tumor
proliferation and survival (55).
These findings suggested a potential role for FASTKD1 CNV in
modulating the composition of immune cells and potentially
influencing the immune response in STAD. In the context of disease,
CNVs have been implicated in a variety of conditions, including
developmental disorders, neurodegenerative diseases (56,57).
In cancer, CNVs may contribute to tumorigenesis by affecting
oncogenes or tumor suppressor genes, disrupting key signaling
pathways or altering the genomic stability of cancer cells
(58,59).
By comparing groups with high and low FASTKD1
expression, the differential immune cell composition between the
two groups can be examined. This analysis aims to uncover potential
differences in the abundance of various immune cell subtypes,
including, but not limited, to T cells (including CD8+ T
cells and CD4+ T cells), B cells, NK cells, macrophages,
neutrophils, DCs and other identifiable immune cell populations.
Notably, a significant association between macrophage infiltration
and survival was detected in patients with STAD. This finding
suggested that the presence or abundance of macrophages in the
tumor microenvironment may have a role in determining patient
outcome (60). In addition.
FASTKD1 expression was revealed to be weakly negatively correlated
with most immune infiltration markers in the TIMER and GEPIA
databases. This finding suggested that, as FASTKD1 expression
increases, the expression or abundance of immune cell infiltration
markers tends to decrease. It is possible that FASTKD1 directly or
indirectly affects the release of immunosuppressive factors or
alters the expression of chemokines that drive immune cell
migration. A comprehensive analysis to investigate the relationship
between FASTKD1 expression and changes in the immune cell landscape
within STAD tumors may provide valuable insights into the potential
role of FASTKD1 in influencing the immune microenvironment.
m6A modification is a common and reversible RNA
modification that has an important role in post-transcriptional
gene regulation. In recent years, extensive research has been
conducted to understand the functional significance of m6A
modification and its impact on various biological processes
(61-63).
In the present study, a significant correlation was observed
between the expression levels of FASTKD1 and two genes, namely
YTHDF1 and LRPPRC. YTHDF1 is a member of the YTH domain family of
RNA-binding proteins that specifically recognizes and binds to
m6A-modified mRNA. As an m6A reader, YTHDF1 has a role in the
regulation of RNA metabolism and translation. It promotes
translation efficiency by interacting with the translation
initiation machinery and recruiting ribosomes to m6A-modified
transcripts. YTHDF1 also influences mRNA decay processes, where it
can either stabilize or promote degradation of m6A-modified
transcripts, depending on the context (64). LRPPRC is a versatile protein
involved in several cellular processes, including mitochondrial
function and RNA metabolism, which has been implicated in the
stabilization and maintenance of mitochondrial transcripts, as well
as the post-transcriptional regulation of nuclear-encoded mRNAs. It
interacts with specific RNA targets, including those involved in
oxidative phosphorylation and mitochondrial biogenesis, to regulate
their processing, stability or translation (65,66).
The present study detected a significant correlation between
FASTKD1 expression and both YTHDF1 and LRPPRC. This correlation
suggested a possible relationship between FASTKD1 and these two
genes in the context of post-transcriptional gene regulation.
Further investigation is required to determine the nature and
functional implications of these correlations, such as whether
FASTKD1 directly interacts with YTHDF1 and LRPPRC, or whether they
share a common regulatory pathway or mechanism.
The present study has some limitations. Firstly, the
association between high FASTKD1 expression and poor prognosis in
patients with STAD lacked validation from clinical trials.
Secondly, despite combining metabolic and genomic signatures to
explore potential biomarkers and underlying mechanisms, the lack of
molecular biology validation represents another limitation of this
study. Future studies should focus on addressing this limitation to
provide more robust evidence for the identified biomarkers and
mechanisms.
In conclusion, the present study demonstrated that
FASTKD1 may be upregulated in STAD, and identified its associations
with clinical characteristics and survival in patients with STAD.
Furthermore, the interaction between FASTKD1, immune infiltration
and m6A modification was investigated. The results indicated that
FASTKD1 is a promising independent diagnostic and prognostic marker
for STAD, and may be a potential target for future molecularly
targeted therapies.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Hubei Province's
Outstanding Medical Academic Leader program, the Foundation for
Innovative Research Team of Hubei Provincial Department of
Education (grant no. T2020025), the Free-exploring Foundation of
Hubei University of Medicine (grant no. FDFR201903), the Innovative
Research Program for Graduates of Hubei University of Medicine
(grant nos. YC2023007 and YC2023035), and the Key Discipline
Project of Hubei University of Medicine.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YY performed the experiments, analyzed the results
and wrote the manuscript. YG assisted in completing
immunohistochemical staining and scoring based on a points system.
ZMH, XSL, YZ, YHZ, ZYL and YXC interpreted the results. YY
collected the clinical samples. ZJP designed and supervised the
project, and revised and finalized the manuscript. YY and YG
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The studies involving human participants were
reviewed and approved by the Ethics Committee of Taihe Hospital
affiliated with Hubei University of Medicine (approval no.
2022KS010). Written informed consent for participation was not
required for this study in accordance with the national legislation
and the institutional requirements. The Institutional Review Board
waived the need for informed consent due to the retrospective
nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, AI tools were
used to improve the readability and language of the manuscript or
to generate images, and subsequently, the authors revised and
edited the content produced by the AI tools as necessary, taking
full responsibility for the ultimate content of the present
manuscript.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Johnston FM and Beckman M: Updates on
management of gastric cancer. Curr Oncol Rep. 21(67)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang FH, Zhang XT, Li YF, Tang L, Qu XJ,
Ying JE, Zhang J, Sun LY, Lin RB, Qiu H, et al: The Chinese society
of clinical oncology (CSCO): Clinical guidelines for the diagnosis
and treatment of gastric cancer, 2021. Cancer Commun (Lond).
41:747–795. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Simarro M, Gimenez-Cassina A, Kedersha N,
Lazaro JB, Adelmant GO, Marto JA, Rhee K, Tisdale S, Danial N,
Benarafa C, et al: Fast kinase domain-containing protein 3 is a
mitochondrial protein essential for cellular respiration. Biochem
Biophys Res Commun. 401:440–466. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Boehm E, Zaganelli S, Maundrell K,
Jourdain AA, Thore S and Martinou JC: FASTKD1 and FASTKD4 have
opposite effects on expression of specific mitochondrial RNAs,
depending upon their endonuclease-like RAP domain. Nucleic Acids
Res. 45:6135–6146. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jourdain AA, Koppen M, Rodley CD,
Maundrell K, Gueguen N, Reynier P, Guaras AM, Enriquez JA, Anderson
P, Simarro M and Martinou JC: A mitochondria-specific isoform of
FASTK is present in mitochondrial RNA granules and regulates gene
expression and function. Cell Rep. 10:1110–1121. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jourdain AA, Popow J, de la Fuente MA,
Martinou JC, Anderson P and Simarro M: The FASTK family of
proteins: Emerging regulators of mitochondrial RNA biology. Nucleic
Acids Res. 45:10941–10947. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Colas E, Perez C, Cabrera S, Pedrola N,
Monge M, Castellvi J, Eyzaguirre F, Gregorio J, Ruiz A, Llaurado M,
et al: Molecular markers of endometrial carcinoma detected in
uterine aspirates. Int J Cancer. 129:2435–2444. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang J, Mi JQ, Debernardi A, Vitte AL,
Emadali A, Meyer JA, Charmpi K, Ycart B, Callanan MB, Carroll WL,
et al: A six gene expression signature defines aggressive subtypes
and predicts outcome in childhood and adult acute lymphoblastic
leukemia. Oncotarget. 6:16527–16542. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Park SJ, Yoon BH, Kim SK and Kim SY:
GENT2: An updated gene expression database for normal and tumor
tissues. BMC Med Genomics. 12 (Suppl 5)(S101)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo
J, Ni Z, Zhang M, Kong X, Hoffman LL, et al: An integrated
transcriptomic and computational analysis for biomarker
identification in gastric cancer. Nucleic Acids Res. 39:1197–1207.
2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li WQ, Hu N, Burton VH, Yang HH, Su H,
Conway CM, Wang L, Wang C, Ding T, Xu Y, et al: PLCE1 mRNA and
protein expression and survival of patients with esophageal
squamous cell carcinoma and gastric adenocarcinoma. Cancer
Epidemiol Biomarkers Prev. 23:1579–1588. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cheng L, Wang P, Yang S, Yang Y and Zhang
Q, Zhang W, Xiao H, Gao H and Zhang Q: Identification of genes with
a correlation between copy number and expression in gastric cancer.
BMC Med Genomics. 5(14)2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang X, Ni Z, Duan Z, Xin Z, Wang H, Tan
J, Wang G and Li F: Overexpression of E2F mRNAs associated with
gastric cancer progression identified by the transcription factor
and miRNA co-regulatory network analysis. PLoS One.
10(e0116979)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A,
Michalowski A and Green JE: A gene expression signature of acquired
chemoresistance to cisplatin and fluorouracil combination
chemotherapy in gastric cancer patients. PLoS One.
6(e16694)2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ooi CH, Ivanova T, Wu J, Lee M, Tan IB,
Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al: Oncogenic
pathway combinations predict clinical prognosis in gastric cancer.
PLoS Genet. 5(e1000676)2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Förster S, Gretschel S, Jöns T, Yashiro M
and Kemmner W: THBS4, a novel stromal molecule of diffuse-type
gastric adenocarcinomas, identified by transcriptome-wide
expression profiling. Mod Pathol. 24:1390–1403. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang G, Hu N, Yang HH, Wang L, Su H, Wang
C, Clifford R, Dawsey EM, Li JM, Ding T, et al: Comparison of
global gene expression of gastric cardia and noncardia cancers from
a high-risk population in China. PLoS One. 8(e63826)2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Busuttil RA, George J, Tothill RW,
Ioculano K, Kowalczyk A, Mitchell C, Lade S, Tan P, Haviv I and
Boussioutas A: A signature predicting poor prognosis in gastric and
ovarian cancer represents a coordinated macrophage and stromal
response. Clin Cancer Res. 20:2761–2772. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46 (D1):D956–D963. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Team RC. R: A language and environment for
statistical computing. MSOR MSOR connections, pp1, 2014.
|
|
26
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14(7)2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang B, Wu Q, Li B, Wang D, Wang L and
Zhou YL: m6A regulator-mediated methylation modification
patterns and tumor microenvironment infiltration characterization
in gastric cancer. Mol Cancer. 19(53)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lánczky A and Győrffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res.
23(e27633)2021.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
An Y and Duan H: The role of m6A RNA
methylation in cancer metabolism. Mol Cancer. 21(14)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu C, Yang Z, Li R, Wu Y, Chi M, Gao S,
Sun X, Meng X and Wang B: Potential roles of N6-methyladenosine
(m6A) in immune cells. J Transl Med. 19(251)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen X, Gong W, Shao X, Shi T, Zhang L,
Dong J, Shi Y, Shen S, Qin J, Jiang Q and Guo B: METTL3-mediated
m6A modification of ATG7 regulates autophagy-GATA4 axis
to promote cellular senescence and osteoarthritis progression. Ann
Rheum Dis. 81:87–99. 2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Marshall KD, Klutho PJ, Song L, Krenz M
and Baines CP: The novel cyclophilin-D-interacting protein FASTKD1
protects cells against oxidative stress-induced cell death. Am J
Physiol Cell Physiol. 317:C584–C599. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ramasubramanian A, Paramasivam A and
Ramani P: FASTK family of genes linked to cancer. Bioinformation.
18:206–213. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nevins JR: Cell cycle targets of the DNA
tumor viruses. Curr Opin Genet Dev. 4:130–134. 1994.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Valimehr S, Sethi A, Shukla M,
Bhattacharyya S, Kazemi M and Rouiller I: Molecular mechanisms
driving and regulating the AAA+ ATPase VCP/p97, an important
therapeutic target for treating cancer, neurological and infectious
diseases. Biomolecules. 13(737)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Sciarrillo R, Wojtuszkiewicz A, Assaraf
YG, Jansen G, Kaspers GJL, Giovannetti E and Cloos J: The role of
alternative splicing in cancer: From oncogenesis to drug
resistance. Drug Resist Updat. 53(100728)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Church AJ, Akkari Y, Deeb K, Kolhe R, Lin
F, Spiteri E, Wolff DJ and Shao L: ACMG Laboratory Quality
Assurance Committee. Electronic address: documents@acmg.net.
Section E6.7-6.12 of the American college of medical genetics and
genomics (ACMG) technical laboratory standards: Cytogenomic studies
of acquired chromosomal abnormalities in solid tumors. Genet Med.
26(101070)2024.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Klaasen SJ, Truong MA, van Jaarsveld RH,
Koprivec I, Štimac V, de Vries SG, Risteski P, Kodba S, Vukušić K,
de Luca KL, et al: Nuclear chromosome locations dictate segregation
error frequencies. Nature. 607:604–609. 2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang M, Sunkel BD, Ray WC and Stanton BZ:
Chromatin structure in cancer. BMC Mol Cell Biol.
23(35)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Barbieri I and Kouzarides T: Role of RNA
modifications in cancer. Nat Rev Cancer. 20:303–322.
2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chang HHY, Pannunzio NR, Adachi N and
Lieber MR: Non-homologous DNA end joining and alternative pathways
to double-strand break repair. Nat Rev Mol Cell Biol. 18:495–506.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sun Y, Liu Y, Ma X and Hu H: The influence
of cell cycle regulation on chemotherapy. Int J Mol Sci.
22(6923)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bowling EA, Wang JH, Gong F, Wu W, Neill
NJ, Kim IS, Tyagi S, Orellana M, Kurley SJ, Dominguez-Vidaña R, et
al: Spliceosome-targeted therapies trigger an antiviral immune
response in triple-negative breast cancer. Cell. 184:384–403.e21.
2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Dimaras H, Corson TW, Cobrinik D, White A,
Zhao J, Munier FL, Abramson DH, Shields CL, Chantada GL, Njuguna F
and Gallie BL: Retinoblastoma. Nat Rev Dis Primers.
1(15021)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ma M, Rodriguez A and Sugimoto K:
Activation of ATR-related protein kinase upon DNA damage
recognition. Curr Genet. 66:327–333. 2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yang H, Zhou C, Dhar A and Pavletich NP:
Mechanism of strand exchange from RecA-DNA synaptic and D-loop
structures. Nature. 586:801–806. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Iliaki S, Beyaert R and Afonina IS:
Polo-like kinase 1 (PLK1) signaling in cancer and beyond. Biochem
Pharmacol. 193(114747)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zelensky A, Kanaar R and Wyman C:
Mediators of homologous DNA pairing. Cold Spring Harb Perspect
Biol. 6(a016451)2014.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kutay U, Jühlen R and Antonin W: Mitotic
disassembly and reassembly of nuclear pore complexes. Trends Cell
Biol. 31:1019–1033. 2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen Y, Jia K, Sun Y, Zhang C, Li Y, Zhang
L, Chen Z, Zhang J, Hu Y, Yuan J, et al: Predicting response to
immunotherapy in gastric cancer via multi-dimensional analyses of
the tumour immune microenvironment. Nat Commun.
13(4851)2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Roy DM, Walsh LA, Desrichard A, Huse JT,
Wu W, Gao J, Bose P, Lee W and Chan TA: Integrated genomics for
pinpointing survival loci within arm-level somatic copy number
alterations. Cancer Cell. 29:737–750. 2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Truty R, Paul J, Kennemer M, Lincoln SE,
Olivares E, Nussbaum RL and Aradhya S: Prevalence and properties of
intragenic copy-number variation in Mendelian disease genes. Genet
Med. 21:114–123. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Dong X, Liu B, Yang L, Wang H, Wu B, Liu
R, Chen H, Chen X, Yu S, Chen B, et al: Clinical exome sequencing
as the first-tier test for diagnosing developmental disorders
covering both CNV and SNV: A Chinese cohort. J Med Genet.
57:558–566. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Gentile G, La Cognata V and Cavallaro S:
The contribution of CNVs to the most common aging-related
neurodegenerative diseases. Aging Clin Exp Res. 33:1187–1195.
2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Malhotra D and Sebat J: CNVs: Harbingers
of a rare variant revolution in psychiatric genetics. Cell.
148:1223–1241. 2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
DeVries AA, Dennis J, Tyrer JP, Peng PC,
Coetzee SG, Reyes AL, Plummer JT, Davis BD, Chen SS, Dezem FS, et
al: Copy number variants are ovarian cancer risk alleles at known
and novel risk loci. J Natl Cancer Inst. 114:1533–1544.
2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Xia Y, Rao L, Yao H, Wang Z, Ning P and
Chen X: Engineering macrophages for cancer immunotherapy and drug
delivery. Adv Mater. 32(e2002054)2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Chen L, Gao Y, Xu S, Yuan J, Wang M, Li T
and Gong J: N6-methyladenosine reader YTHDF family in biological
processes: Structures, roles, and mechanisms. Front Immunol.
14(1162607)2023.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Liu L, Li H, Hu D, Wang Y, Shao W, Zhong
J, Yang S, Liu J and Zhang J: Insights into N6-methyladenosine and
programmed cell death in cancer. Mol Cancer. 21(32)2022.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wu Y, Wang Z, Shen J, Yan W, Xiang S, Liu
H and Huang W: The role of m6A methylation in osteosarcoma
biological processes and its potential clinical value. Hum
Genomics. 16(12)2022.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Hu J, Qiu D, Yu A, Hu J, Deng H, Li H, Yi
Z, Chen J and Zu X: YTHDF1 is a potential pan-cancer biomarker for
prognosis and immunotherapy. Front Oncol. 11(607224)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wei WS, Wang N, Deng MH, Dong P, Liu JY,
Xiang Z, Li XD, Li ZY, Liu ZH, Peng YL, et al: LRPPRC regulates
redox homeostasis via the circANKHD1/FOXM1 axis to enhance bladder
urothelial carcinoma tumorigenesis. Redox Biol.
48(102201)2021.PubMed/NCBI View Article : Google Scholar : (Epub ahead of
print).
|
|
66
|
Cui J, Wang L, Ren X, Zhang Y and Zhang H:
LRPPRC: A multifunctional protein involved in energy metabolism and
human disease. Front Physiol. 10(595)2019.PubMed/NCBI View Article : Google Scholar
|