Introduction
Hypercholesterolemia is a well-established risk
factor for atherosclerotic cardiovascular disease (ASCVD), and
ASCVD is one of the leading causes of mortality worldwide (1). It was estimated by The World Health
Organization that 17.9 million individuals died from cardiovascular
disease in 2019, and 85% of these deaths were due to a heart attack
or stroke (2). Previous studies
have confirmed that high levels of total cholesterol (TC) and
low-density lipoprotein cholesterol (LDL-C) in the serum of adult
humans and atherosclerotic mice are associated with ASCVD (3,4).
Therefore, the discovery and development of
anti-hypercholesterolemic drugs is essential to medical
science.
Sodium sulphate is the effective ingredient of
mirabilite, which is a type of mineral used as a drug in
traditional Chinese medicine (5,6).
Mirabilite is also a purgative (7), although the mechanism of its laxative
and diarrhea-inducing effects requires further exploration. It is
known that the entry of excessive quantities of bile acids into the
colon can cause diarrhea (8).
Also, the absorption of bile acids in the terminal ileum and colon
is essential to the enterohepatic circulation of bile acids
(9), and resection of the terminal
ileum can cause bile acid diarrhea (8,10).
Studies have also confirmed that bile acid malabsorption causes
diarrhea (10,11). However, it is unclear whether
mirabilite increases the level of bile acids in the colon. Bile
acids induce the expression of fibroblast growth factor (FGF)15/19
in enterocytes by stimulating the activation of farnesoid X
receptor (FXR) in these cells (12). FGF15/19 is released into the blood
and interacts with FGF receptor 4 (FGFR4) and Klotho β (KLB) on the
surface of hepatocytes to downregulate the expression of the
cytochrome P450 family 7 subfamily A member 1 (Cyp7a1) gene,
which encodes the rate-limiting enzyme in bile acid synthesis
(13-15).
It is hypothesised that if mirabilite reduces the absorption of
bile acids in the intestine, it may increase the conversion of
cholesterols to bile acids via inhibition of the FGF15/19 signaling
pathway in hepatocytes.
The gut microbiota has been confirmed to be key in
regulating the health of the host, particularly with regard to
glucose and lipid metabolism (16). Previous studies have reported that
the gut microbiota also impacts the cholesterol and bile acid
metabolism of the host (17,18).
Gut microbial-derived metabolites, including short chain fatty
acids, primary and secondary bile acids and trimethylamine N-oxide,
have been found to have important functions in the maintenance of
cardiovascular health (19). A
recent study demonstrated that plant proteins can ameliorate
hypercholesterolemia in hamsters by regulating the gut microbiota
(20). However, it remains unclear
if sodium sulphate regulates the metabolism of cholesterol and bile
acids via modulation of the gut microbiota of the host.
In the present study, hypercholesterolemic mouse
models were generated by feeding a high-cholesterol diet (HCD) to
C57BL/6 mice. Three different doses of sodium sulphate were
administered to the mice to study the efficiency of sodium sulphate
as an anti-hypercholesterolemic agent and the underlying
mechanisms.
Materials and methods
Mice
All mouse experimental procedures were approved by
The Guangdong Pharmaceutical University Experimental Animal Ethics
Committee (Guangzhou, China; approval no. gdpulacspf2017030-1) and
are reported according to the ARRIVE guidelines. A total of 50 male
C57BL/6 mice (age, 7 weeks; body weight, ~24 g) were purchased from
Hunan Lex Jingda Laboratory Animal Co., Ltd. The mice were housed
in a specific pathogen-free animal facility with a 12-h light/dark
cycle and 60-65% humidity, at 25˚C, and with free access to food
and water. After 1 week of acclimatization, 40 mice were fed an HCD
(Dyets, Inc.; cat. no. ASHF3; containing 22.60% protein, 45.20%
carbohydrate, 20.10% fat and 1.25% cholesterol) and the remaining
10 mice continued to be fed a normal food diet (NFD; Beijing Keao
Xieli Feed Co., Ltd.; cat. no. 2212; containing 23.07% protein,
65.08% carbohydrate and 11.85% fat). After 4 weeks, the mice fed
the HCD were divided into four groups (n=10/group), based on the TC
concentration observed in the serum from blood collected from the
tail vein, such that the TC level was similar in each group
(Fig. S1A and B). The dosage of sodium sulphate used in
the present study was based on the sodium sulphate content in
mirabilite administered to mice and rats in a previous study
(6). In three of the HCD groups,
mice were intragastrically treated with sodium sulphate (Damao
Chemical Regent Factory; cat. no. 7757-82-6) at a low dose (LSS;
158.5 mg/kg/day; aqueous solute, 0.01 ml/g/mouse), middle dose
(MSS; 317.0 mg/kg/day; aqueous solute, 0.01 ml/g/mouse) and high
dose (HSS; 634.0 mg/kg/day; aqueous solute, 0.01 ml/g/mouse), along
with the HCD. As such, these three groups were designated the HCD +
LSS, HCD + MSS and HCD + HSS groups, respectively. The fourth HCD
group was fed the HCD only and was designated the HCD group. The
mice fed the NFD were used as the control group (CON group). Mice
from the CON and HCD groups were intragastrically administered an
equal volume of water (0.01 ml/g/mouse). The body weight of the
mice was measured once a week. After 3 weeks of sodium sulphate
administration, blood was collected from the orbital vein of the
mice following anesthesia with 3% isoflurane. After blood
collection, the mice were sacrificed via cervical dislocation.
Animal death was confirmed through the stoppage of breathing. The
hepatic tissues and intestines of the mice were then collected for
biochemical, histological and molecular analyses.
Blood and hepatic biochemical profile
assays
The concentrations of TC and triglycerides (TG) in
the serum and hepatic tissues, and the concentrations of
high-density lipoprotein cholesterol (HDL-C), LDL-C, total bile
acid (TBA), alanine aminotransferase (ALT) and aspartate
aminotransferase (AST) in the serum were measured according to the
manufacturer's protocols for each kit. The kits for measuring the
concentrations of TC (cat. no. A111-1-1), TG (cat. no. A110-1-1),
HDL-C (cat. no. A112-1-1), LDL-C (cat. no. A113-1-1), TBA (cat. no.
E003-2-1), ALT (cat. no. C009-2-1) and AST (cat. no. C010-2-1) were
purchased from Nanjing Jiancheng Bioengineering Institute.
Haematoxylin and eosin (H&E)
staining
Mouse hepatic tissues were fixed in 4%
paraformaldehyde at 4˚C overnight and then embedded in paraffin for
H&E staining. The 4-µm paraffin sections were stained with
haematoxylin (cat. no. H9627; Sigma-Aldrich; Merck KGaA) for 3 min
followed by eosin (cat. no. E4009; Sigma-Aldrich; Merck KGaA) for
20 sec, both at room temperature. The images were captured with a
PerkinElmer Automated Quantitative Pathology System (PerkinElmer,
Inc.).
Transcriptome analysis
Hepatic tissues from each group of mice were fresh
frozen in liquid nitrogen and then stored at -80˚C. RNA extraction,
library construction, sequencing and transcriptome analysis were
all conducted by Guangzhou Gene Denovo Biotechnology Co., Ltd.
Briefly, total RNA from each sample was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The RNA quality was evaluated using an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc.) and then checked via
RNase-free agarose gel electrophoresis. Following extraction,
eukaryotic mRNA was enriched using oligo(dT) beads (New England
BioLabs, Inc.). The enriched mRNA molecules were fragmented in
fragmentation buffer, and then reverse transcribed into cDNA using
random primers (New England BioLabs, Inc.). The cDNA fragments were
subsequently purified using a QiaQuick PCR extraction kit (Qiagen
China Co., Ltd.), followed by end-repaired poly(A) addition and
then ligation to Illumina sequencing adapters. The HiSeq Rapid
Cluster Kit v2 (PE-402-4002) and HiSeq Rapid SBS Kit v2
(FC-402-4023) were used. The ligation products were separated by
size via agarose gel electrophoresis, then PCR amplified and
finally sequenced using an Illumina HiSeq 2500 System (Illumina,
Inc.). Then, an ABI StepOnePlus Real Time PCR System (hermo Fisher
Scientific, Inc.) was used to detect library concentration. The
concentration was >5 ng/µl. The reads were then filtered using
fastp (version 0.18.0) (21) and
differential expression analysis was performed using DESeq2
software (version 1.20.0) (22).
Differentially expressed mRNAs with a false discovery rate (FDR)
<0.05 and absolute fold change ≥2 were considered included in
subsequent analyses.
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a
public pathway-related database used to analyze identified
significantly enriched metabolic pathways or signal transduction
pathways of differentially expressed genes (DEGs) compared with the
whole genome background (23). The
calculated P-values underwent FDR correction, and KEGG pathways
with FDR ≤0.05 were defined as significantly enriched pathways of
the DEGs.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from each hepatic tissue and
ileal sample harvested from the experimental mice using TRIzol
reagent, and then subjected to RT using the PrimeScript™ RT Reagent
kit (Takara Bio, Inc.) at 37˚C for 15 min followed by 85˚C for 5
sec. qPCR was performed using the SYBR Premix Ex Taq kit (Takara
Bio, Inc.) and the LightCycler 480II System (Roche Diagnostics).
The thermocycling conditions were as follows: 95˚C for 30 sec; and
then 40 cycles of 95˚C for 5 sec, 60˚C for 20 sec and 65˚C for 15
sec. GAPDH was used as the internal reference. All primers used for
qPCR in the present study are listed in Table SI. The 2-ΔΔCq method
was used to quantify the relative transcriptional level of each
gene (24).
Western blotting
Hepatic tissues were lysed in
Radio-Immunoprecipitation Assay lysis buffer (Dalian Meilun Biology
Technology Co., Ltd.), and then centrifuged at 13,680 x g at 4˚C
for 30 min to harvest the supernatant. The protein concentration in
the supernatant was measured using a BCA kit (cat. no. P0011;
Beyotime Institute of Biotechnology). Then, equal amounts of
protein (32 µg/lane) were separated using SDS-PAGE (10% gel) and
subsequently transferred to a PVDF membrane. The PVDF membrane was
blocked with 5% skimmed milk in Tris-buffered saline Tween 20
(0.1%) buffer for 1 h at room temperature, then incubated with
primary antibodies at 4˚C overnight. The primary
antibodies comprised: Rabbit anti-GAPDH (1:1,000; cat. no. 2118S;
CST Biological Reagents Co., Ltd.), mouse anti-CYP7A1 (1:1,000;
cat. no. 2683295; MilliporeSigma), rabbit anti-hydroxy-δ-5-steroid
dehydrogenase, 3 β- and steroid δ-isomerase 7 (HSD3B7; 1:1,000;
cat. no. ab190223; Abcam), rabbit anti-aldo-keto reductase family 1
member D1 (AKR1D1; 1:1,000; cat. no. ab101393; Abcam), rabbit
anti-acyl-CoA oxidase 2 (1:1,000; cat. no. ab197808; Abcam), rabbit
anti-hydroxysteroid 17-β dehydrogenase 4 (HSD17B4; 1:1,000; cat.
no. ab97971; Abcam), rabbit anti-CYP27A1 (1:2,000; cat. no.
ab126785; Abcam), rabbit anti-CYP39A1 (1:1,000; cat. no. ab129334;
Abcam), rabbit anti-isopentenyl-diphosphate δ isomerase 1 (IDI1;
1:3,000; cat. no. ab97448; Abcam), rabbit anti-lanosterol synthase
[anti-oxidosqualene-lanosterol cyclase (OSC); 1:1,000; cat. no.
ab80364; Abcam], rabbit anti-farnesyl diphosphate
farnesyltransferase 1 (FDFT1; 1:1,000; cat. no. ab195046; Abcam),
rabbit anti-farnesyl diphosphate synthase (FDPS; 1:2,000; cat. no.
ab153805; Abcam), rabbit anti-mevalonate diphosphate decarboxylase
(MVD; 1:1,000; cat. no. ab96226; Abcam), rabbit anti-mevalonate
kinase (MVK; 1:1,000; cat. no. ab154515; Abcam), rabbit
anti-3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR; 1:1,000; cat.
no. ab174830; Abcam), rabbit anti-low density lipoprotein receptor
(LDLR; 1:1,000; cat. no. ab52818; Abcam), rabbit anti-c-Jun
N-terminal kinase (JNK; 1:1,000; cat. no. 9252s; CST Biological
Reagents Co., Ltd.), mouse anti-phosphorylated (p)-JNK (1:2,000;
cat. no. 9255s; CST Biological Reagents Co., Ltd.), rabbit
anti-p-insulin receptor substrate 1 (p-IRS1; Tyr608) mouse
(1:1,000; cat. no. 09-432; MilliporeSigma), rabbit anti-IRS-1
(1:1,000; cat. no. 2382S; CST Biological Reagents Co., Ltd.),
rabbit anti-p-AKT (Ser473) (1:1,000; cat. no. 4060S; CST Biological
Reagents Co., Ltd.), rabbit anti-AKT (1:1,000; cat. no. 4685S; CST
Biological Reagents Co., Ltd.), mouse anti-tribbles pseudokinase 3
(TRB3; 1:100; cat. no. sc-390242; Santa Cruz Biotechnology, Inc.),
rabbit anti-c-Jun (1:1,000; cat. no. ab32137; Abcam), mouse
anti-p-c-Jun (1:200; cat. no. sc-822; Santa Cruz Biotechnology,
Inc.) and goat anti-KLB (0.5 µg/ml; cat. no. AF5889; R&D
Systems China Co., Ltd.). Then, the PVDF membrane was washed in
Tris-buffered saline Tween 20 (0.1%) buffer for 1 h at room
temperature and subsequently incubated with horseradish peroxidase
(HRP)-labeled secondary antibodies, including HRP-goat anti-rabbit
IgG (1:5,000; cat. no. os0701; Earthox Life Sciences), HRP-donkey
anti-goat IgG (1:2,000; cat. no. ab6885; Abcam) and HRP-goat
anti-mouse IgG (1:2,000; cat. no. ab6789; Abcam) at room
temperature for 1 h. Finally, the signals were detected using
Enhanced Chemiluminescence ECL solution (cat. no. MA0186-2;
Meilunbio), and quantification of the bands was conducted using
ImageJ software (version 1.53a; National Institutes of Health).
16S ribosomal (r)DNA gene
analysis
Fecal samples collected the day before tissue
harvesting were flash frozen in liquid nitrogen after collection
from each mouse and stored at -80˚C until use. The extraction of
bacterial DNA from the fecal samples, the PCR amplification of
bacterial 16S rDNA genes, sequencing and analysis were conducted by
Guangzhou Gene Denovo Biotechnology Co., Ltd. All experimental
procedures were performed as previously described (25).
Statistical analysis
Statistical differences were determined using SPSS
software (version 23.0; IBM Corp.). Data are presented as the mean
± SEM. One-way ANOVA followed by Tukey's post hoc test was
conducted to analyze differences among the groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Sodium sulphate ameliorates
hypercholesterolemia in mice fed an HCD
To evaluate the effects of sodium sulphate on
hypercholesterolemia, 8-week-old male C57BL/6 mice were fed an HCD
for 4 weeks, and then the TC concentration in the mice serum was
determined (Fig. S1A). After 4
weeks of feeding with the HCD, the serum TC concentration was
significantly higher compared with that of the mice in the CON
group (Fig. S1B). The mice fed an
HCD were then divided into four groups, with matching of the serum
TC concentration in each group. Then, mice in the HCD + LSS, HCD +
MSS and HCD + HSS groups were treated with sodium sulphate for 3
weeks (Fig. S1A). The body weight
and food intake of mice in the CON, HCD, HCD + LSS, HCD + MSS and
HCD + HSS groups exhibited no significant differences during these
3 weeks (Fig. S1C and D). Although the average daily defecation
mass of mice fed an HCD was lower than that of the CON group, it
was higher in the sodium sulphate treated mice compared with the
CON and HCD groups (Fig. S1E).
However, these differences in defecation mass were not found to be
significant, and the fecal pellets of the mice in the sodium
sulphate groups were normal.
Although the serum TC concentration of the mice in
the HCD group was significantly higher than that in the CON group,
administration of sodium sulphate significantly reduced the serum
TC concentration in mice fed an HCD (Fig. 1A). However, the serum TG
concentration of the mice in each group was similar (Fig. 1B). The TC level in the liver was
significantly increased in the mice from the HCD group compared
with the CON group (Fig. 1C). In
addition, the TC level in the liver exhibited a downward trend in
the mice from the HCD + LSS, HCD + MSS and HCD + HSS groups
compared with the HCD group, but this reduction was not significant
(Fig. 1C). The TG level in the
liver was significantly higher in mice fed an HCD compared with
those fed an NFD, and the administration of sodium sulphate did not
reduce it (Fig. 1D). The serum
LDL-C concentration did not significant differ among the mice of
the five groups (Fig. 1E).
However, the serum HDL-C concentrations in the mice from the HCD +
LSS, HCD + MSS and HCD + HSS groups were significantly higher than
those in the mice from the HCD group (Fig. 1F). In addition, the LDL-C/HDL-C
ratio was significantly reduced in the serum of the mice from the
HCD + LSS and HCD + HSS groups compared with the HCD group
(Fig. 1G). The serum TBA
concentration in the mice from the HCD group was significantly
reduced compared that in the mice from the CON group, and the serum
TBA concentrations in the HCD + LSS, HCD + MSS and HCD + HSS groups
were slightly but not significantly lower than the serum TBA
concentration in the HCD group (Fig.
1H). In addition, when the serum ALT and AST concentrations
were compared among the mice in all groups, no significant
differences were detected (Fig. 1I
and J). These results indicate
that sodium sulphate may ameliorate the hypercholesterolemia in
mice induced by an HCD.
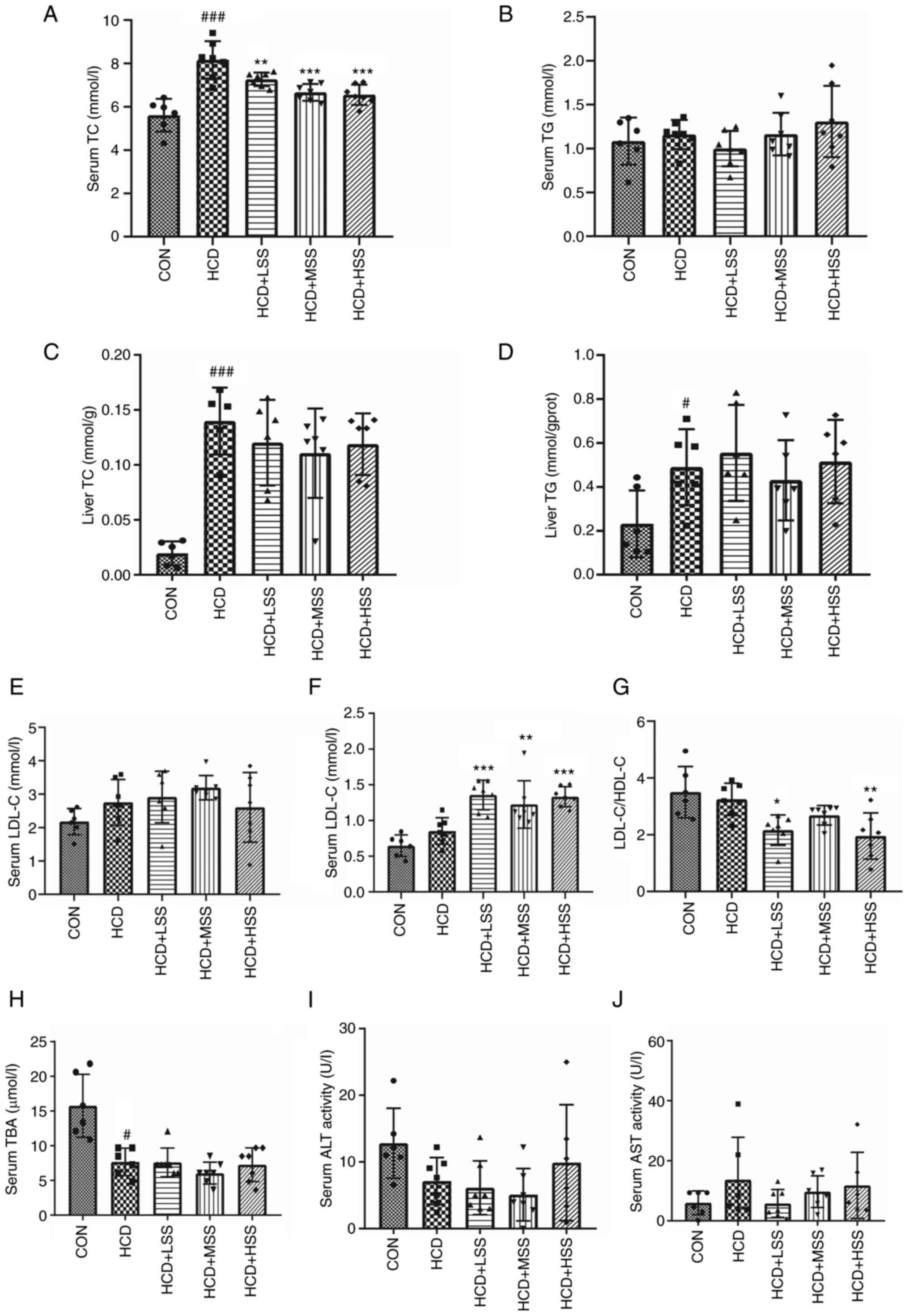 | Figure 1Effects of sodium sulphate on mice
fed an HCD. Serum concentrations of (A) TC and (B) TG in mice from
the five study groups: CON, HCD, HCD + LSS, HCD + MSS and HCD +
HSS. Levels of (C) TC and (D) TG in the livers of the mice. (E)
LDL-C concentration, (F) HDL-C concentration and (G) LDL-C/HDL-C
ratio in the serum of the mice. Serum concentrations of (H) TBA,
(I) ALT and (J) AST in the mice. #P<0.05,
###P<0.001 vs. the CON group; *P<0.05,
**P<0.01, ***P<0.001 vs. the HCD group.
HCD, high cholesterol diet; TC, total cholesterol; TG,
triglycerides; CON, control; LSS, low dose of sodium sulphate; MSS,
middle dose of sodium sulphate; HSS, high dose of sodium sulphate;
LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density
lipoprotein cholesterol; TBA, total bile acid; ALT, alanine
aminotransferase; AST, aspartate aminotransferase; gprot, g of
protein. |
Since the liver is the major organ involved in
cholesterol metabolism (26), the
morphology and histology of the livers of the mice in the five
groups were assessed. The morphology of the livers in mice fed with
an HCD with or without sodium sulphate appeared slightly paler
compared with those in the CON group (Fig. S2A). The gallbladders from mice in
the HCD group were dark red, and the administration of sodium
sulphate improved the colour and increased the size of the
gallbladders from these mice (Fig.
S2A). The liver weight of mice in the HCD group was
significantly reduced compared with that in the CON group, although
the reduction was slight, and the liver weights of mice in the
sodium sulphate administration groups exhibited no significant
differences compared with the HCD group (Fig. S2B). The liver/body weight ratio of
mice fed an HCD was significantly lower than that of mice fed an
NCD, and the administration of sodium sulphate did not increase the
liver/body weight ratio of these mice (Fig. S2C). The results of H&E
staining demonstrated that excessive quantities of lipid droplets
accumulated in the liver tissues of mice from the HCD group, and
the haematoxylin staining intensity of the nuclei in certain
hepatocytes of this group was weaker (Fig. S2D). The weak nuclear staining
indicated that the hepatocytes were damaged (27). Following the administration of
sodium sulphate to mice fed an HCD, excessive lipid accumulation
was still detected in the hepatocytes, but the nuclear staining in
most hepatocytes of these mice was normal (Fig. S2D). These results demonstrated
that, although sodium sulphate did not mitigate non-alcoholic fatty
liver disease (NAFLD) in mice fed an HCD, it protected hepatocytes
against HCD-induced damage.
Sodium sulphate upregulates the
hepatic expression of bile acid synthesis-associated genes in mice
fed an HCD
RNA-sequencing (seq) technology was used to compare
the transcriptional profiles of hepatic tissues from mice in the
CON, HCD and HCD + MSS groups. There were 148 upregulated and 230
downregulated genes in the hepatic tissues from the HCD group
compared with the CON group (Fig.
S3A and B). The mRNA
expression levels of 116 genes were significantly increased and
those of 127 genes were significantly decreased in the hepatic
tissues from the HCD + MSS group compared with the HCD group
(Fig. S3A and C). The expression levels of 203 genes
were higher and those of 286 genes were lower in the hepatic
tissues from the HCD + MSS group compared with the CON group
(Fig. S3A and D). The RNA-seq results demonstrated that
genes associated with bile acid synthesis, such as Cyp7a1,
were upregulated in the livers of the HCD + MSS group compared with
the HCD group (Fig. 2A). Since
bile acid transporters play important roles in the regulation of
bile acid metabolism (28), the
transcriptional levels of genes encoding bile acid transporters in
the liver were also analysed. The RNA-seq results demonstrated that
the mRNA level of solute carrier organic anion transporter family
member 1b2 (Slco1b2) was notably reduced in the hepatic
tissues from the HCD group compared with the CON group, but there
was no significant change in Slco1b2 in the HCD + MSS group
compared with the HCD group (Fig.
S3E).
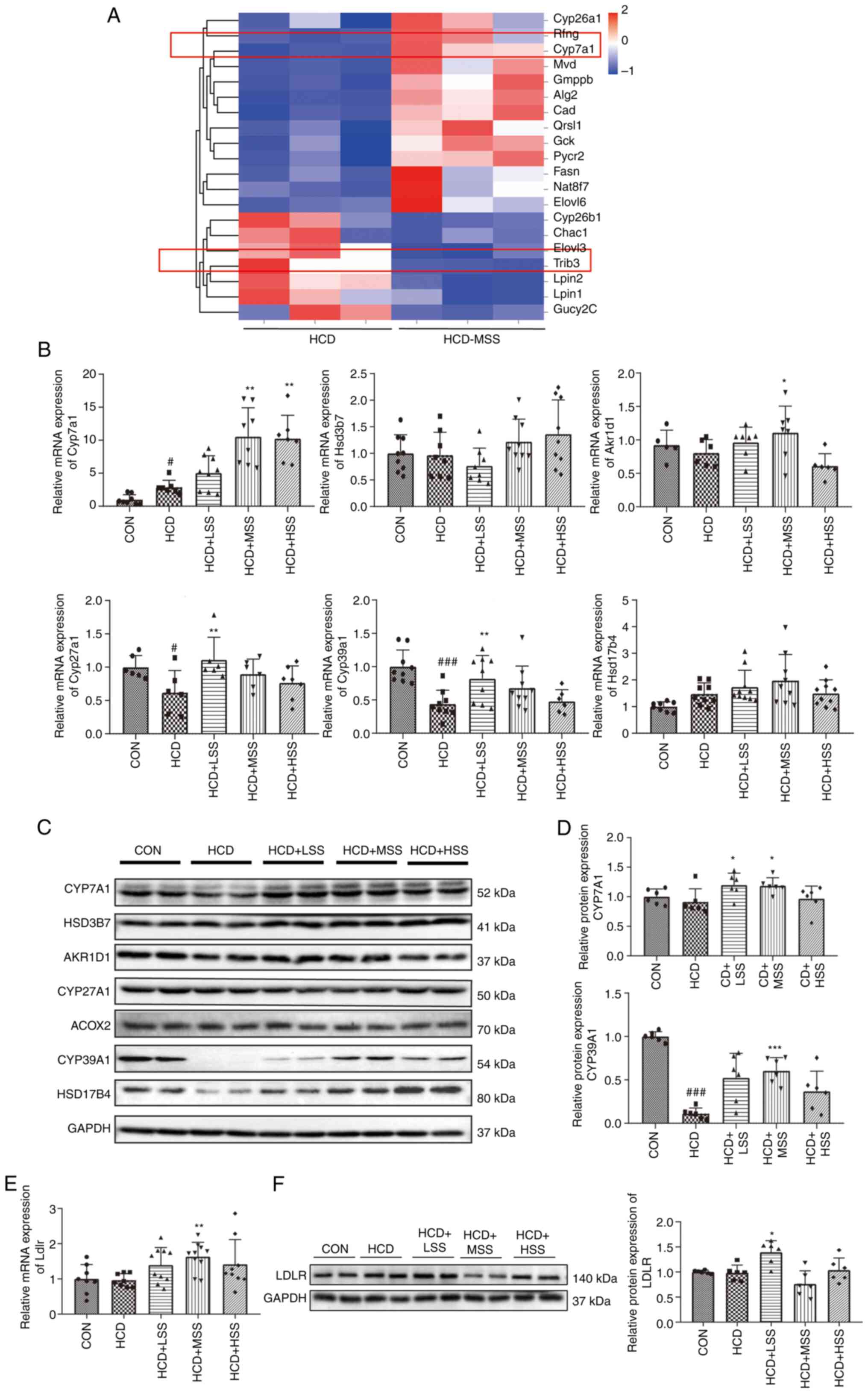 | Figure 2Effect of sodium sulphate on the
hepatic expression of genes associated with bile acid synthesis in
mice fed an HCD. (A) Heatmap of genes associated with glucose and
lipid metabolism in the liver tissues of mice in the HCD and HCD +
MSS groups. (B) Relative mRNA expression levels of Cyp7a1,
Hsd3b7, Akr1d1, Cyp27a1, Cyp39a1 and
Hsd17b4 in the liver tissues of mice from five groups: CON,
HCD, HCD + LSS, HCD + MSS and HCD + HSS. (C) Western blotting
results for CYP7A1, HSD3B7, AKR1D1, CYP27A1, ACOX2, CYP39A1 and
HSD17B4 in the liver tissues of mice from the five groups. (D)
Semi-quantitative analysis of the CYP7A1 and CYP39A1 western
blotting results. (E) Relative mRNA expression levels of
Ldlr in the liver tissues of mice from the five groups. (F)
Western blotting results for LDLR in the liver tissues from mice
from the five groups. The right panel shows the semi-quantitative
analysis. GAPDH was used as the internal control for all
experiments. #P<0.05, ###P<0.001 vs.
the CON group; *P<0.05, **P<0.01,
***P<0.001 vs. the HCD group. HCD, high cholesterol
diet; CON, control; LSS, low dose of sodium sulphate; MSS, middle
dose of sodium sulphate; HSS, high dose of sodium sulphate;
Cyp7/27/39a1, cytochrome P450 family 7/27/39 subfamily A member 1;
HSD3B7, hydroxy-δ-5-steroid dehydrogenase, 3 β- and steroid
δ-isomerase 7; AKR1D1, aldo-keto reductase family 1 member D1;
HSD17B4, hydroxysteroid 17-β dehydrogenase 4; ACOX2, acyl-CoA
oxidase 2; LDLR, low density lipoprotein receptor. |
The expression levels of genes associated with bile
acid synthesis were further tested via RT-qPCR and western
blotting. Cyp7a1 expression was increased at the mRNA level
in the hepatic tissues from the HCD group compared with the CON
group (Fig. 2B). However, the
CYP7A1 protein expression level was slightly lower in the livers
from the HCD group compared with the CON group, although this
reduction was not significant (Fig.
2C and D). The Cyp7a1
mRNA levels were significantly higher in the livers from the HCD +
MSS and HCD + HSS groups compared with the HCD group (Fig. 2B), and the CYP7A1 protein levels
were significantly increased in the liver tissues from the HCD +
LSS and HCD + MSS groups compared with the HCD group (Fig. 2C and D). The mRNA levels of Akr1d1 in
the HCD + MSS group and Cyp27a1 in the HCD + LSS group were
significantly upregulated compared with those in the HCD group
(Fig. 2B), but no marked change in
the AKR1D1 and CYP27A1 protein levels was observed among the five
groups (Fig. 2C). The mRNA and
protein expression levels of CYP39A1 were significantly reduced in
the hepatic tissues from the HCD group compared with the CON group
(Fig. 2B-D). In addition, the
hepatic Cyp39a1 mRNA expression levels were significantly
upregulated in the HCD + LSS group compared with the HCD group
(Fig. 2B), and the CYP39A1 protein
expression level was significantly higher in the hepatic tissues
from mice in the HCD + MSS group compared with the HCD group
(Fig. 2C and D). No significant difference in the
Ldlr mRNA level was detected between the hepatic tissues of
the HCD and CON groups (Fig. 2E).
However, the Ldlr transcriptional level was significantly
higher in the hepatic tissues from mice in the HCD + MSS group
compared with the HCD group (Fig.
2E). The Ldlr mRNA levels in the livers of mice from the
HCD + LSS and HCD + HSS groups were also increased compared with
the Ldlr mRNA level in the CON group, but this increase was
not significant (Fig. 2E). The
western blotting results demonstrated that the LDLR protein level
in the hepatic tissues of mice from the HCD + LSS group was also
significantly increased compared with that in the HCD group
(Fig. 2F). These results suggest
that sodium sulphate might alleviate hypercholesterolemia in mice
fed an HCD via upregulation of the expression of genes that encode
enzymes that catalyze bile acid production, including Cyp7a1
and Cyp39a1, and upregulation of the expression of
Ldlr, which takes up excessive LDL-C from the blood into the
hepatocytes of mice.
The hepatic expression levels of genes associated
with cholesterol synthesis were also assessed. The mRNA expression
levels of 3-hydroxy-3-methylglutaryl-CoA synthase 1, Hmgcr,
Mvk, Mvd, Idi1, Fdps, Fdft1,
squalene epoxidase, Osc, 7-dehydrocholesterol reductase and
Cyp51a1 in hepatic tissues from the HCD group were
significantly reduced 2 to >10-fold compared with those in the
CON group (Fig. S4A), and the
levels of most of these mRNAs were slightly increased after sodium
sulphate administration, some significantly, namely Mvd,
Idi1, Fdps and Cyp51a1 (Fig. S4A). However, the Fdft1 mRNA
levels in the hepatic tissues of mice from the HCD + MSS and HCD +
HSS groups were markedly lower compared with those in the HCD group
(Fig. S4A). At the protein level,
the expression of HMGCR, the rate-limiting enzyme of cholesterol
biosynthesis (29,30), was not significantly different
among the five groups. However, the MVK, MVD, IDI1, FDPS, FDFT1 and
LSS levels were all notably downregulated in the hepatic tissues
from the HCD group compared with the CON group, and were not
affected by sodium sulphate administration (Fig. S4B).
Sodium sulphate improves insulin
resistance in the livers of mice fed an HCD
The RNA-seq results demonstrated that the expression
of Trib3, the mRNA that encodes TRB3, was significantly
reduced in the livers of mice from the HCD + MSS group compared
with the HCD group (Fig. 2A).
Increased Trib3 expression is known to cause insulin
resistance in hepatocytes in vivo and in vitro
(31-34).
KEGG analysis of the mRNAs in the livers of the mice that were
differentially expressed between the HCD and the HCD + MSS groups
indicated that ‘Insulin signaling pathway’ and ‘Insulin resistance’
were among the top 10 pathways that affected the HCD group
following sodium sulphate administration (Fig. 3A). The results of RT-qPCR and
western blotting analysis demonstrated that Trib3 expression
was significantly increased in the hepatic tissues of mice from the
HCD group compared with the CON group, at the mRNA and protein
levels (Fig. 3B). The Trib3
mRNA levels were significantly reduced in the hepatic tissues of
mice fed an HCD following the administration of sodium sulphate
(Fig. 3B). In addition, the TRB3
protein levels were significantly lower in the hepatic tissues of
mice from the HCD + LSS and HCD + MSS groups compared with the HCD
group (Fig. 3B).
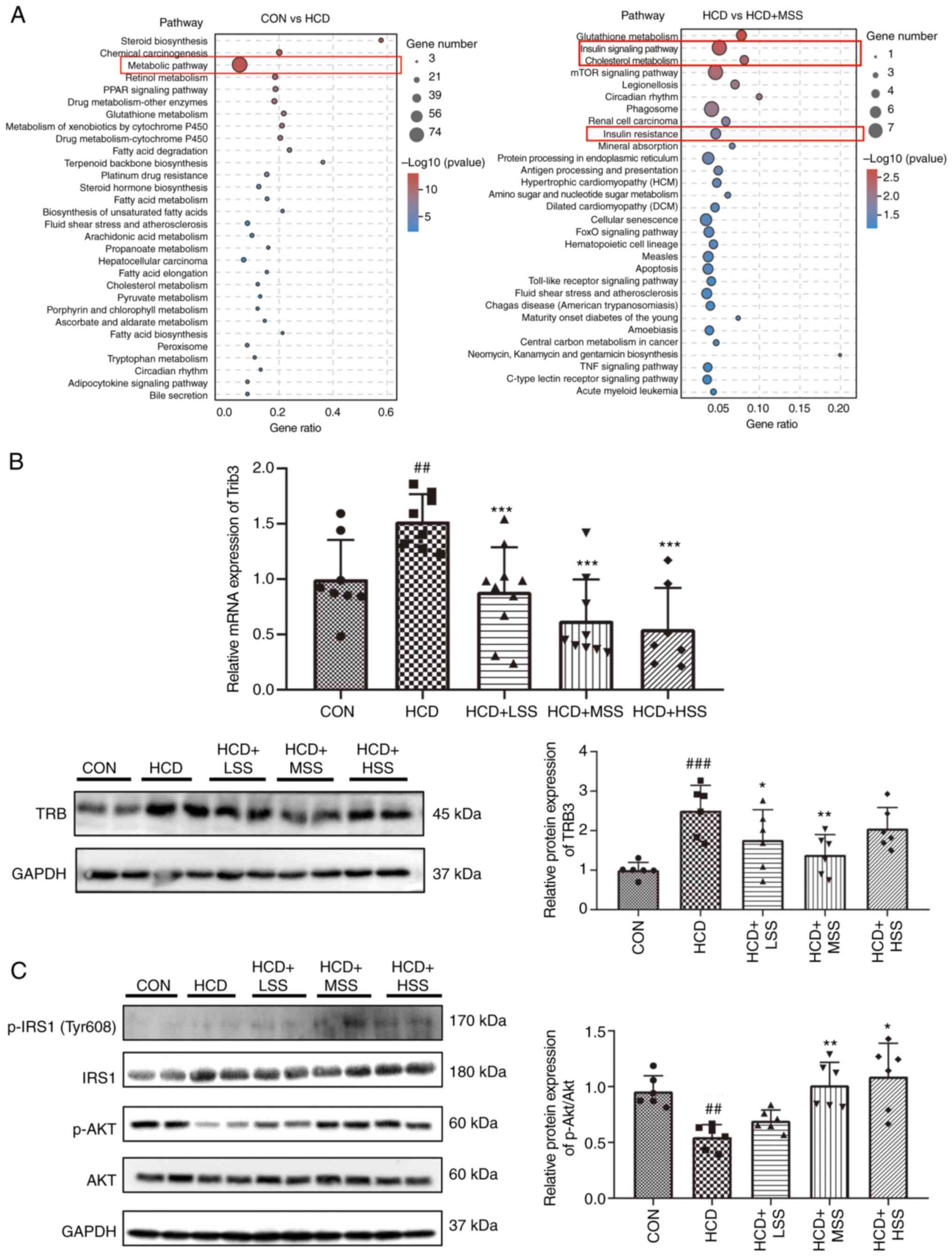 | Figure 3Effect of sodium sulphate on the
insulin signaling pathway in the liver tissues of mice fed an HCD.
(A) Kyoto Encyclopedia of Genes and Genomes analysis of the
biological pathways of differently expressed mRNAs in the livers of
mice from the CON vs. HCD and HCD vs. HCD + MSS groups. (B) RT-qPCR
and western blotting results of the Trib3 mRNA and TRB3
protein expression levels in the liver tissues of mice from the
CON, HCD, HCD + LSS, HCD + MSS and HCD + HSS groups, respectively.
(C) Western blotting results showing the protein expression levels
of p-IRS1 (Tyr608), IRS1, p-AKT and AKT in the liver tissues of
mice from the CON, HCD, HCD + LSS, HCD + MSS and HCD + HSS groups.
The right panel shows the semi-quantitative analysis of the
p-AKT/AKT ratio. GAPDH was used as the internal control for western
blotting and RT-qPCR. ##P<0.01,
###P<0.001 vs. the CON group; *P<0.05,
**P<0.01, ***P<0.001 vs. the HCD group.
HCD, high cholesterol diet; CON, control; LSS, low dose of sodium
sulphate; MSS, middle dose of sodium sulphate; HSS, high dose of
sodium sulphate; RT-qPCR, reverse transcription-quantitative PCR;
Trib3/TRB3, tribbles pseudokinase 3; p-, phosphorylated; IRS1,
insulin receptor substrate 1. |
Furthermore, the activation and protein levels of
components of the insulin signal pathways were evaluated by the
western blotting. The p-IRS1 (Tyr608) protein levels in the hepatic
tissues from the HCD + LSS, HCD + MSS and HCD + HSS groups were
higher compared with those in the CON and HCD groups (Fig. 3C). The p-AKT levels in the hepatic
tissues of mice from the HCD group were lower compared with those
in the CON group, and the administration of sodium sulphate to
HCD-fed mice increased these levels (Fig. 3C). The ratio of p-AKT/AKT was
significantly lower in liver tissue from the HCD group compared
with the CON group, and was significantly increased in the HCD +
MSS and HCD + HSS groups compared with the HCD group (Fig. 3C). These results indicate that
sodium sulphate attenuated the hepatic insulin resistance in mice
fed an HCD by inhibiting the expression of Trib3 in
hepatocytes.
Sodium sulphate inhibits the
FGF15/FGF4-Klb/JNK/c-Jun signaling pathway in the hepatocytes of
mice fed an HCD
A number of studies have confirmed that the
reduction of FGF15 expression in the ileum can induce the
expression of Cyp7a1 in hepatocytes to increase the
conversion of cholesterol to bile acid (13,35,36).
Therefore, the Fgf15 mRNA expression levels in the ileum of
mice from the five groups were assessed using RT-qPCR. The
Fgf15 transcriptional level in the HCD group was slightly
lower than that in the CON group, but the reduction was not
significant (Fig. 4A). The
Fgf15 mRNA levels were lower in the HCD + LSS, HCD + MSS and
HCD + HSS groups compared with the HCD group, but these changes
were also not significant (Fig.
4A). However, it is speculated that sodium sulphate may inhibit
the expression of FGF15 in the ileum of mice fed an HCD. In the
hepatic tissues, the mRNA expression level of Fgfr4, which
encodes the receptor for FGF15 (15,37),
was significantly reduced in the HCD group compared with the CON
group, and was unaffected by the administration of sodium sulphate
(Fig. 4B). No significant
differences in the mRNA and protein expression levels of KLB, which
is the co-receptor of FGFR4(14),
were detected in the hepatic tissues from mice in the CON and HCD
groups (Fig. 4C and D). However, the Klb mRNA levels
were significantly downregulated in the hepatic tissues from mice
fed an HCD following the administration of sodium sulphate, and the
KLB protein levels were also significantly lower in the hepatic
tissues of mice from the HCD + MSS and HCD + HSS groups compared
with the HCD group (Fig. 4C and
D). These results demonstrate that
the FGF15 signal in the hepatocytes of HCD fed mice treated with
sodium sulphate might weaken due to the downregulation of FGF15 in
the ileum and the reduction of KLB expression in the liver of these
mice.
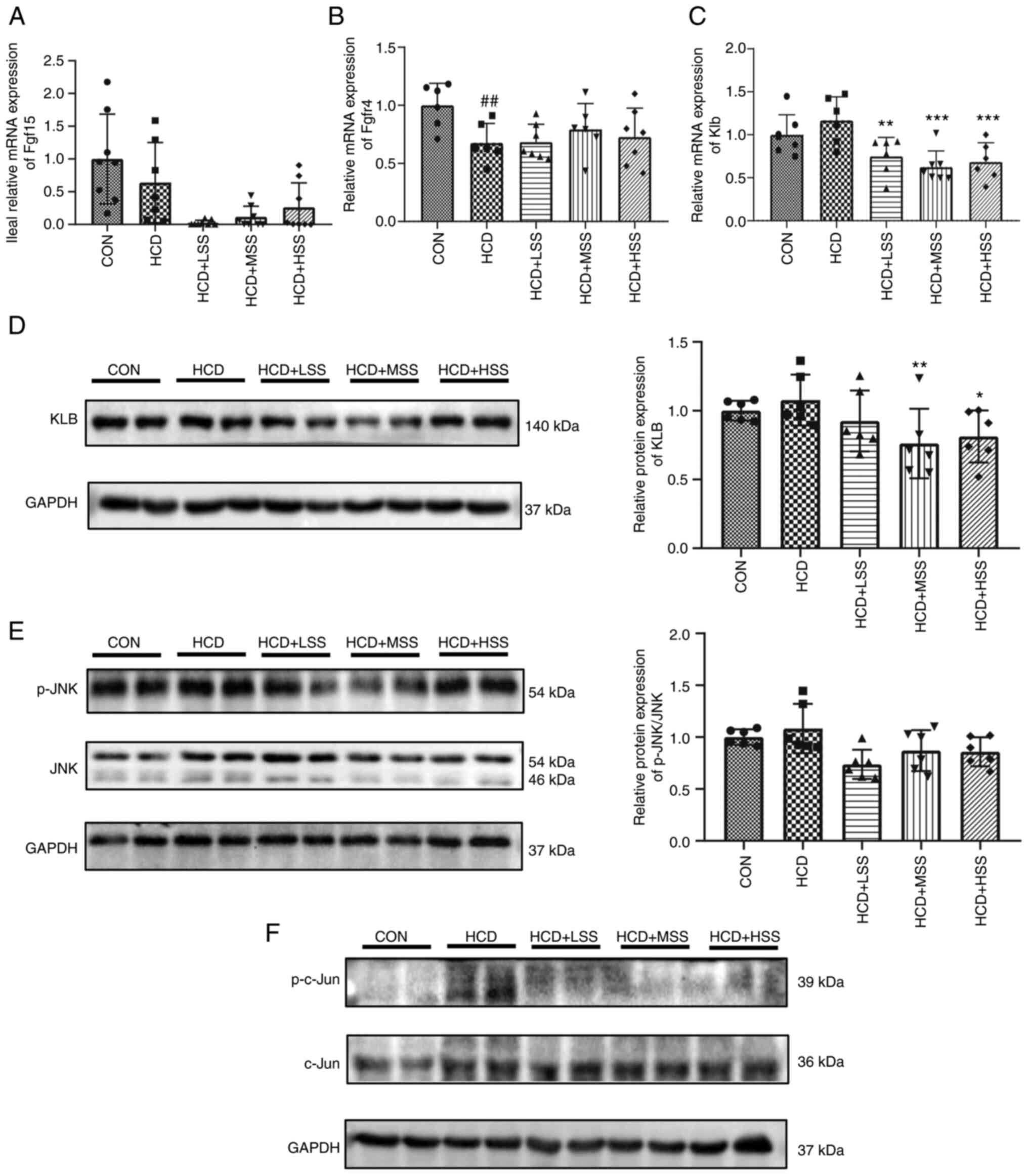 | Figure 4Effect of sodium sulphate on the
FGF15/FGFR4-Klb/JNK signaling pathway in the livers of mice fed an
HCD. Reverse transcription-quantitative PCR results of the relative
mRNA expression levels of (A) Fgf15 in the ileum and of (B)
Fgfr4 and (C) Klb in the liver tissues of mice from
the five study groups: CON, HCD, HCD + LSS, HCD + MSS and HCD +
HSS. (D) Western blotting results showing the protein expression
level of KLB in the liver tissues of mice from the five groups.
GAPDH was used as the internal control. The right panel shows the
semi-quantitative analysis. (E) The p-JNK and JNK protein
expression levels in the livers of mice from the five groups. The
right panel shows the semi-quantitative analysis of the p-JNK/JNK
ratio. (F) Western blotting results showing the p-c-Jun and c-Jun
protein expression levels in the liver tissues of mice from the
five groups. GAPDH was used as the internal control.
##P<0.01 vs. the CON group; *P<0.05,
**P<0.01, ***P<0.001 vs. the HCD group.
HCD, high cholesterol diet; CON, control; LSS, low dose of sodium
sulphate; MSS, middle dose of sodium sulphate; HSS, high dose of
sodium sulphate; FGF, fibroblast growth factor; FGFR, FGF receptor;
KLB, Klotho β; JNK, c-Jun N-terminal kinase; p-,
phosphorylated. |
JNK is an important kinase that is activated by
FGF15 signaling to inhibit the transcription of Cyp7a1 in
hepatocytes (38,39). Activated JNK suppresses the
expression of Cyp7a1 via the activation of c-Jun (40). Therefore, the activation and
expression levels of JNK and its target, c-Jun, were further
assessed using western blotting. The p-JNK/JNK ratio was slightly
higher in the hepatic tissues of mice from the HCD group compared
with the CON group, but this increase was not significant, and the
administration of sodium sulphate reduced this ratio, although this
reduction was also not significant (Fig. 4E). The p-c-Jun level was notably
increased in the liver tissue of the HCD group compared with the
CON group, and was decreased following administration of sodium
sulphate (Fig. 4F). These results
indicate that sodium sulphate may upregulate Cyp7a1
expression via inhibition of the FGF15/FGFR4-KLB/JNK/c-Jun
signaling pathway to promote the conversion of cholesterol to bile
acid.
Sodium sulphate changes the
composition and function of the gut microbiota in mice fed an
HCD
The gut microbiota has been demonstrated to have an
important role in regulating the lipid metabolism of the host
(41). Therefore, 16S rDNA
sequencing was performed to analyze the composition and function of
the gut microbiota of mice from the CON, HCD, HCD + LSS, HCD + MSS
and HCD + HSS groups. The Shannon rarefaction curves of each group
reached the saturation plateau (Fig.
5A), indicating that the sequence coverage of the fecal samples
was sufficient to describe the composition of the gut microbiota of
all groups. Principal coordinate analysis demonstrated that the
five groups were significantly distinguished (Fig. 5B). At the phylum level, the
proportional abundance of Bacteroidetes was significantly
lower in the HCD group compared with the CON group, and was
significantly reduced in the HCD + LSS and HCD + MSS groups
compared with the HCD group (Fig.
5C and D). The proportional
abundance of Proteobacteria was significantly higher in the
HCD group compared with the CON group, and was significantly
reduced in the HCD + LSS group compared with the HCD group
(Fig. 5C and D). Furthermore, the proportional
abundance of Firmicutes was significantly increased in the
sodium sulphate treated groups compared with the HCD group
(Fig. 5C and D).
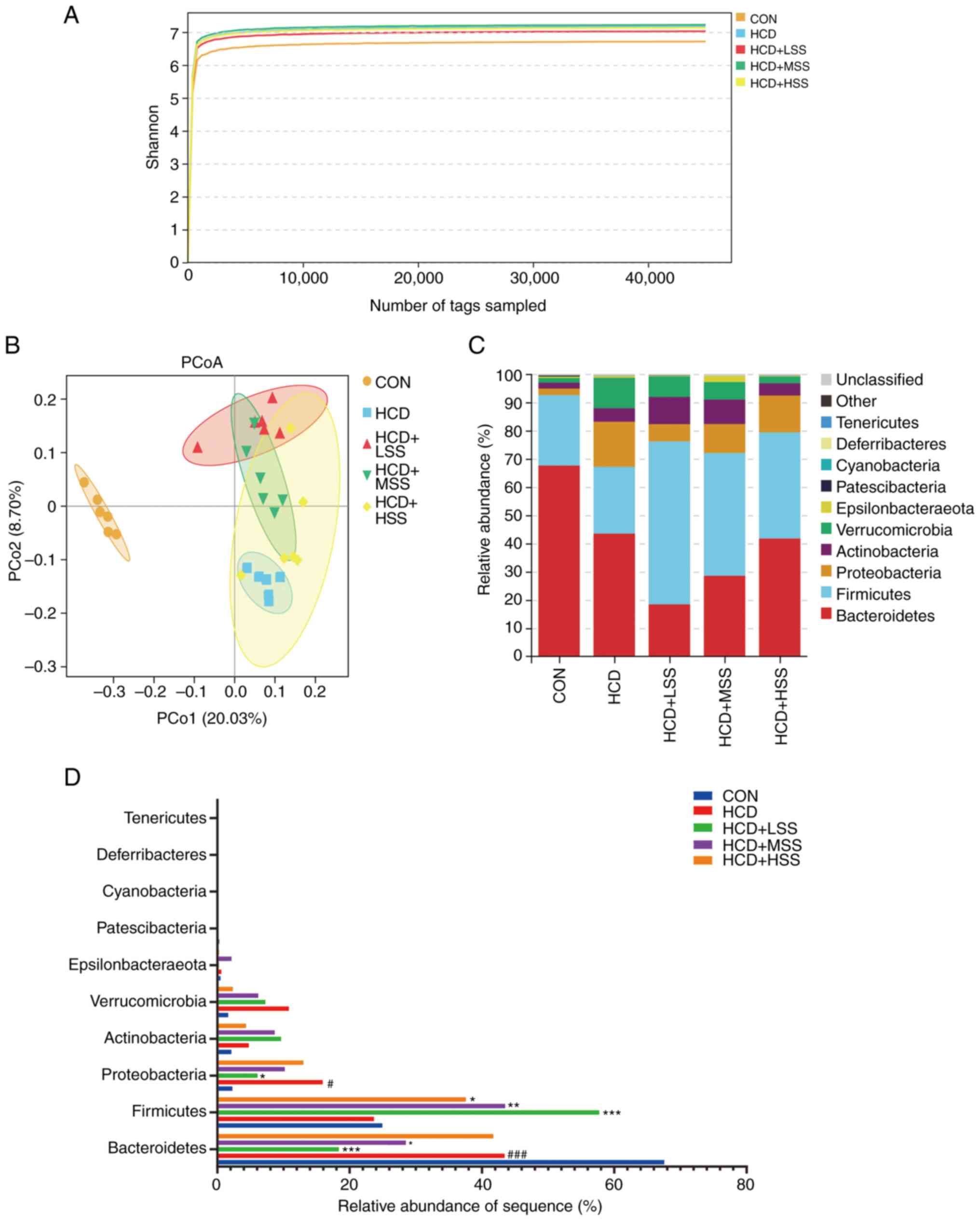 | Figure 5Sodium sulphate changed the relative
abundance of the gut microbiota in mice fed an HCD. (A) Shannon
rarefaction curves for the five study groups: CON, HCD, HCD + LSS,
HCD + MSS and HCD + HSS. (B) The PCoA analysis of the gut
microbiota from mice in the five groups. (C) Relative abundance of
the gut microbiota at the phylum level from mice in the five
groups. The different colors indicate different flora. (D) Bar
chart of the proportional abundance of the gut microbiota at the
phylum level in mice from the five groups. #P<0.05,
###P<0.001 vs. the CON group; *P<0.05,
**P<0.01, ***P<0.001 vs. the HCD group.
HCD, high cholesterol diet; CON, control; LSS, low dose of sodium
sulphate; MSS, middle dose of sodium sulphate; HSS, high dose of
sodium sulphate; PCoA, principal coordinate analysis. |
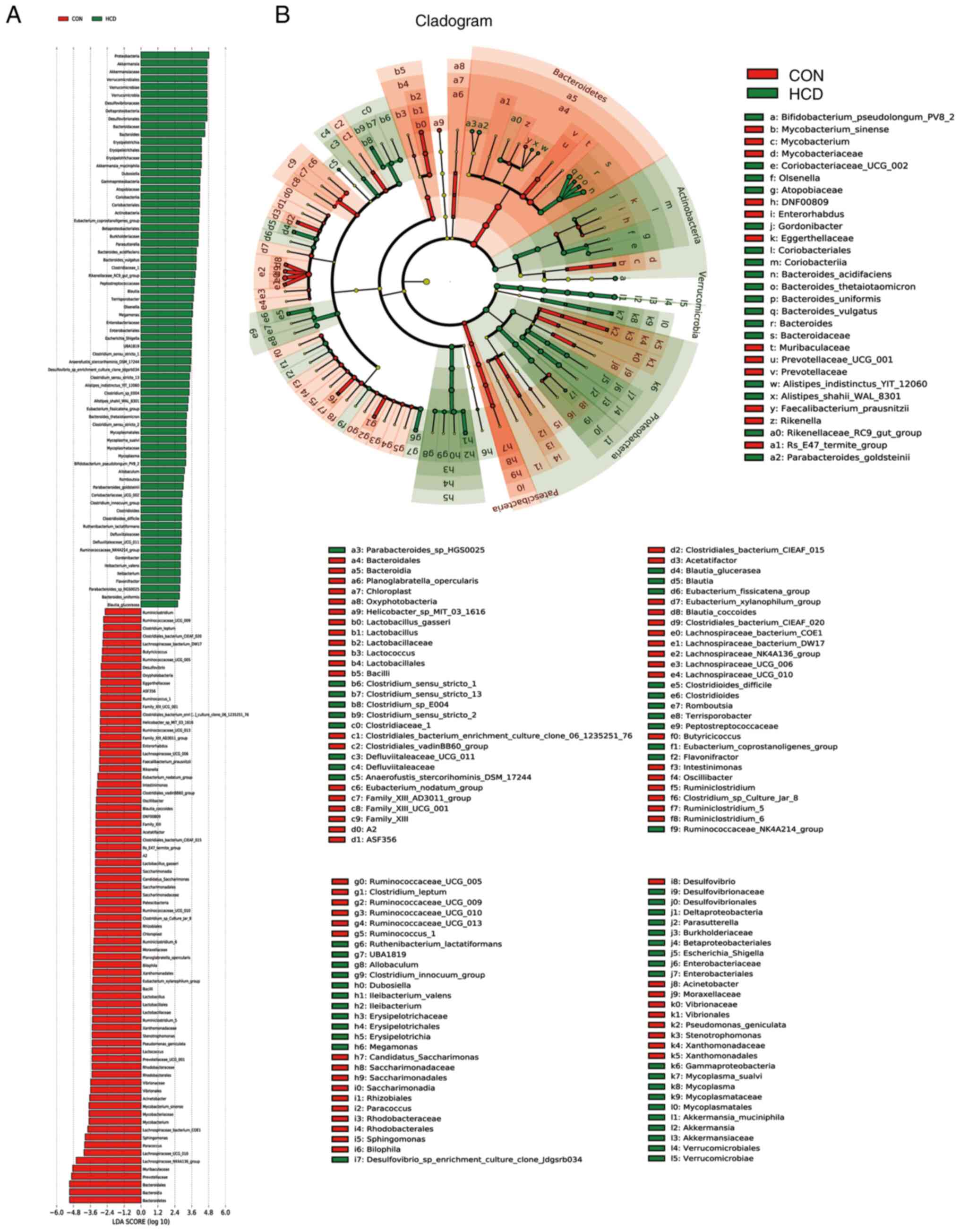
The results of the LEFse analysis demonstrated that
there were 147 bacterial taxa that differed in abundance between
the CON and HCD groups, with 76 predominant in the CON group and 71
predominant in the HCD group (Fig.
6). There were 115 bacterial taxa that differed in abundance
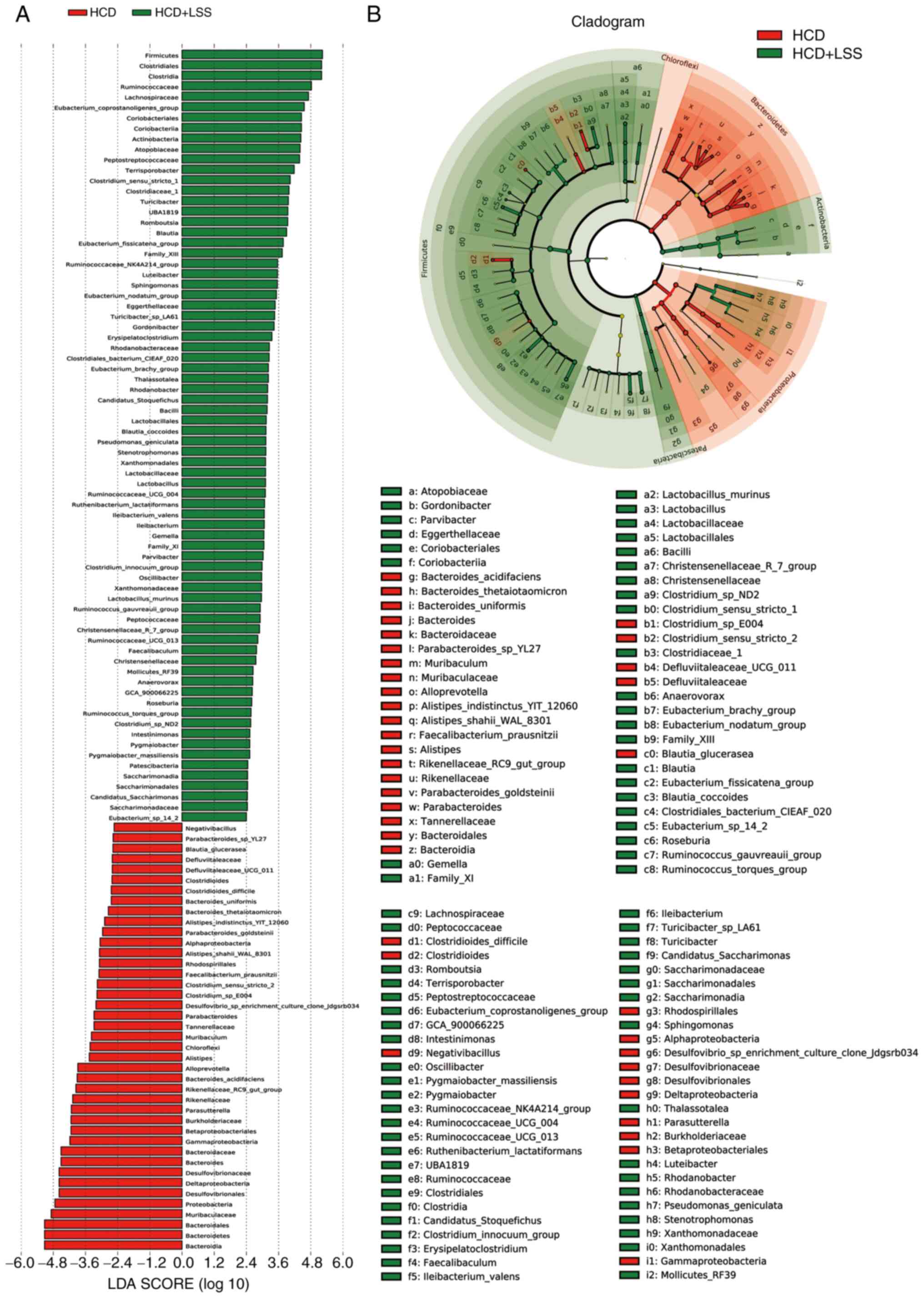
between the HCD and HCD + LSS groups, with 41 predominant in the
HCD group and 74 predominant in the HCD + LSS group (Fig. 7). There were 91 bacterial taxa that
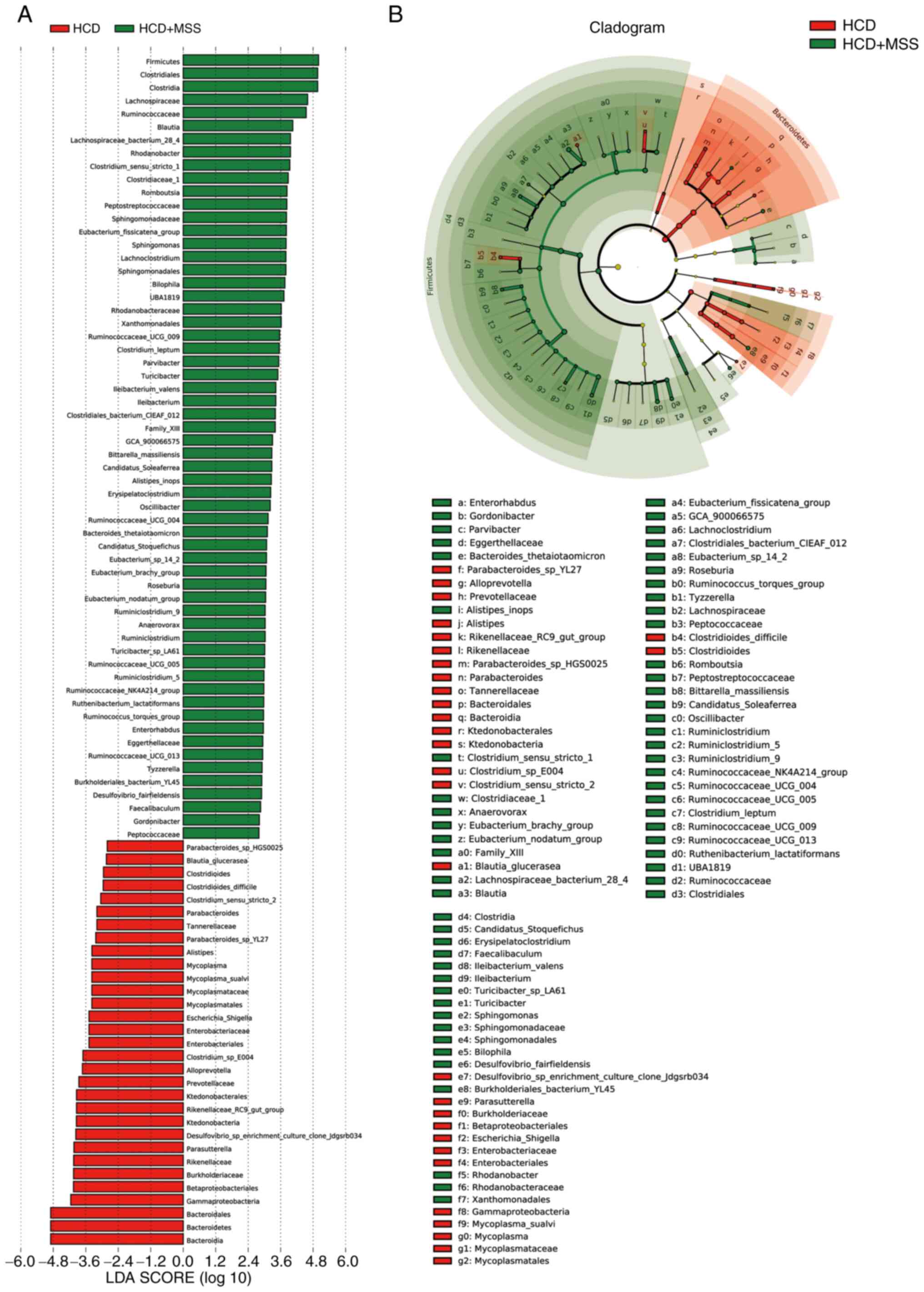
differed in abundance between the HCD and HCD + MSS groups, with 31
predominant in the HCD group and 60 predominant in the HCD + MSS
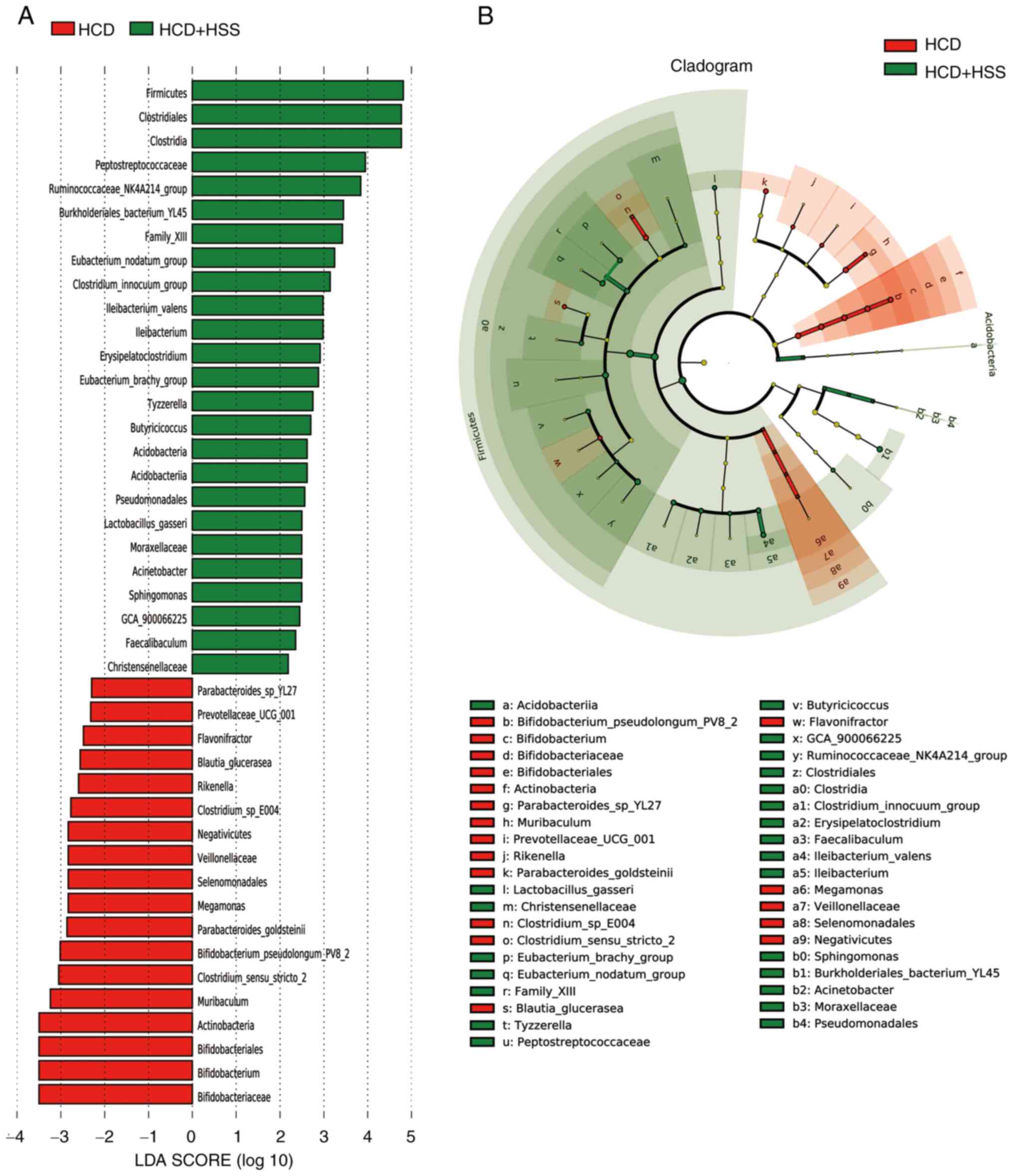
group (Fig. 8). There were 43
bacterial taxa that differed in abundance between the HCD and HCD +
HSS groups, with 18 predominant in the HCD group and 25 predominant
in the HCD+HSS group (Fig. 9).
Compared with the HCD group, there were 13 predominant bacterial
taxa in all three groups treated with sodium sulphate, including:
Phylum, Firmicutes; order, Clostridiales; class,
Clostridia; family, Peptostreptococcaceae; genus,
Ruminococcaceae_NK4A214_group; family, Family_XIII;
genus, Eubacterium_nodatum_group; species,
Ileibacterium_valens; genus, Ileibacterium; genus,
Erysipelatoclostridium; genus,
Eubacterium_brachy_group; genus, Sphingomonas; and
genus, Faecalibaculum (Fig.
7, Fig. 8 and Fig. 9).
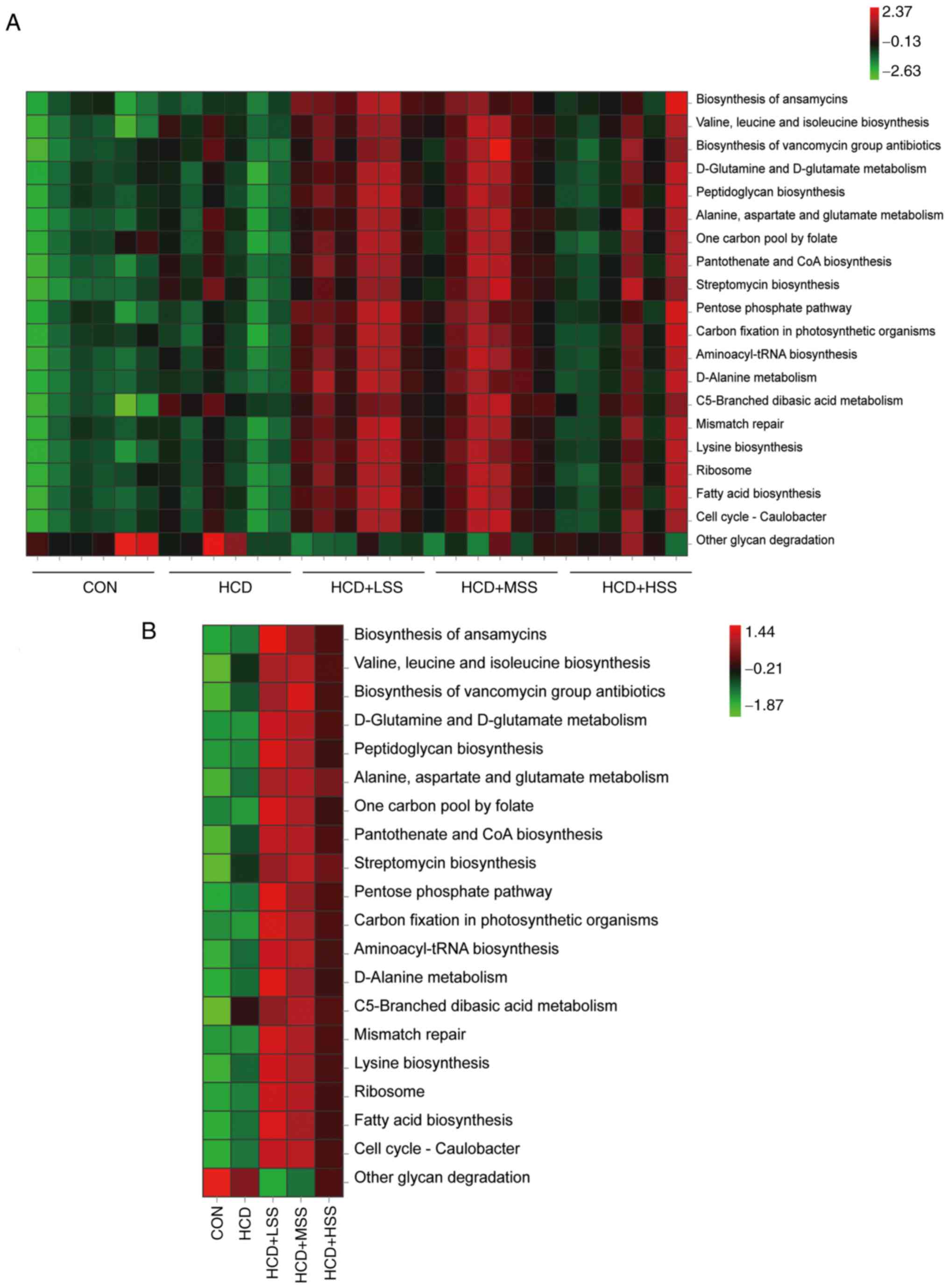
The top 20 altered pathways in the KEGG pathway
analysis are shown in Fig. 10.
These included 19 pathways that were significantly increased in all
three sodium sulphate treated groups compared with the CON and HCD
groups, including ‘Biosynthesis of ansamycins’, ‘Valine, leucine
and isoleucine biosynthesis’, ‘Biosynthesis of vancomycin group
antibiotics’, ‘D-Glutamine and D-glutamate metabolism’,
‘Peptidoglycan biosynthesis’, ‘Alanine, aspartate and glutamate
metabolism’, ‘One carbon pool by folate’, ‘Pantothenate and CoA
biosynthesis’, ‘Streptomycin biosynthesis’, ‘Pentose phosphate
pathway’, ‘Carbon fixation in photosynthetic organism’,
‘Aminoacyl-tRNA biosynthesis’, ‘D-Alanine metabolism’, ‘C5-Branched
dibasic acid metabolism’, ‘Mismatch repair’, ‘Lysine biosynthesis’,
‘Ribosome’, ‘Fatty acid biosynthesis’ and ‘Cell cycle-Caulobacter’.
Most of these pathways were also increased in the HCD group
compared to the CON group, with the exception of ‘D-Glutamine and
D-glutamate metabolism’, ‘One carbon pool by folate’, ‘Carbon
fixation in photosynthetic organism’ and ‘Mismatch repair’. The
‘Other glycan degradation’ pathway was significantly decreased in
all three of the sodium sulphate treated groups compared with the
CON and HCD groups, and also decreased in the HCD group compared
with the CON group.
Discussion
In the present study, a novel role of sodium
sulphate in the amelioration of hypercholesterolemia in mice fed an
HCD was uncovered. Firstly, it was confirmed that the
administration of sodium sulphate reduced the TC concentration and
LDL-C/HDL-C ratio in the serum of mice fed an HCD. It was also
found that sodium sulphate upregulated the expression of
Cyp7a1, Cyp39a1 and Ldlr in the hepatic
tissues of mice fed an HCD to increase the conversion and uptake of
cholesterol in hepatocytes. Subsequently, it was demonstrated that
sodium sulphate attenuated the insulin resistance in the livers of
mice fed an HCD via downregulation of the expression of
Trib3 to increase the activation of AKT in hepatocytes.
Finally, it was shown that sodium sulphate regulated the expression
of Cyp7a1 via inhibition of the FGF15/FGFR4-KLB/JNK/c-Jun
signaling pathway in the livers of mice fed an HCD. These results
indicate the function and mechanisms of sodium sulphate in the
reduction of hypercholesterolemia in mice fed an HCD.
Increased TC levels causes >2.6 million deaths
every year worldwide, and one-third of coronary heart disease cases
are caused by an elevated serum TC concentration (42). An increase in serum LDL-C
concentration is also a risk factor of ASCVD (43). The serum LDL-C level in patients
with homozygous familial hypercholesterolemia patients is highly
elevated and accelerates premature ASCVD (44). In the present study, sodium
sulphate significantly reduced the serum concentration of TC and
the LDL-C/HDL-C ratio in mice that were administered an HCD. These
results indicate that sodium sulphate may be used to control and
treat atherosclerosis and ASCVD. Mirabilite has been used in
traditional Chinese medicine for nearly 2,000 years (6). Mirabilite is one of the four herbs of
the traditional Chinese formulation Da-Cheng-Qi-Tang that is used
to treat atherosclerosis (45,46).
However, it is unclear which herb of the Da-Cheng-Qi-Tang has an
anti-atherosclerotic function. The results of the present study
indicate that sodium sulphate, the effective ingredient of
mirabilite, might be active against atherosclerosis since it
attenuates hypercholesterolemia in mice.
CYP7A1 plays an important role in the maintenance
of the homeostasis of cholesterol and bile acid in vivo
(47). Cyp7a1-/-
mice exhibit a marked reduction in bile acid synthesis (48) and exacerbated alcohol-induced
hepatic inflammation and injury (49). A number of studies have
demonstrated that the upregulation of Cyp7a1 in the liver
reduces the serum cholesterol concentration in rat and mouse
models. For example, it was reported that diosgenin attenuated
hypercholesterolemia in rats fed a high-fat diet (HFD) via
regulation of the scavenger receptor class B type
1/carboxylesterase-1/CYP7A1/FXR pathway, and Cyp7a1 was
significantly upregulated by diosgenin in the livers of HFD-fed
rats (50). In addition, it has
been found that flavonoids from mulberry leaves and their active
metabolite, quercetin reduce excessive cholesterol accumulation in
rats with orotic acid-induced NAFLD, and upregulate the expression
of Cyp7a1 in the hepatic tissue of these rats (51). Furthermore, in another study tomato
seed oil reduced the concentration of TC, TG and LDL-C and the
LDL-C/HDL-C ratio in the plasma of mice fed an HFD by increasing
the expression of hepatic peroxisome proliferator-activated
receptor α, acyl-CoA-dehydrogenase long chain, CYP7A1, liver X
receptor α, ATP binding cassette subfamily A member 1 and scavenger
receptor class B type 1(52).
Cyp39a1 is one of the key genes in the synthesis of bile
acid via the mevalonate pathway (53). It has been shown that the
activation of pregnane X receptor induces hypercholesterolemia in
wild-type mice and accelerates atherosclerosis in
ApoE-/- mice, while significantly repressing the
Cyp39a1 expression level in the livers of these mice
(54). Hence, Cyp39a1 is
also essential for the metabolic elimination of cholesterol in
vivo. In the present study, the expression of Cyp7a1 and
Cyp39a1 in the hepatic tissues of mice fed an HCD was
significantly upregulated by the administration of sodium sulphate,
indicating that sodium sulphate enhances the conversion of
cholesterols to bile acids in the mouse hepatocytes. The
administration of sodium sulphate also increased hepatic
Ldlr expression in the mice fed an HCD, and the upregulation
of Ldlr or increase in LDLR activity results in the removal
of excessive LDL-C from the circulation (55). The increase in the conversion of
cholesterols to bile acids could also explain the changes in the
composition and function of the gut microbiota in the present study
since bile acids are important regulators of microbiota (56).
An increase in Trib3 expression has been
found to induce insulin resistance in the hepatocytes, myotubes and
other types of cells (31,57-59).
It has been reported that TRB3 protein expression is increased in
the livers of elderly rats and is associated with insulin
resistance (57). It has also been
demonstrated that TRB3 is an endogenous inhibitor of AKT that
inhibits the insulin signaling pathway (31). In addition, lipotoxicity has been
shown to induce the upregulation of TRB3 and COP1, and thereby
induce the degradation of sirtuin1, resulting in insulin resistance
in hepatocytes in vivo and in vitro (58). Furthermore, in another study,
TRPM2-activated Ca2+ signaling aggravated endothelial
insulin resistance via the PERK/ATF4/TRB3 cascade in HFD-induced
obese mice (59). In the present
study, the administration of sodium sulphate significantly
downregulated Trib3 expression, which was significantly
increased in the livers of mice fed an HCD, and alleviated the
insulin resistance in the hepatic tissues caused by the HCD. These
results indicate that sodium sulphate mitigates the insulin
resistance induced by increased TRB3 expression in the liver and
other tissues. The insulin resistance index will be determined in
future studies to confirm the ability of sodium sulphate to
ameliorate insulin resistance in mice fed an HCD.
Previous studies have confirmed that the FGF15/19
signaling pathway plays an important role in regulating
Cyp7a1 expression in hepatocytes. For example, in one study,
blocking the ileum bile salt transporter SLC10A2 in HFD-fed Syrian
golden hamsters significantly suppressed FGF15/19 expression in the
ileum, and it was found that the reduced FGF15/19 signaling in
hepatocytes upregulated the expression of Cyp7a1 in the
hepatic tissues of these animals via reduction of the
phosphorylation of ERK1/2 and JNK in hepatocytes (38). In addition, it was reported that
chronic overexpression of FGF21 in mice significantly upregulated
the expression of Cyp7a1, as FGF21 antagonised the function
of FGF15/19 as an inhibitor of Cyp7a1 expression in
hepatocytes (60). In the present
study, the transcriptional levels of FGF15 were very low in the
ileum of HCD-fed mice following the administration of sodium
sulphate. We hypothesize that sodium sulphate inhibits the
absorption of bile acid in the ileum of mice, then reduces the
expression of FGF15 in these tissues, since the serum TBA
concentrations in the serum of the sodium sulphate-treated mice
were lower than those in the HCD group, albeit not significantly.
The downregulation of FGF15 in the ileum of mice by sodium sulphate
may be associated with the increase in Cyp7a1 expression in
hepatocytes.
FGF15/19 binds to FGFR4 and its co-receptor, KLB,
on hepatic membranes to inhibit Cyp7a1 expression in
hepatocytes (61,62). In a previous study,
FGFR4-/- mice exhibited upregulated hepatic
Cyp7a1 expression and elevated bile-acid excretion (63). In another study, it was reported
that the expression of Cyp7a1 and Cyp8b1 was highly
upregulated in the livers of Klb knockout mice, and the
synthesis and excretion of bile acid were also notably increased in
these mice (64). Therefore, the
downregulation of Klb in the livers of mice could be another
mechanism by which sodium sulphate increases the expression of
Cyp7a1 in hepatocytes and elevates the synthesis of bile
acid in mice fed an HCD. It has been confirmed that FGF15/19
signaling downregulates the expression of Cyp7a1 via the
ERK1/2 and JNK pathways in hepatocytes (65). In the present study, the
phosphorylation of JNK and its major target, c-Jun, was
downregulated in the livers of mice fed an HCD after the
administration of sodium sulphate for 3 weeks. Therefore, we
hypothesize that sodium sulphate ameliorates the
hypercholesterolemia in mice caused by HCD via inhibition of the
FGF15/FGFR4-KLB/JNK/c-Jun signaling pathway in hepatocytes. The
inhibition of this pathway might affect the gut microbiota due to
the increase in the production of bile acids in the liver, although
it is unclear whether sodium sulphate inhibits the
FGF15/FGFR4-KLB/JNK/c-Jun pathway directly or via the regulation of
the gut microbiota. In addition, JNK activation might promote the
serine phosphorylation of IRS1 to reduce the p-AKT/AKT ratio in the
insulin signaling pathway in hepatocytes (66). Hence, inhibition of the
FGF15/FGFR4-KLB/JNK/c-Jun pathway might be a mechanism by which
sodium sulphate increases the p-AKT/AKT ratio in the insulin signal
pathway in hepatocytes.
In the present study, the Firmicutes phylum
was significantly increased in mice treated with sodium sulphate. A
previous study found that oatmeal reduced the levels of TC and
LDL-C in patients with hypercholesterolemia, and the
Firmicutes phylum was also significantly increased in the
fecal microbiota of these patients (67). It has also been reported that the
gut microbiota in ob/ob mice have upregulated KEGG pathways
associated with the breakdown of indigestible dietary
polysaccharides, such as butanoate metabolism, galactose metabolism
and starch/sucrose metabolism, to increase the energy harvest for
the host (68). In the present
study, the KEGG analysis demonstrated that the pathways for
‘Peptidoglycan biosynthesis’ and ‘Carbon fixation in photosynthetic
organism’ were increased, and the pathway for ‘Other glycan
degradation’ was decreased in mice following treatment with sodium
sulphate. We hypothesize that the changes in the composition and
functions of the gut microbiota in mice treated with sodium
sulphate might reduce the extraction of energy from food by the
host. Furthermore, the reduced metabolic extraction might improve
the reduced p-AKT/AKT ratio in the insulin signaling pathway in the
hepatocytes of mice fed an HCD.
In conclusion, sodium sulphate mitigated the
hypercholesterolemia and hepatic insulin resistance in mice fed an
HCD (Fig. 11). Sodium sulphate
also downregulated the highly expressed Trib3 in the
hepatocytes of HCD-fed mice to alleviate hepatic insulin resistance
in these mice. In the enterocytes of the ileum from mice fed an
HCD, the expression of FGF15 was downregulated by the
administration of sodium sulphate. In addition, the hepatic
expression of KLB, which is the co-receptor of FGFR4 in the binding
of FGF15, was also significantly downregulated by sodium sulphate,
and the activation of JNK, which is downstream of FGF15/FGFR4, was
also reduced. Furthermore, the activation of c-Jun, which is the
major target of p-JNK, was also reduced, and the expression of
Cyp7a1, which is inhibited by p-c-Jun, was significantly
increased to enhance the conversion of cholesterols to bile acids
in mice fed an HCD. In future, it would be worthwhile to explore
the ability of sodium sulphate to alleviate the serum levels of TC
and LDL-C in animal models of familial hypercholesterolemia.
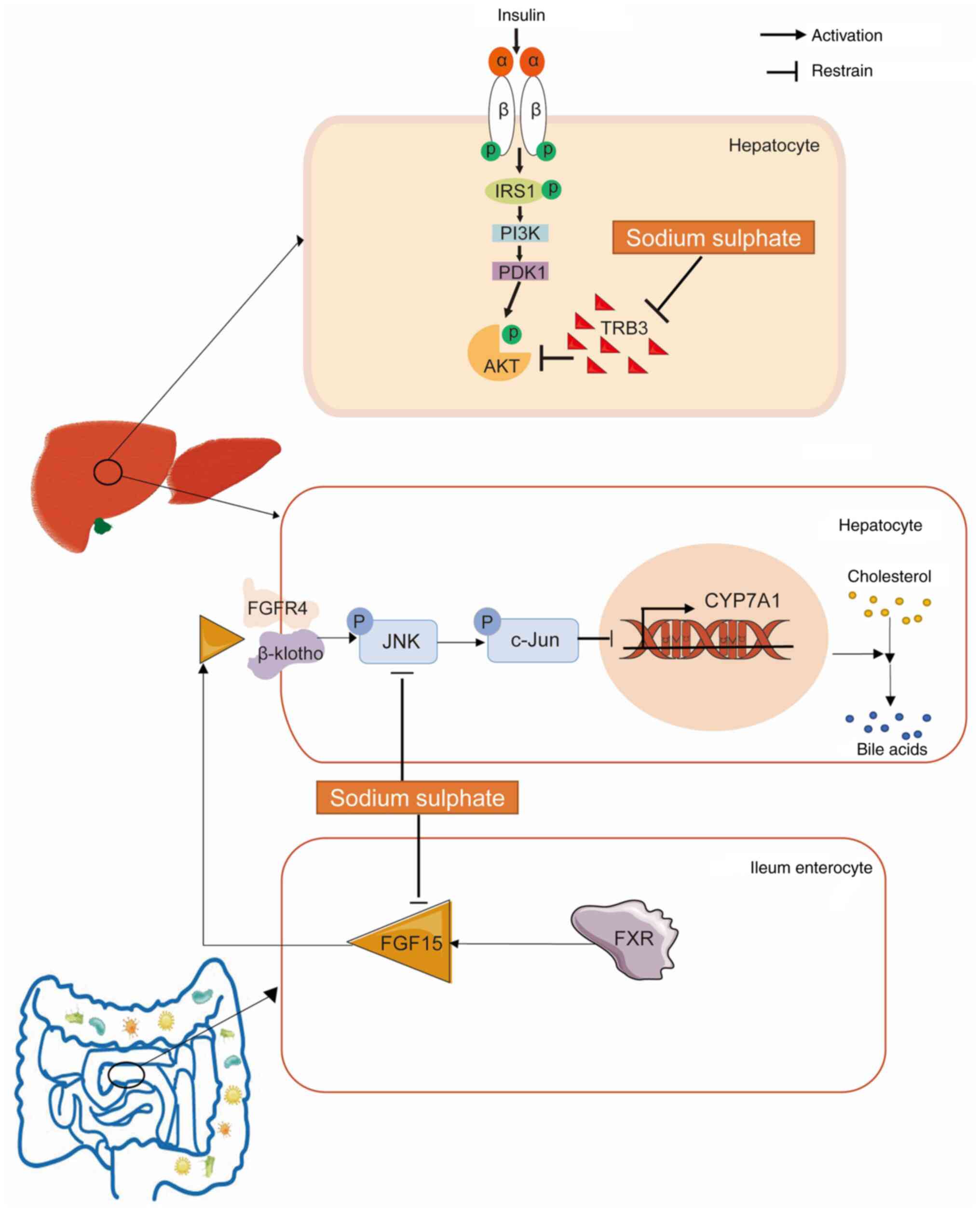 | Figure 11Sodium sulphate ameliorates
hypercholesterolemia and hepatic insulin resistance in mice fed an
HCD. Sodium sulphate inhibits the highly expressed TRB3 in the
hepatocytes of mice fed an HCD, to attenuate the hepatic insulin
resistance of these mice. In the enterocytes of the ileum, FGF15
expression is downregulated following the administration of sodium
sulphate to mice fed an HCD. In addition, the hepatic expression of
KLB, which is the co-receptor of FGFR4 for FGF15 binding, is
significantly downregulated by sodium sulphate, and reduces the
activation of JNK, which is downstream of the FGF15/FGFR4-KLB
signaling pathway. The activation of c-Jun, which is the major
target of p-JNK, is notably reduced, and the expression of
Cyp7a1, which is inhibited by p-c-Jun, is significantly
increased, which enhances the conversion of cholesterols to bile
acids in these mice. HCD, high cholesterol diet; TRB3, tribbles
homolog 3; FGF, fibroblast growth factor; FGFR, FGF receptor; KLB,
Klotho β; JNK, c-Jun N-terminal kinase; p-, phosphorylated; IRS1,
insulin receptor substrate 1; PDK1, 3-phosphoinositide-dependent
kinase 1; FXR, farnesoid X receptor. |
Supplementary Material
Generation of hypercholesterolemic
mouse models and the administration of sodium sulphate to
hypercholesterolemic mice. (A) Overview of the experimental design.
(B) Serum TC concentration in mice after 4 weeks of high
cholesterol administration. (C) Body weight curves and (D) average
daily food intake curves of mice from the CON, HCD, HCD + LSS, HCD
+ MSS, and HCD + HSS groups during the administration of sodium
sulphate. (E) Average daily defecation mass of mice from the five
groups. ###P<0.001 vs. the CON group. CON, control;
HCD, high cholesterol diet; TC, total cholesterol; LSS, low dose of
sodium sulphate; MSS, middle dose of sodium sulphate; HSS, high
dose of sodium sulphate.
Effect of sodium sulphate on the
morphology and histology of the livers from mice fed an HCD. (A)
Representative photographic images of the livers of mice from the
five study groups: CON, HCD, HCD + LSS, HCD + MSS, and HCD + HSS.
(B) Liver weight and (C) liver/body weight ratio of mice from the
five groups. (D) Representative images of haematoxylin and eosin
stained liver tissue sections from mice in the five groups. Scale
bar, 40 μm. ##P<0.01, # #
#P<0.001 vs. the CON group. CON, control; HCD, high
cholesterol diet; LSS, low dose of sodium sulphate; MSS, middle
dose of sodium sulphate; HSS, high dose of sodium sulphate.
Comparison of the hepatic mRNA
expression profiles in mice from the CON, HCD and HCD + MSS groups
determined via RNA-sequencing technology. (A) Graph showing the
numbers of differentially expressed mRNAs in the liver tissues of
mice between the CON vs. HCD, CON vs. HCD + MSS, and HCD vs. HCD +
MSS groups. Heatmap showing the differ-entially expressed mRNAs in
the liver tissues of mice between the (B) CON and HCD, (C) HCD and
HCD + MSS groups and (D) CON and HCD + MSS groups. (E) Heatmap
showing the differentially expressed mRNAs encoding bile acid
transporters in the livers of mice from the CON, HCD and HCD + nMSS
groups. CON, control; HCD, high cholesterol diet; MSS, middle dose
of sodium sulphate.
Effect of sodium sulphate on the
expression of genes associated with cholesterol synthesis in the
liver tissues of mice fed an HCD. (A) Graphs showing the relative
mRNA expression levels of Hmgcs1, Hmgcr, Mvk, Mvd, Idi1, Fdps,
Fdft1, Sqle, Lss, Dhcr7 and Cyp51a1 in the liver tissues from mice
in the CON, HCD, HCD + LSS, HCD + MSS and HCD + HSS groups. (B)
Western blotting results showing the protein expression levels of
HMGCR, MVK, MVD, IDI1, FDPS, FDFT1 and LSS in the liver tissues of
mice from the five groups. ##P<0.01,
###P<0.001 vs. the CON group; *P<0.05,
**P<0.01 vs. the HCD group. CON, control; HCD, high
cholesterol diet; LSS, low dose of sodium sulphate; MSS, middle
dose of sodium sulphate; HSS, high dose of sodium sulphate; Hmgcs1,
3-hydroxy-3-methylglutaryl-CoA synthase 1; Hmgcr,
3-hydroxy-3-methylglutaryl-CoA reductase; Mvk, mevalonate kinase;
Mvd, mevalonate diphosphate decarboxylase; Idi1,
isopentenyl-diphosphate δ isomerase 1; Fdps, farnesyl diphosphate
synthase; Fdft1, farnesyl diphosphate farnesyltransferase 1; Sqle,
squalene epoxidase; Oss, oxidosqualene-lanosterol cyclase
(lanosterol synthase); Dhcr7, 7-dehydrocholesterol reductase;
Cyp51a1, cytochrome P450 family member 51 subfamily member A member
1.
Primer sequences used for quantitative
PCR.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the National Natural
Science Foundation of China (grant nos. 81830113, 81803912 and
82171855); National key R & D plan ‘Research on modernization
of traditional Chinese medicine’ (grant no. 2018YFC1704200); Major
basic and applied basic research projects of Guangdong Province of
China (grant no. 2019B030302005); the Guangdong Basic and Applied
Basic Research Foundation (grant no. 2021A1515012383); the Opening
Foundation of the Key Laboratory of Regenerative Biology, Guangzhou
Institutes of Biomedicine and Health, Chinese Academy of Sciences
(grant no. KLRB201807); the Science and Technology Planning Project
of Guangzhou City (grant no. 201803010069); and the Science and
Technology Project of Yue-Xiu District of Guangzhou (grant no.
2018-WS-011).
Availability of data and materials
The RNA-seq data is available from https://www.ncbi.nlm.nih.gov/sra/PRJNA774883. The
other datasets generated or analysed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
ZL, YY and JG designed the project and conceived
the manuscript. YY, CY and SY analysed and interpreted the results
of mRNA sequencing and the 16S rDNA sequences. YY and ZL wrote the
draft of the manuscript and revised it critically. SY, HR, YY and
CY established the mouse model and administered sodium sulphate.
YY, SY, ZL, HR, CY, HW, TZ, FY, YN, LC, QH and QS performed the
morphological, biochemical and molecular experiments, including
serum and liver biochemical profiles, western blotting, RT-qPCR and
H&E staining. SY, HR and CY created the figures and table. ZL
and YY confirm the authenticity of all the raw data. All authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The mouse experiments were approved by The
Guangdong Pharmaceutical University Experimental Animal Ethics
Committee (approval no. gdpulacspf2017030-1; Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ruotsalainen AK, Mäkinen P and
Ylä-Herttuala S: Novel RNAi-based therapies for atherosclerosis.
Curr Atheroscler Rep. 23(45)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
World Health Organization (WHO):
Cardiovascular diseases (CVDs). WHO, Geneva, 2021. https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds).
Accessed June 11, 2021.
|
|
3
|
Zhang M, Deng Q and Wang L, Huang Z, Zhou
M, Li Y, Zhao Z, Zhang Y and Wang L: Prevalence of dyslipidemia and
achievement of low-density lipoprotein cholesterol targets in
Chinese adults: A nationally representative survey of 163,641
adults. Int J Cardiol. 260:196–203. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Meng XD, Yao HH, Wang LM, Yu M, Shi S,
Yuan ZX and Liu J: Knockdown of GAS5 inhibits atherosclerosis
progression via reducing EZH2-mediated ABCA1 transcription in
ApoE(-/-) mice. Mol Ther Nucl Acids. 19:84–96. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tseng SH, Lee HH, Chen LG, Wu CH and Wang
CC: Effects of three purgative decoctions on inflammatory
mediators. J Ethnopharmacol. 105:118–124. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhong XG, Zheng FJ, Li YH, Xu H, Wang Q,
Liu YC, Liu M, Wu RH, Gao YS, Zhang SJ, et al: Specific link
between lung and large intestine: A new perspective on neuropeptide
secretion in lung with herbal laxative stimulation. Evid Based
Complement Alternat Med. 2013(547837)2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sun H, Zhang AH, Zhang HL, Zhou XH, Wang
XQ, Liu L and Wang XJ: Ultra-performance liquid chromatography/mass
spectrometry technology and high-throughput metabolomics for
deciphering the preventive mechanism of mirabilite on colorectal
cancer via the modulation of complex metabolic networks. RSC Adv.
9:35356–35363. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mottacki N, Simrén M and Bajor A: Review
article: Bile acid diarrhoea-pathogenesis, diagnosis and
management. Aliment Pharmacol Ther. 43:884–898. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jahnel J, Fickert P, Hauer AC, Högenauer
C, Avian A and Trauner M: Inflammatory bowel disease alters
intestinal bile acid transporter expression. Drug Metab Dispos.
42:1423–1431. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Camilleri M and Vijayvargiya P: The role
of bile acids in chronic diarrhea. Am J Gastroenterol.
115:1596–1603. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shin A, Camilleri M, Vijayvargiya P,
Busciglio I, Burton D, Ryks M, Rhoten D, Lueke A, Saenger A,
Girtman A and Zinsmeister AR: Bowel functions, fecal unconjugated
primary and secondary bile acids, and colonic transit in patients
with irritable bowel syndrome. Clin Gastroenterol Hepatol.
11:1270–1275.e1271. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kim YC, Seok S, Zhang Y, Ma J, Kong B, Guo
G, Kemper B and Kemper JK: Intestinal FGF15/19 physiologically
repress hepatic lipogenesis in the late fed-state by activating SHP
and DNMT3A. Nat Commun. 11(5969)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fiorucci S, Distrutti E, Carino A,
Zampella A and Biagioli M: Bile acids and their receptors in
metabolic disorders. Prog Lipid Res. 82(101094)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu X, Ge H, Lemon B, Weiszmann J, Gupte J,
Hawkins N, Li X, Tang J, Lindberg R and Li Y: Selective activation
of FGFR4 by an FGF19 variant does not improve glucose metabolism in
ob/ob mice. Proc Natl Acad Sci USA. 106:14379–14384.
2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Williams CM, Calderon JH, Hock E, Jimenez
Y, Barringer K, Carbonaro M, Molina-Portela MDP, Thurston G, Li Z
and Daly C: Monomeric/dimeric forms of Fgf15/FGF19 show
differential activity in hepatocyte proliferation and metabolic
function. FASEB J. 35(e21286)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
de Vos WM, Tilg H, Van Hul M and Cani PD:
Gut microbiome and health: Mechanistic insights. Gut. 71:1020–1032.
2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kriaa A, Bourgin M, Potiron A, Mkaouar H,
Jablaoui A, Gérard P, Maguin E and Rhimi M: Microbial impact on
cholesterol and bile acid metabolism: Current status and future
prospects. J Lipid Res. 60:323–332. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Le Roy T, Lécuyer E, Chassaing B, Rhimi M,
Lhomme M, Boudebbouze S, Ichou F, Barceló JH, Huby T, Guerin M, et
al: The intestinal microbiota regulates host cholesterol
homeostasis. BMC Biol. 17(94)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Vourakis M, Mayer G and Rousseau G: The
role of gut microbiota on cholesterol metabolism in
atherosclerosis. Int J Mol Sci. 22(8074)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tong LT, Xiao T, Wang L, Lu C, Liu L, Zhou
X, Wang A, Qin W and Wang F: Plant protein reduces serum
cholesterol levels in hypercholesterolemia hamsters by modulating
the compositions of gut microbiota and metabolites. iScience.
24(103435)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen S, Zhou Y, Chen Y and Gu J: fastp: An
ultra-fast all-in-one FASTQ preprocessor. Bioinformatics.
34:i884–i890. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucl Acids Res. 36:D480–D484. 2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhuri D, Gurkan H, Eker D, Karal Y,
Yalcintepe S, Atli E, Demir S and Atli EI: Investigation on the
effects of modifying genes on the spinal muscular atrophy
phenotype. Global Med Gene. 9:226–236. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lei Z, Wu H, Yang Y, Hu Q, Lei Y, Liu W,
Nie Y, Yang L, Zhang X, Yang C, et al: Ovariectomy impaired hepatic
glucose and lipid homeostasis and altered the gut microbiota in
mice with different diets. Front Endocrinol (Lausanne).
12(708838)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lennernäs H and Fager G: Pharmacodynamics
and pharmacokinetics of the HMG-CoA reductase inhibitors. Clin
Pharmacokinet. 32:403–425. 1997.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu A, Jin H, Dirsch O, Deng M, Huang H,
Bröcker-Preuss M and Dahmen U: Release of danger signals during
ischemic storage of the liver: A potential marker of organ damage?
Mediators Inflamm. 2010(436145)2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tan D, Ling L, Qin L, Lu Y, Wu D and He Y:
Rosiglitazone induces hepatocyte injury by increasing DCA
accumulation through OATP1A4 inhibiting in mice. Arab J Chem.
16(105142)2023.
|
|
29
|
Khan AA, Sundar P, Natarajan B, Gupta V,
Arige V, Reddy SS, Barthwal MK and Mahapatra NR: An
evolutionarily-conserved promoter allele governs HMG-CoA reductase
expression in spontaneously hypertensive rat. J Mol Cell Cardiol.
158:140–152. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhong S, Li L, Liang N, Zhang L, Xu X,
Chen S and Yin H: Acetaldehyde Dehydrogenase 2 regulates HMG-CoA
reductase stability and cholesterol synthesis in the liver. Red
Biol. 41(101919)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cheng KK, Iglesias MA, Lam KS, Wang Y,
Sweeney G, Zhu W, Vanhoutte PM, Kraegen EW and Xu A: APPL1
potentiates insulin-mediated inhibition of hepatic glucose
production and alleviates diabetes via Akt activation in mice. Cell
Metab. 9:417–427. 2009.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Du K, Herzig S, Kulkarni RN and Montminy
M: TRB3: A tribbles homolog that inhibits Akt/PKB activation by
insulin in liver. Science. 300:1574–1577. 2003.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Koo SH, Satoh H, Herzig S, Lee CH, Hedrick
S, Kulkarni R, Evans RM, Olefsky J and Montminy M: PGC-1 promotes
insulin resistance in liver through PPAR-alpha-dependent induction
of TRB-3. Nat Med. 10:530–534. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Lei Z, Yang L, Yang Y, Yang J, Niu Z,
Zhang X, Song Q, Lei Y, Wu H and Guo J: Activation of Wnt/β-catenin
pathway causes insulin resistance and increases lipogenesis in
HepG2 cells via regulation of endoplasmic reticulum stress. Biochem
Biophys Res Commun. 526:764–771. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang Z, Du Z, Liu Q, Wu T, Tang Q, Zhang
J, Huang C, Huang Y, Li R, Li Y, et al: Glucagon-like peptide 1
analogue prevents cholesterol gallstone formation by modulating
intestinal farnesoid X receptor activity. Metabolism.
118(154728)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang F, Zhao C, Yang M, Zhang L, Wei R,
Meng K, Bao Y, Zhang L and Zheng J: Four citrus flavanones exert
atherosclerosis alleviation effects in apoE(-/-) mice via different
metabolic and signaling pathways. J Agric Food Chem. 69:5226–5237.
2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Gulfo J, Rotondo F, de León CG,
Cornide-Petronio ME, Fuster C, Gracia-Sancho J, Jiménez-Castro MB
and Peralta C: FGF15 improves outcomes after brain dead donor liver
transplantation with steatotic and non-steatotic grafts in rats. J
Hepatol. 73:1131–1143. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ge MX, Niu WX, Ren JF, Cai SY, Yu DK, Liu
HT, Zhang N, Zhang YX, Wang YC, Shao RG, et al: A novel ASBT
inhibitor, IMB17-15, repressed nonalcoholic fatty liver disease
development in high-fat diet-fed Syrian golden hamsters. Acta Pharm
Sin. 40:895–907. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jung D, York JP, Wang L, Yang C, Zhang A,
Francis HL, Webb P, McKeehan WL, Alpini G, Lesage GD, et al:
FXR-induced secretion of FGF15/19 inhibits CYP27 expression in
cholangiocytes through p38 kinase pathway. Pflugers Arch.
466:1011–1019. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gupta S, Stravitz RT, Dent P and Hylemon
PB: Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene
expression by bile acids in primary rat hepatocytes is mediated by
the c-Jun N-terminal kinase pathway. J Biol Chem. 276:15816–15822.
2001.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Schoeler M and Caesar R: Dietary lipids,
gut microbiota and lipid metabolism. Rev Endocr Metab Disord.
20:461–472. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Peters SA, Singhateh Y, Mackay D, Huxley
RR and Woodward M: Total cholesterol as a risk factor for coronary
heart disease and stroke in women compared with men: A systematic
review and meta-analysis. Atherosclerosis. 248:123–131.
2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Cao J, Remaley AT, Guan W, Devaraj S and
Tsai MY: Performance of novel low-density lipoprotein-cholesterol
calculation methods in predicting clinical and subclinical
atherosclerotic cardiovascular disease risk: The multi-ethnic study
of atherosclerosis. Atherosclerosis. 327:1–4. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wu Y, Jiang L, Zhang H, Cheng S, Wen W, Xu
L, Zhang F, Yang Y, Wang L and Chen J: Integrated analysis of
microRNA and mRNA expression profiles in homozygous familial
hypercholesterolemia patients and validation of atherosclerosis
associated critical regulatory network. Genomics. 113:2572–2582.
2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ha KT, Kim JK, Lee YC and Kim CH:
Inhibitory effect of Daesungki-Tang on the invasiveness potential
of hepatocellular carcinoma through inhibition of matrix
metalloproteinase-2 and -9 activities. Toxic App Pharmacol.
200:1–6. 2004.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chung HJ, Kim DW, Maruyama I and Tani T:
Effects of traditional Chinese formulations on rat carotid artery
injured by balloon endothelial denudation. Am J Chin Med.
31:201–212. 2003.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chiang JY: Bile acids: Regulation of
synthesis. J Lipid Res. 50:1955–1966. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Schwarz M, Russell DW, Dietschy JM and
Turley SD: Marked reduction in bile acid synthesis in cholesterol
7alpha-hydroxylase-deficient mice does not lead to diminished
tissue cholesterol turnover or to hypercholesterolemia. J Lipid
Res. 39:1833–1843. 1998.PubMed/NCBI
|
|
49
|
Donepudi AC, Ferrell JM, Boehme S, Choi HS
and Chiang JYL: Deficiency of cholesterol 7α-hydroxylase in bile
acid synthesis exacerbates alcohol-induced liver injury in mice.
Hepatol Commun. 2:99–112. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yu L, Lu H, Yang X, Li R, Shi J, Yu Y, Ma
C, Sun F, Zhang S and Zhang F: Diosgenin alleviates
hypercholesterolemia via SRB1/CES-1/CYP7A1/FXR pathway in high-fat
diet-fed rats. Toxicol Appl Pharmacol. 412(115388)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hu Y, Xu J, Chen Q, Liu M, Wang S, Yu H,
Zhang Y and Wang T: Regulation effects of total flavonoids in Morus
alba L. on hepatic cholesterol disorders in orotic acid induced
NAFLD rats. BMC Complement Med Ther. 20(257)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
He WS, Li L, Rui J, Li J, Sun Y, Cui D and
Xu B: Tomato seed oil attenuates hyperlipidemia and modulates gut
microbiota in C57BL/6J mice. Food Funct. 11:4275–4290.
2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhang Y, Liu Y, Duan J, Wang H, Zhang Y,
Qiao K and Wang J: Cholesterol depletion sensitizes gallbladder
cancer to cisplatin by impairing DNA damage response. Cell Cycle.
18:3337–3350. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhou C, King N, Chen KY and Breslow JL:
Activation of PXR induces hypercholesterolemia in wild-type and
accelerates atherosclerosis in apoE deficient mice. J Lipid Res.
50:2004–2013. 2009.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Tiwari V and Khokhar M: Mechanism of
action of anti-hypercholesterolemia drugs and their resistance. Eur
J Pharmacol. 741:156–170. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Fuchs CD, Paumgartner G, Mlitz V, Kunczer
V, Halilbasic E, Leditznig N, Wahlström A, Ståhlman M, Thüringer A,
Kashofer K, et al: Colesevelam attenuates cholestatic liver and
bile duct injury in Mdr2(-/-) mice by modulating composition,
signalling and excretion of faecal bile acids. Gut. 67:1683–1691.
2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Gaspar RC, Muñoz VR, Nakandakari S, Vieira
RFL, da Conceição LR, de Oliveira F, Crisol BM, da Silva ASR,
Cintra DE, de Moura LP, et al: Aging is associated with increased
TRB3, ER stress, and hepatic glucose production in the liver of
rats. Exp Gerontol. 139(111021)2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ren X, Chen N, Chen Y, Liu W and Hu Y:
TRB3 stimulates SIRT1 degradation and induces insulin resistance by
lipotoxicity via COP1. Exp Cell Res. 382(111428)2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Sun L, Liu YL, Ye F, Xie JW, Zeng JW, Qin
L, Xue J, Wang YT, Guo KM, Ma MM, et al: Free fatty acid-induced
H(2)O(2) activates TRPM2 to aggravate endothelial insulin
resistance via Ca(2+)-dependent PERK/ATF4/TRB3 cascade in obese
mice. Free Radic Biol Med. 143:288–299. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zhang J, Gupte J, Gong Y, Weiszmann J,
Zhang Y, Lee KJ, Richards WG and Li Y: Chronic over-expression of
fibroblast growth factor 21 increases bile acid biosynthesis by
opposing FGF15/19 ACTION. EBioMedicine. 15:173–183. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Fu T, Kim YC, Byun S, Kim DH, Seok S,
Suino-Powell K, Xu HE, Kemper B and Kemper JK: FXR primes the liver
for intestinal FGF15 signaling by transient induction of β-Klotho.
Mol Endocrinol. 30:92–103. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Kliewer SA and Mangelsdorf DJ: Bile acids
as hormones: The FXR-FGF15/19 Pathway. Dig Dis. 33:327–331.
2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Yu C, Wang F, Kan M, Jin C, Jones RB,
Weinstein M, Deng CX and McKeehan WL: Elevated cholesterol
metabolism and bile acid synthesis in mice lacking membrane
tyrosine kinase receptor FGFR4. J Biol Chem. 275:15482–15489.
2000.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Ito S, Fujimori T, Furuya A, Satoh J and
Nabeshima Y and Nabeshima Y: Impaired negative feedback suppression
of bile acid synthesis in mice lacking betaKlotho. J Clin Invest.
115:2202–2208. 2005.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Ge MX, Shao RG and He HW: Advances in
understanding the regulatory mechanism of cholesterol
7α-hydroxylase. Biochem Pharmacol. 164:152–164. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Samuel VT and Shulman GI: Mechanisms for
insulin resistance: Common threads and missing links. Cell.
148:852–871. 2012.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Ye M, Sun J, Chen Y, Ren Q, Li Z, Zhao Y,
Pan Y and Xue H: Oatmeal induced gut microbiota alteration and its
relationship with improved lipid profiles: A secondary analysis of
a randomized clinical trial. Nutr Metab (Lond).
17(85)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini
V, Mardis ER and Gordon JI: An obesity-associated gut microbiome
with increased capacity for energy harvest. Nature. 444:1027–1031.
2006.PubMed/NCBI View Article : Google Scholar
|