Introduction
Hearing loss is the most prevalent neurosensory
disorder in humans. The World Health Organization stated in its
World Report on Hearing that >1.5 billion individuals presently
experience various levels of hearing impairment. Alarmingly, this
number is projected to increase to 2.5 billion by the year
2050(1). The reported incidence of
hearing loss in newborns ranges from 1 to 2 per 1,000 births, with
a genetic cause identified in more than half of these cases
(2,3). Early detection and intervention are
crucial for hearing loss in newborns. Hearing loss is one of the
most common congenital anomalies among newborns, and if it is not
diagnosed and treated in a timely manner, it can severely impact
the language, social and cognitive development of a child (4). Since the 1990s, the implementation of
universal newborn hearing screening (UNHS) programs has played a
crucial role in the early detection, diagnosis, and intervention of
hearing loss, leading to significant social benefits (4,5). The
UNHS program is globally acknowledged as a highly successful public
health initiative. However, this program has limitations.
Traditional UNHS may not identify children with late-onset or
progressive sensorineural hearing loss, potentially negating the
benefits of early intervention and improved outcomes (6).
In 2006, Morton and Nance (7) pioneered the integration of
molecular-level genetic screening into standard newborn hearing
assessments. This innovative approach entails the collection of
umbilical cord blood or heel-stick blood samples from newborns
shortly after birth, typically within the first 3 days, to identify
both their susceptibility to deafness and common genetic factors
contributing to it (7). Several
studies have demonstrated that combining newborn hearing and
genetic screening significantly enhances early detection rates of
hearing impairments in newborns (8-10),
thereby facilitating earlier personalized and targeted counseling
and intervention measures.
The ‘combined screening’ approach has demonstrated
clear advantages in clinical practice, but it has yet to be fully
elucidated in the context of evidence-based medicine. Therefore,
this systematic review and meta-analysis aimed to determine the
rate of hearing screening pass and genetic screening failure (UNHS
pass/genetic failure) for hearing impairment in China. Furthermore,
the present study aimed to explore the benefits of combined newborn
hearing and genetic screening, particularly for the early diagnosis
and intervention of hearing loss patients at genetic risk. Although
previous research has revealed the potential of combined screening,
further exploration is required to determine how to implement this
strategy most effectively to maximize its clinical benefits
(11). Specifically, it remains
unclear which genetic mutations should be prioritized in standard
screening protocols and how to implement these screenings in
resource-limited settings. Additionally, further research is needed
on the specific impact of combined screening on the success rate of
early interventions and how it may improve outcomes for hearing and
language development in the long term. By addressing these
questions, the present study seeks not only to improve the rate of
early identification of newborn hearing loss but also to explore
how early diagnosis and timely interventions can provide the best
developmental prospects for affected children.
Materials and methods
Protocol registration
The present systematic review and meta-analysis
protocol was registered at the International Platform of Registered
Systematic Review and Meta-analysis Protocols (INPLASY;
registration no. INPLASY202440035; https://inplasy.com/?s=INPLASY202440035),
DOI:10.37766/inplasy2024.4.0035.
Literature inclusion and exclusion
criteria
The inclusion criteria were as follows: i) Original
research; ii) the subjects were newborns; iii) the research
detection technology was a combination of neonatal hearing and
genetic screening; iv) no <3 genes were screened; v) the
original data were complete, and relevant data could be extracted
directly or indirectly for statistical analysis; and vi) the study
was published in English.
The exclusion criteria were as follows: i) Duplicate
published studies; ii) incomplete data or unavailable data; iii)
animal experiments; iv) case reports, reviews and systematic
reviews; v) non-universal neonatal hearing and genetic screening;
and vi) fewer than 3 genes were screened.
Search strategy
In the present meta-analysis, the PubMed (https://pubmed.ncbi.nlm.nih.gov/), Embase
(https://www.embase.com/) and Cochrane Library
databases (https://www.cochranelibrary.com/) were searched from
inception to September 2023. The search terms used were ‘newborn’
‘neonate’ AND ‘hearing loss’ ‘deaf’ AND ‘hearing screening’ AND
‘genetic screening’.
Literature screening and data
extraction
Specifically, two researchers conducted the
literature search, data screening and data extraction processes.
Any disagreements were resolved by consulting a third party. The
following data were extracted from the included studies: Basic
information from the literature, the sample size of the study, the
sex and weight of the study population, and the number of
individuals who passed the hearing screening but failed the genetic
screening, including gap junction protein beta 2 (GJB2), solute
carrier family 26 member 4 (SLC26A4) and mitochondrially encoded
12S RRNA (MT-RNR1). The basic information of the document included
the title of the document, the first author, and the year of
publication.
Literature quality assessment
The 11-item, cross-sectional Research Quality
Evaluation Scale recommended by the Agency for Health Research and
Quality (AHRQ) in the United States was used to evaluate the
quality of the included studies. Responses of ‘yes’, ‘no’ and ‘not
clear’ were scored as ‘1’, ‘0’ and ‘0’, respectively. The total
score ranges from 0 to 11 points. Scores from 0 to 3 are classified
as low quality, scores from 4 to 7 are classified as medium
quality, and scores from 8 to 11 are classified as high quality
(12). The meta-analysis was
performed in accordance with the Preferred Reporting Items for
Systematic Reviews and Meta-Analyses (PRISMA) guidelines (13).
Data synthesis and statistical
analysis
All the data were processed with the statistical
software STATA 15.1 (StataCorp LP). For the synthesis analysis of
the primary outcomes, the weighted mean effect size and its 95%
confidence interval (CI) for each study were calculated to estimate
the overall impact of combined screening on the detection rate of
newborn hearing loss. When multiple studies reported the same
outcome measures, forest plots were utilized to visually display
the effect sizes and their CIs for each study, as well as the total
effect size after combining all studies. A heterogeneity test
result of P>0.1 and I2<50% indicated homogeneity
across; P<0.1 and I2>50% indicated heterogeneity.
In cases of heterogeneity, sensitivity analysis was performed to
identify the sources of heterogeneity, and a random effects model
was applied. This meta-analysis used a random effects model to
summarize the effects. The funnel plot method and Egger's test were
used to investigate publication bias (14).
Results
Literature search results
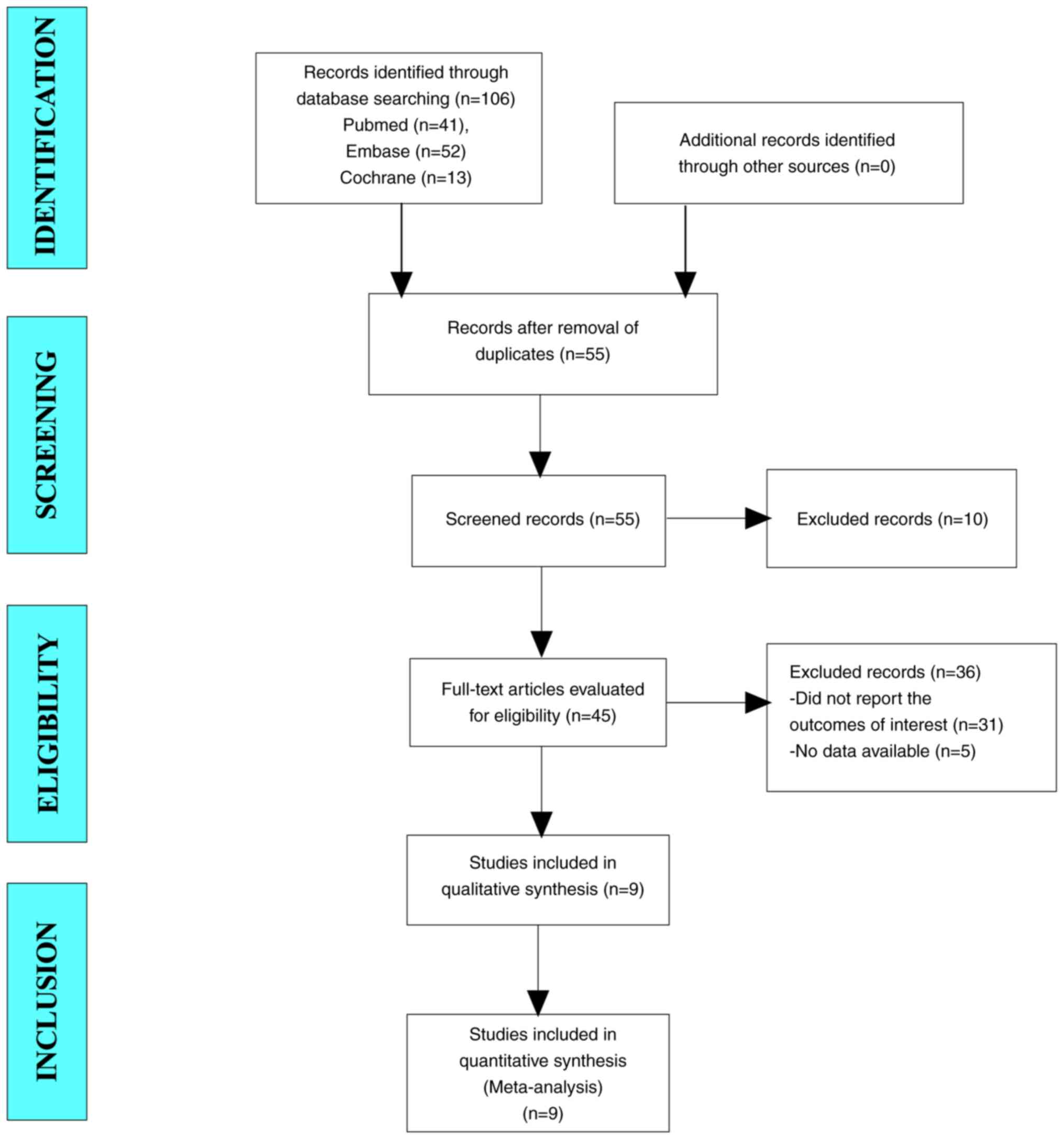
A total of 106 articles were initially retrieved
from the literature search. After excluding duplicate trials, 55
trials remained for screening. After reading the titles and
abstracts, a total of 45 articles were included for full-text
screening. Ultimately, nine studies were included in the
meta-analysis (Fig. 1).
Baseline characteristics and quality
assessment of the included studies
A total of nine cross-sectional studies were
included in this meta-analysis (15-23).
The sample sizes ranged from 1,716 to 180,469, and there were a
total of 377,688 participants. Among the studies describing the sex
distribution, there were 89,701 male and 81,188 female
participants, indicating that the sex distribution was even. The
mean body weight ranged from 3,145.9 to 3,294.6 g (Table I). The AHRQ scores used for quality
assessment were all above eight (Table II).
 | Table IBaseline characteristics and quality
assessment of the included studies. |
Table I
Baseline characteristics and quality
assessment of the included studies.
| First author | Region | Study design | Sample size | Sex
(male/female) | Age (days) | Body weight (g) | Passed hearing
screening | (Refs.) |
|---|
| Wang et al,
2011 | China | Cross-sectional
study | 14,913 | 8,294/6,664 | <28 | 3,236.4±527.3 | 812 | (15) |
| Zhang et al,
2012 | China | Cross-sectional
study | 10,043 | 5,433/4,610 | <28 | 3,145.9±433.1 | 362 | (16) |
| Zhang et al,
2013 | China | Cross-sectional
study | 58,397 | 30,819/27,578 | <28 | / | 770 | (17) |
| Peng et al,
2016 | China | Cross-sectional
study | 9,317 | 4,567/4,750 | <3 | / | 129 | (18) |
| Wu et al,
2017 | China | Cross-sectional
study | 2,024 | / | <28 | / | 42 | (19) |
| Lu et al,
2018 | China | Cross-sectional
study | 1,716 | 837/879 | <28 | / | 58 | (20) |
| Dai et al,
2019 | China | Cross-sectional
study | 1,80,469 | / | <3 | / | 1,915 | (21) |
| Luo et al,
2022 | China | Cross-sectional
study | 24,349 | / | <28 | / | 316 | (22) |
| Wen et al
(1), 2023 | China | Cross-sectional
study | 41,690 | 21,676/20,014 | <3 | 3,294.6±488.8 | 236 | (23) |
| Wen et al
(2), 2023 | China | Cross-sectional
study | 34,770 | 18,075/16,693 | <3 | 3,285.6±490.2 | 187 | (23) |
 | Table IIQuality assessment of the included
studies. |
Table II
Quality assessment of the included
studies.
| First author | A | B | C | D | E | F | G | H | I | J | K | Total | (Refs.) |
|---|
| Wang et
al | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | (15) |
| Zhang et
al | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 9 | (16) |
| Zhang et
al | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | (17) |
| Peng et
al | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 9 | (18) |
| Wu et
al | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 8 | (19) |
| Lu et
al | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | (20) |
| Dai et
al | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | (21) |
| Luo et
al | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 9 | (22) |
| Wen et al
(1) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | (23) |
| Wen et al
(2) | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | (23) |
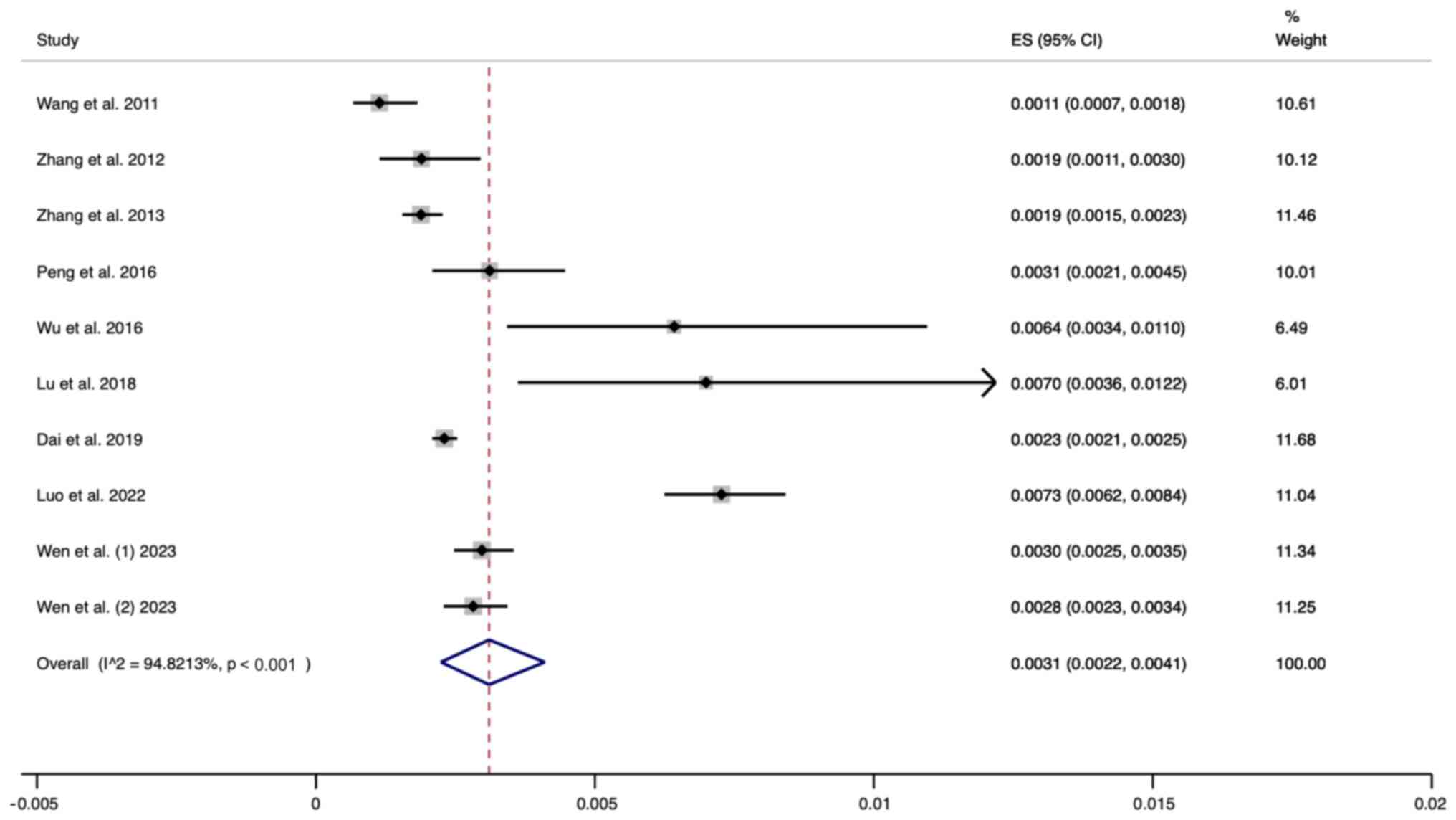
Results of meta-analysis. Prevalence
of UNHS pass/genetic screening failure in neonates
A total of ten studies were included in this
analysis. The random effects model was used for analysis. The
pooled results revealed that the prevalence of passing the UNHS
while failing genetic screening was 0.0031 (95% CI, 0.0022-0.0041)
(Fig. 2).
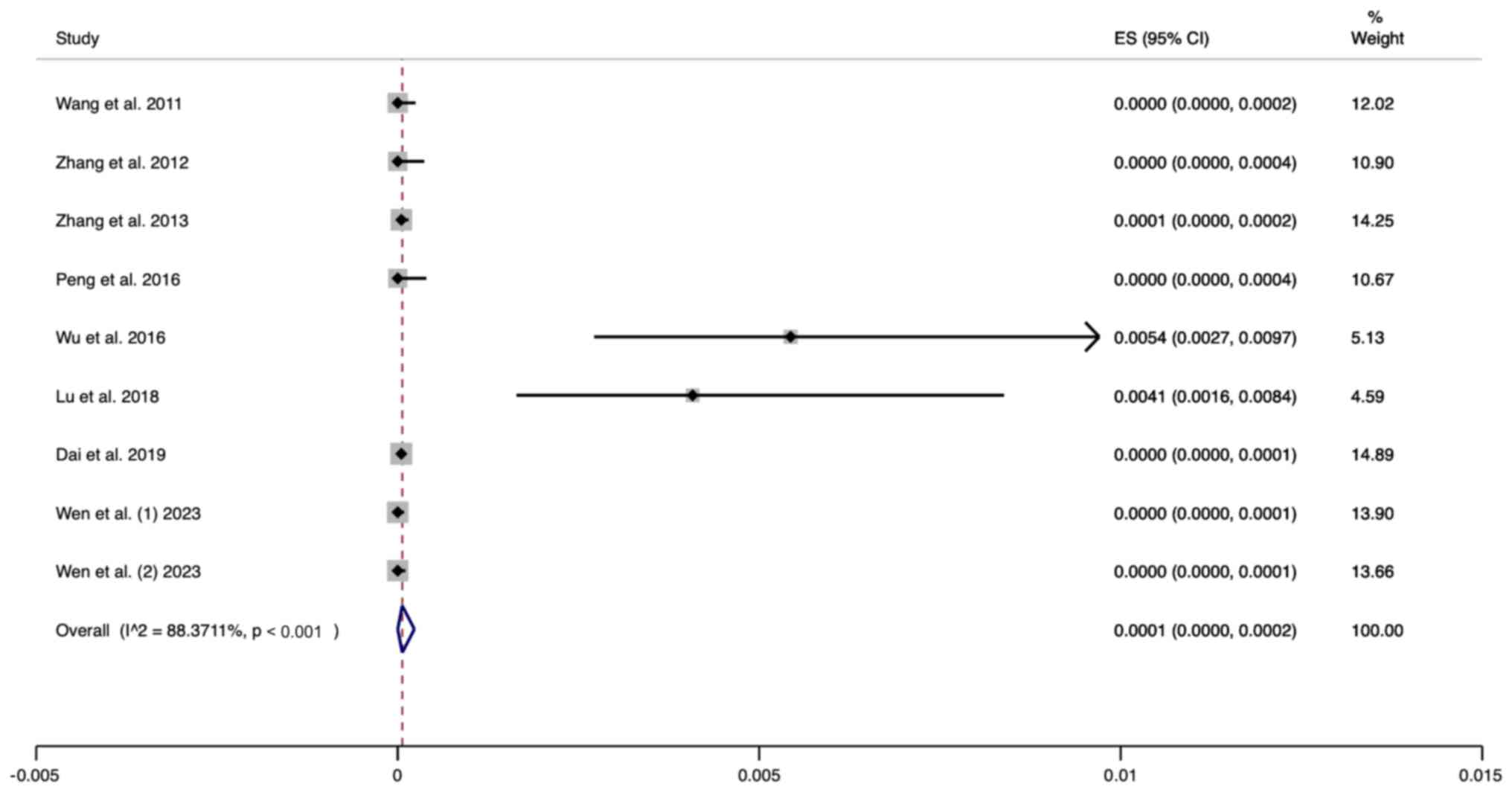
Prevalence of UNHS pass/GJB2 variant
screening failure in neonates
A total of nine studies were included in the present
meta analysis. The random effects model was used for analysis. The
pooled results revealed that the prevalence of passing the UNHS and
failing the GJB2 variant screening was 0.0001 (95% CI,
0.0000-0.0002) (Fig. 3).
Prevalence of NHS pass/SLC26A4 variant
screening failure in neonates
A total of nine studies were included in this
analysis. The random effects model was used for analysis. The
pooled results revealed that the prevalence of passing the UNHS and
failing the SLC26A4 variant screening was 0.0000 (95% CI,
0.0000-0.0000) (Fig. 4).
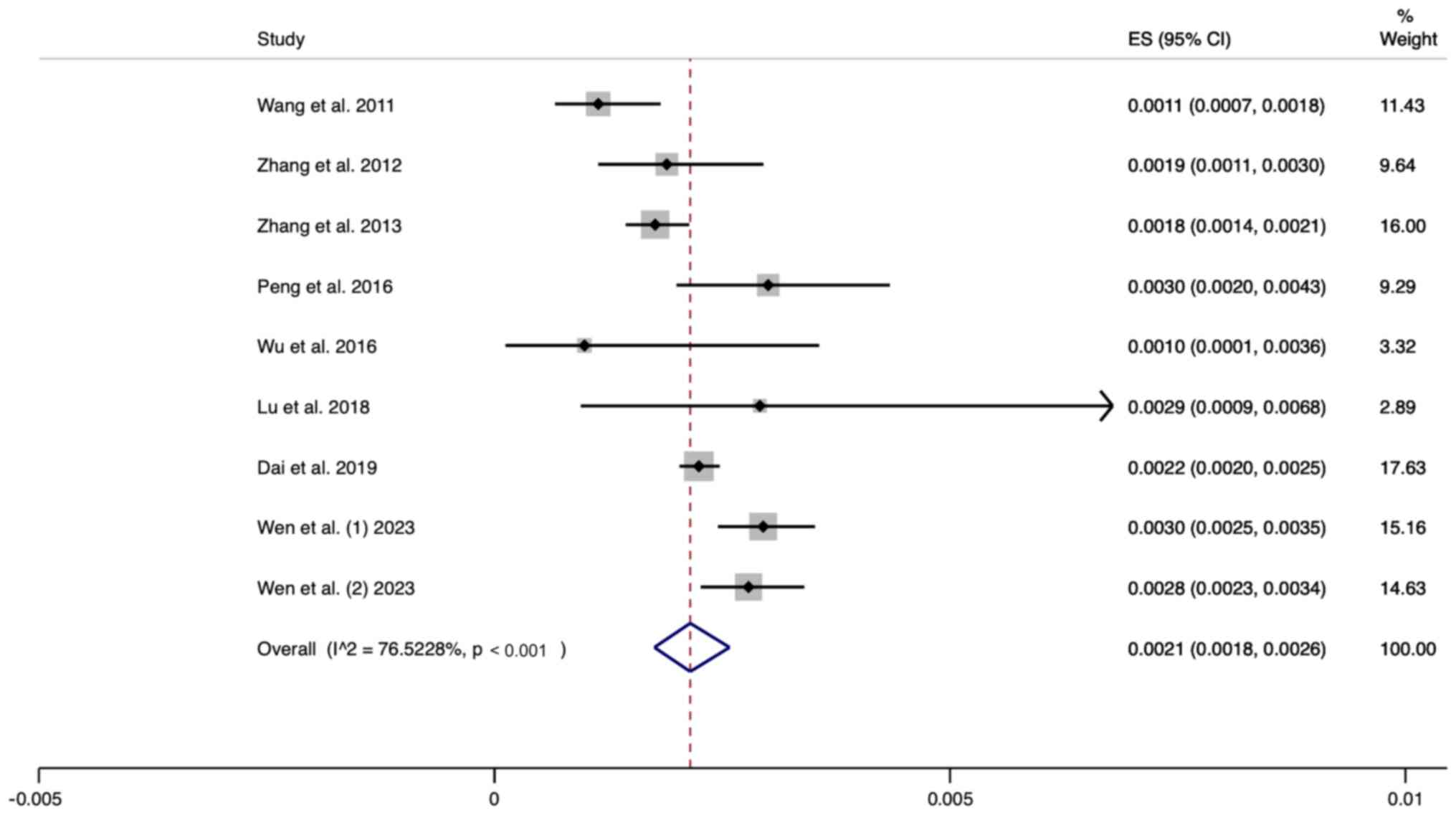
Prevalence of NHS pass/MT-RNR1 variant
screening failure in neonates
A total of nine studies were included in this
analysis. The random effects model was used for analysis. The
pooled results revealed that the prevalence of passing the UNHS and
failing the MT-RNR1 variant screening was 0.0021 (95% CI,
0.0018-0.0026) (Fig. 5).
Sensitivity analysis
A sensitivity analysis was performed to exclude each
of these trials one by one and then a pooled analysis of the
remaining trials was performed. The results revealed that no
individual study had a significant effect on the pooled outcomes,
indicating that the results of the meta-analysis were stable and
reliable.
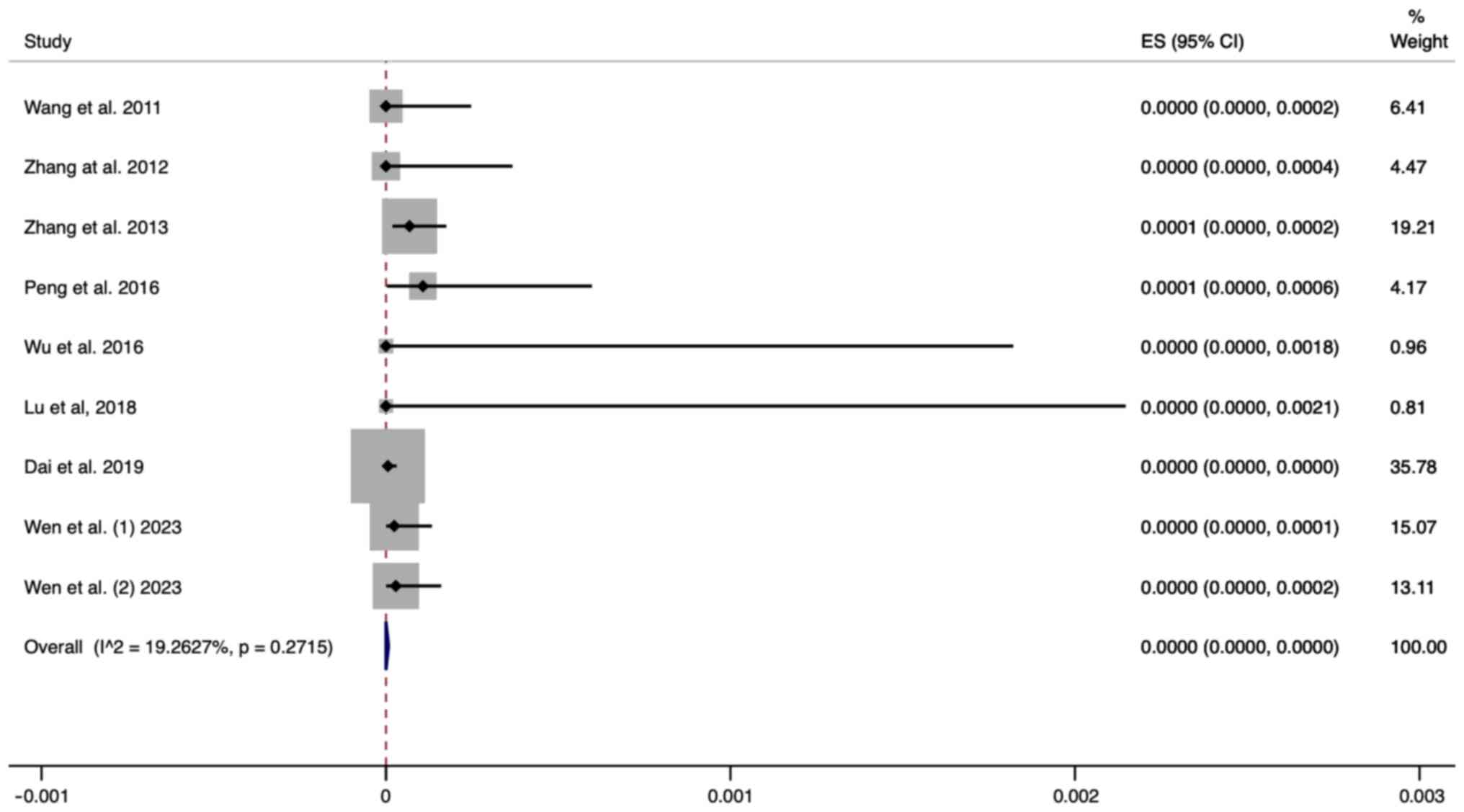
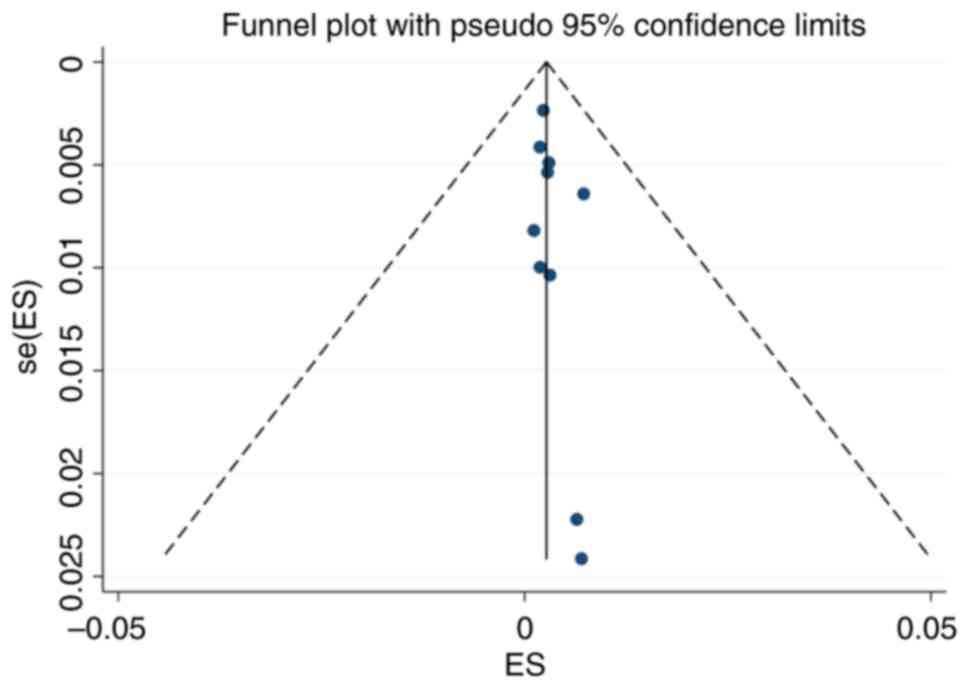
Publication bias
The funnel plot for the included studies is
basically symmetrical. The P-value of Egger's test was 0.267,
indicating that no significant publication bias was found in the
present study (Fig. 6).
Discussion
In 2007, Wang et al (24) initially outlined the protocol and
strategy for concurrent screening of hearing and genetic factors of
newborns. They recommended a combined approach for prelingual
hearing loss, high-risk children with delayed-onset hearing issues,
and carriers of genes associated with deafness. Additionally, they
emphasized the importance of incorporating regular follow-up and
monitoring. This approach has since evolved into a robust and
effective screening strategy (24). The present meta-analysis included a
total of nine articles and investigated 377,688 newborns. The
authors calculated the rate of passing the hearing screening while
failing the genetic screening and explored the advantages of
combined newborn hearing and genetic screening.
The present study took the perspective of
evidence-based medicine to explore the advantages of combined
newborn hearing and genetic screening. The results of the
meta-analysis revealed that the rate of passing the hearing
screening while failing the genetic screening was 0.31%. Among
these cases, the rate of passing the hearing screening while
failing the mitochondrial gene screening was 0.21%, while it was
0.01% for the GJB2 gene screening, and 0.00% for the SLC26A4 gene
screening. In 2019, researchers analyzed 16 studies on neonatal
hearing and genetic screening and discovered that the weighted
average of the failure rate of hearing screening and genetic
screening was 1.4% (25), which
was higher than the combined rate of 0.22% obtained from the random
effects model meta-analysis in the present study. These main
differences can be summarized in the following three points. i)
Scope of genetic screening: The previous studies did not limit the
scope of genetic screening and included a wide range of studies
with varying qualities. The studies included in their research
included screening strategies ranging from single-gene single-locus
studies to studies involving multiple genes and 20 loci. ii)
Diverse study populations: Previous studies did not have uniform
requirements for study subjects, including various screening
populations. Some of the studies in the literature focused only on
pediatric patients with confirmed diagnoses of sensorineural
hearing loss. iii) Meta-analysis methodology: Previous studies did
not use a standardized meta-analysis method. They directly merged
the effect rates of each independent sample, simply calculating
their arithmetic averages, without considering the weight of each
original study. By contrast, the present study strictly established
inclusion and exclusion criteria, including data from universally
conducted newborn hearing and genetic screening, rather than a
specific population. A rigorous evaluation of literature
heterogeneity and publication bias was also conducted, ensuring
greater stability of the results. Due to the different genetic
screening strategies used in different studies, the rates of
hearing screening and genetic screening failure varied somewhat
between these groups, ranging from 0 to 0.67% (26). Currently, a variety of genetic
screening strategies have been adopted both domestically and
internationally. These included single-gene single-locus screening,
screening of three genes and four loci, screening of three genes
with multiple loci, screening of four genes with nine loci, and
screening of four genes with 20 loci, among others. To meet the
statistical requirements and reduce heterogeneity, the present
study included only studies in which at least three
deafness-related genes were screened during the meta-analysis
process. This approach enhances the credibility of the
meta-analysis results and increases the clinical reference value of
the present study.
The present study revealed that the rate of passing
the hearing screening, while failing the genetic screening was
0.31%. This suggests that combined screening can additionally
detect 0.31% of newborns carrying mutations in genes associated
with hearing loss, which cannot be detected through physical
screening alone. Among these patients, 0.21% passed the hearing
screening, while failing the mitochondrial gene screening. This
indicates that combined screening can additionally detect 0.21% of
newborns carrying pathogenic variations in mitochondrial genes.
This approach is equivalent to detecting 1 newborn carrying a
pathogenic mitochondrial gene variation for every 500 newborns
screened through the combined approach. Notably, newborns with
mitochondrial gene mutations are missed by pure hearing screening,
and they are at significant risk of irreversible hearing loss when
exposed to aminoglycoside medications. Therefore, the application
of a combined screening strategy, along with knowledge of genetic
screening results and associated risks, allows for the effective
avoidance of exposure to specific medications, thus preserving
excellent hearing and preventing tragic outcomes such as
‘ototoxicity from a single dose’. This approach serves as a
preventive measure against adverse events. Combining screening with
regular follow-up and monitoring is a potent strategy for the early
detection of prelingual hearing loss, identifying individuals
sensitive to medications, identifying late-onset high-risk
children, or identifying carriers of genes associated with
deafness. It also guides appropriate intervention plans, enabling
early personalized targeted interventions (27,28).
Furthermore, the simplicity and cost-effectiveness of this method
make it a viable option in both resource-rich and resource-limited
settings, ensuring a broader implementation potential.
Additionally, its non-invasive nature and rapid feedback of results
enhance patient compliance and facilitate early diagnosis, which
are crucial for effective intervention.
Therefore, the concurrent newborn hearing and
genetic screening offer the following advantages over traditional
newborn-hearing screening methods: i) Early identification of
genetic hearing loss: By integrating genetic screening, it is
possible to identify genetic hearing loss early on, that external
hearing tests may not detect. This is because some genetic hearing
losses may not manifest at birth or may appear as late-onset
hearing loss. ii) Targeted intervention: Understanding the specific
causes of newborn genetic hearing loss can help doctors and parents
develop more personalized and targeted intervention measures, such
as treatment strategies for specific gene mutations. iii) Avoiding
environmental risks: For hearing loss caused by certain genetic
variations, specific environmental factors (such as certain
medications) may exacerbate hearing loss. Early discovery of these
genetic risks through genetic screening can prevent exposure to
these environmental risks, thus protecting hearing. iv) Reducing
unnecessary re-screening and anxiety: Traditional hearing screening
may require multiple tests to determine hearing status, especially
for newborns who fail the initial screening but pass subsequent
tests. Completing hearing and genetic screening all at once can
reduce parental anxiety and the repetitive consumption of medical
resources. v) Long-term monitoring and intervention planning: For
genetic hearing loss, even if the initial screening results are
normal, hearing loss may develop over time. Genetic screening
results can provide a basis for long-term monitoring and timely
intervention for these children.
Of note, the present study also has several
limitations: i) Despite incorporating multiple studies in this
meta-analysis, the small sample sizes of some studies might have
impacted the reliability and generalizability of the results.
Studies with small sample sizes are often more susceptible to
random errors, which could lead to biased outcomes. Identifying
such biases is crucial for interpreting the overall results of a
meta-analysis. Therefore, a random effects model was used to pool
the results of various studies, considering the heterogeneity among
studies to mitigate the impact of biases that might arise from
small study groups on the overall conclusions. Additionally, a
sensitivity analysis was conducted by systematically excluding
small-sample studies to observe their impact on the overall
conclusions; however, the results of the sensitivity analysis
revealed that the conclusions of this meta-analysis are robust.
Future research should consider recruiting larger sample sizes to
enhance the reliability of the findings and reduce the risk of
bias. ii) The present study aimed to conduct a comprehensive
analysis of the effectiveness of combined newborn hearing and
genetic screening. However, the limitations in the data of the
present study must be acknowledged and considered as a preventive
reason for the authors not conducting subgroup analyses. Future
research should endeavor to collect more comprehensive data,
including but not limited to the gestational age at birth and
different types of genetic mutations in newborns. This would enable
researchers to carry out more detailed subgroup analyses, thereby
enhancing the understanding of how the benefits of combined
screening may vary among different populations and how screening
strategies could be optimized based on the specific needs of
certain subgroups. iii) The present study, focused on the initial
results of newborn hearing and genetic screening rather than
long-term follow-up data. Considering that the subjects were
newborns, screenings are usually conducted shortly after birth,
aiming to promptly identify potential hearing impairments and
genetic risks. Therefore, the traditional concept of ‘follow-up
rates’ does not apply to the present study; the present study
concentrated on the analysis of the data at the time of screening.
Among newborns with positive initial screening results, the
proportion who underwent further diagnostic tests may be low. This
is indeed a matter of concern, as it could affect the accuracy of
the estimates of the prevalence of hearing loss or genetic
mutations. In future work, ensuring comprehensive follow-up and
assessment for individuals with positive initial screenings will be
crucial to improving the quality of research. Additionally, due to
the studies included not focusing on the types of hearing loss,
this research was unable to further analyze the differences among
various types of hearing loss. Future work necessitates the conduct
of large-scale cross-sectional studies to delve into the variations
in genetic screening results across different types of hearing
loss. Such investigations will contribute to a deeper understanding
of the underlying mechanisms and guide more precise intervention
measures.
In summary, the results of the present study
indicated that the rate of passing the hearing screening while
failing the genetic screening is 0.31%. Specifically, the rate of
passing the hearing screening while failing the MT-RNR1 screening
was 0.21%, and for the GJB2 screening, it was 0.01%. Combined
screening has a significant advantage over pure hearing screening,
especially in terms of identifying newborns with mitochondrial gene
mutations, which increases the sensitivity of these patients to
certain medications. Given these findings, the integration of
genetic screening into newborn hearing screening programs is
recommended, especially for infants with a family history of
genetic hearing loss. Additionally, future research should focus on
evaluating the effectiveness of combined screening across diverse
populations, exploring new genetic markers to enhance screening
accuracy, and further investigating the long-term impact of this
strategy on the hearing development and language abilities of
children, and overall quality of life for them and their families.
Through these efforts, the aim in the present study was to improve
early diagnosis and intervention measures for newborn hearing loss,
thereby improving the health and well-being of affected
children.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by Mianyang City Health
Commission 2019 Research Project Grant (grant no. 201910).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
KP, ZS and JLi analyzed the data and wrote the
manuscript. YW and JLu participated in the literature review and
figure drawing. The genetic test for deafness was completed by JL,
YW and ZS. DZ and AW participated in the data extraction and
literature quality assessment. PX, DZ and AW conducted the data
collection and analysis. TL and LL participated in figure drawing
and data analysis. KP and PX confirm the authenticity of all the
raw data. PX conceived the study. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nieman CL and McMahon CM: The World Health
Organization's world report on hearing: A call to action for
hearing care providers. J Laryngol Otol. 134:377–378.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fortnum HM, Summerfield AQ, Marshall DH,
Davis AC and Bamford JM: Prevalence of permanent childhood hearing
impairment in the United Kingdom and implications for universal
neonatal hearing screening: Questionnaire based ascertainment
study. BMJ. 323:536–540. 2001.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kennedy C and McCann D: Universal neonatal
hearing screening moving from evidence to practice. Arch Dis Child
Fetal Neonatal Ed. 89:F378–F383. 2004.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tobe RG, Mori R, Huang L, Xu L, Han D and
Shibuya K: Cost-effectiveness analysis of a national neonatal
hearing screening program in China: Conditions for the scale-up.
PLoS One. 8(e51990)2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang LH, Zhang L, Tobe RY, Qi FH, Sun L,
Teng Y, Ke QL, Mai F, Zhang XF, Zhang M, et al: Cost-effectiveness
analysis of neonatal hearing screening program in China: Should
universal screening be prioritized? BMC Health Serv Res.
12(97)2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Young NM, Reilly BK and Burke L:
Limitations of universal newborn hearing screening in early
identification of pediatric cochlear implant candidates. Arch
Otolaryngol Head Neck Surg. 137:230–234. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Morton CC and Nance WE: Newborn hearing
screening-a silent revolution. N Engl J Med. 354:2151–2164.
2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang Q, Xiang J, Sun J, Yang Y, Guan J,
Wang D, Song C, Guo L, Wang H, Chen Y, et al: Nationwide population
genetic screening improves outcomes of newborn screening for
hearing loss in China. Genet Med. 21:2231–2238. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tang X, Liu L, Liang S, Liang M, Liao T,
Luo S, Yan T and Chen J: Concurrent newborn hearing and genetic
screening in a Multi-ethnic population in South China. Front
Pediatr. 9(734300)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Barendregt JJ, Doi SA, Lee YY, Norman RE
and Vos T: Meta-analysis of prevalence. J Epidemiol Community
Health. 67:974–978. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yáñez-Baeza C, Aguilera-Eguía RA,
Fuentes-Barría H and Roco-Videla Á: Importance of the PRISMA
guideline. Nutr Hosp. 40:670–675. 2023.PubMed/NCBI View Article : Google Scholar : (In Spanish).
|
|
12
|
Zhu QW, Li MT, Zhuang X, Chen K, Xu WQ,
Jiang YH and Qin G: Assessment of hearing screening combined with
limited and expanded genetic screening for newborns in Nantong,
China. JAMA Netw Open. 4(e2125544)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zeng X, Liu Z, Wang J and Zeng X: Combined
hearing screening and genetic screening of deafness among Hakka
newborns in China. Int J Pediatr Otorhinolaryngol.
136(110120)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang QJ, Zhao YL, Rao SQ, Guo YF, He Y,
Lan L, Yang WY, Zheng QY, Ruben RJ, Han DY and Shen Y: Newborn
hearing concurrent gene screening can improve care for hearing
loss: A study on 14,913 Chinese newborns. Int J Pediatr
Otorhinolaryngol. 75:535–542. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang Z, Ding W, Liu X, Xu B, Du W, Nan S
and Guo Y: Auditory screening concurrent deafness predisposing
genes screening in 10,043 neonates in Gansu province, China. Int J
Pediatr Otorhinolaryngol. 76:984–988. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang J, Wang P, Han B, Ding Y, Pan L, Zou
J, Liu H, Pang X, Liu E, Wang H, et al: Newborn hearing concurrent
genetic screening for hearing impairment-a clinical practice in
58,397 neonates in Tianjin, China. Int J Pediatr Otorhinolaryngol.
77:1929–1935. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Peng Q, Huang S, Liang Y, Ma K, Li S, Yang
L, Li W, Ma Q, Liu Q, Zhong B and Lu X: Concurrent genetic and
standard screening for hearing impairment in 9317 Southern Chinese
Newborns. Genet Test Mol Biomarkers. 20:603–608. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu CC, Tsai CH, Hung CC, Lin YH, Lin YH,
Huang FL, Tsao PN, Su YN, Lee YL, Hsieh WS and Hsu CJ: Newborn
genetic screening for hearing impairment: A population-based
longitudinal study. Genet Med. 19:6–12. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lu CY, Tsao PN, Ke YY, Lin YH, Lin YH,
Hung CC, Su YN, Hsu WC, Hsieh WS, Huang LM, et al: Concurrent
hearing, genetic, and cytomegalovirus screening in Newborns,
Taiwan. J Pediatr. 199:144–50.e1. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dai P, Huang LH, Wang GJ, Gao X, Qu CY,
Chen XW, Ma FR, Zhang J, Xing WL, Xi SY, et al: Concurrent hearing
and genetic screening of 180,469 neonates with Follow-up in
Beijing, China. Am J Hum Genet. 105:803–812. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Luo H, Yang Y, Wang X, Xu F, Huang C, Liu
D, Zhang L, Huang T, Ma P, Lu Q, et al: Concurrent newborn hearing
and genetic screening of common hearing loss variants with
bloodspot-based targeted next generation sequencing in Jiangxi
province. Front Pediatr. 10(1020519)2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wen C, Yang X, Cheng X, Zhang W, Li Y,
Wang J, Wang C, Ruan Y, Zhao L, Lu H, et al: Optimized concurrent
hearing and genetic screening in Beijing, China: A cross-sectional
study. Biosci Trends. 17:148–159. 2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang QJ, Zhao YL, Lan L, Zhao C, Han MK
and Han DY: Studies of the strategy for newborn gene screening.
Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 42:809–813.
2007.PubMed/NCBI(In Chinese).
|
|
25
|
D'Aguillo C, Bressler S, Yan D, Mittal R,
Fifer R, Blanton SH and Liu X: Genetic screening as an adjunct to
universal newborn hearing screening: Literature review and
implications for non-congenital pre-lingual hearing loss. Int J
Audiol. 58:834–850. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Nivoloni Kde A, da Silva-Costa SM, Pomilio
MC, Pereira T, Lopes Kde C, de Moraes VC, Alexandrino F, de
Oliveira CA and Sartorato EL: Newborn hearing screening and genetic
testing in 8974 Brazilian neonates. Int J Pediatr Otorhinolaryngol.
74:926–929. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Han B, Zong L, Li Q, Zhang Z, Wang D, Lan
L, Zhang J, Zhao Y and Wang Q: Newborn genetic screening for high
risk deafness-associated mutations with a new Tetra-primer ARMS PCR
kit. Int J Pediatr Otorhinolaryngol. 77:1440–1445. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guo L, Xiang J, Sun L, Yan X, Yang J, Wu
H, Guo K, Peng J, Xie X, Yin Y, et al: Concurrent hearing and
genetic screening in a general newborn population. Hum Genet.
139:521–530. 2020.PubMed/NCBI View Article : Google Scholar
|