Introduction
An extended and severe inflammatory response may
cause damage to bone and cartilage in the inflammatory autoimmune
disease rheumatoid arthritis (RA) (1). The pathophysiology of RA is complex,
involving immune cells, and environmental and genetic factors
(2,3). Among them, macrophages are crucial to
the pathophysiology of RA. Notably, macrophages induce the
translocation of a high number of immune cells to the joints, where
they secrete inflammatory cytokines to maintain an inflammatory
environment (4,5). In different settings, macrophages
develop into two distinct phenotypes; namely, M1 (pro-inflammatory)
and M2 (anti-inflammatory) macrophages. In some cases, M1 and M2
macrophages may alternate between types, in order to control the
inflammatory response of the body (6,7).
Notably, the prognosis of patients with RA has been reported to be
associated with the biological properties of macrophages (8). Targeted alterations in macrophage
activity and inflammatory state may exhibit potential in the
treatment of patients with RA.
Alterations in gene expression that are heritable
and reversible are referred to as epigenetics. Epigenetic
modifications may occur in factors, such as metal, organic
pollutants and air particles, associated with signaling between the
environment and gene. Notably, epigenetics may affect the
expression of inflammatory and matrix degradation pathways, which
are strongly associated with the onset of RA (9-11).
The results of previous studies have revealed that epigenetic
reprogramming and epigenetic enzymes modify the inflammatory state
of macrophages (12,13). mRNA molecules most commonly undergo
N6-methyladenosine (m6A) RNA methylation, which is a type of
epitranscriptomic alteration (14,15).
Methyltransferases (writers), demethylases (erasers) and
m6A-associated binding proteins (readers) are responsible for
processing m6A alterations (16,17).
The results of previous studies have identified the significance of
m6A methylation alterations in immune regulation and inflammatory
response mechanisms (18-20).
However, the effects of m6A methylation alterations on the clinical
diagnosis of RA and their specific roles in macrophage polarization
remain to be fully elucidated. Thus, the present study aimed to
determine the potential association between symptoms of RA,
clinical markers and m6A methylation modifier enzyme genes. In
addition, the present study aimed to investigate whether genes
encoding m6A methylation modifier enzymes may control the
inflammatory state and promote distinct pathways of macrophage
development. Collectively, the results of the present study may
uncover novel treatment targets, and could provide a theoretical
basis for the role of m6A methylation modifier enzyme genes in
macrophage polarization in RA.
Materials and methods
Database analysis of m6A methylation
and RA macrophages
Differentially expressed genes in RA macrophages
(compared with healthy control macrophages) were obtained from the
GSE97779 dataset using the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/)
(21). The GSE97779 dataset
consisted of nine samples of synovial fluid-derived macrophages
obtained from patients with RA, and five samples of macrophages
obtained from healthy control (HC) human monocytes. The Affymetrix
microarray platform was utilized for data analysis. The GEO2R
backend uses the established Bioconductor R software package
(Bioconductor 3.18; Fred Hutchinson Cancer Research Center) to
transform and analyze GEO data and the results are presented as a
genetic table sorted by significance that can be visualized via GEO
Profile graphs (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html).
The m6A2Target database (http://rm2target.canceromics.org/) provided the m6A
methylation modifier enzyme genes. Among the 21 m6A methylation
modifier enzyme genes, eight writers (METTL3, ZC3H13, METTL14,
RBM15B, CBLL1, WTAP, RBM15 and KIAA1429), two erasers (FTO and
ALKBH5) and 11 readers (YTHDC1, YTHDC2, ELAVL1, YTHDF1, LRPPRC,
YTHDF2, FMR1, YTHDF3, HNRNPC, HNRNPA2B1 and IGF2BP1) were
selected.
Patients
RA and HC peripheral blood samples were collected to
determine the mRNA expression levels of m6A. In addition, synovial
tissue was collected from RA and HC knee joints to verify the
protein expression levels of METTL14. The patients with RA had not
received any treatment prior to blood or synovial tissue
collection. The patients with RA from whom synovium was collected
during surgery included one man and three women, with a mean ± SD
age of 45.05±10.82 years. The HC volunteers from whom synovium was
collected during arthroscopy also included one man and three women,
with a mean ± SD age 40.89±11.62 years. The synovial tissue samples
were all obtained from the Department of Orthopedics, The First
Affiliated Hospital of Anhui University of Chinese Medicine (Hefei,
China). The participants for synovial tissue collection were
recruited between December 2022 and March 2024.
Details of the participants from whom blood samples
were collected are included in Table
SI. The participants for blood collection were recruited
between January 2022 and August 2023. Patients with RA (30 cases)
were admitted to the Rheumatology and Immunology Department, The
First Affiliated Hospital of Anhui University of Chinese Medicine,
and HC individuals (30 cases) were selected from the Physical
Examination Center of The First Affiliated Hospital of Anhui
University of Chinese Medicine.
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Anhui University of
Chinese Medicine (ethics approval no. 2019AH-12), and all patients
provided written informed consent. Notably, there were no
statistically significant differences in age or sex between the RA
and HC groups. RA disease activity was distinguished according to
the Disease Activity Score-28 (DAS28) of the patients (22). 2.6< DAS28 <5.10 indicates low
disease activity, whereas DAS28 ≥5.10 indicates high disease
activity. The pain perception of patients with RA was evaluated by
visual analogue scale (VAS) (23).
The correlation between METTL14 and clinical immunoinflammatory
indicators was analyzed to determine the potential association
between METTL14 and immune inflammation. The patients with RA were
divided into eight subgroups according to the presence or absence
of clinical symptoms. These subgroups include the joint tenderness
group (20 cases), the non-joint tenderness group (10 cases), the
joint swelling group (13 cases), the non-joint swelling group (17
cases), the joint morning stiffness group (17 cases), the non-joint
morning stiffness group (13 cases), the limitation of joint motion
group (16 cases) and the normal joint activity group (14 cases).
White blood cells, red blood cells, hemoglobin, hematocrit,
platelets, mean platelet volume, platelet distribution width,
procalcitonin, rheumatoid factor (RF), anti-cyclic peptide
containing citrulline, C-reactive protein (CRP), immunoglobulin A,
immunoglobulin G (IgG), immunoglobulin M and serum amyloid A were
measured using a fully automated biochemical analyzer (LTS 008AS;
Hitachi, Inc.). Erythrocyte sedimentation rate was measured using
an automated erythrocyte sedimentation rate analyzer (Roller 20;
Alifax, Inc.).
Reverse transcription-quantitative
(RT-q)PCR
Peripheral venous blood samples (3-5 ml) were
collected from the subjects, lymphocyte separation fluid (cat. no.
1308106078; Shanghai Kelaman Reagent Co., Ltd.) was added, and
samples were centrifuged at 2,500 x g for 20 min 18˚C. After
centrifugation, there was obvious stratification, and the white
membrane between the plasma and separation solution was considered
the mononuclear cell layer. These cells were added to a clean 15-ml
centrifuge tube, washed with 10 ml PBS and centrifuged at room
temperature and 250 x g for 10 min. The supernatant was discarded,
the cells were resuspended in 5 ml PBS and centrifuged again at
room temperature and 250 x g for 10 min. Following removal of the
supernatant, the collected cells were considered mononuclear cells.
TRIzol® reagent (cat. no. 15596026; Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract RNA from the cells.
RMBase v2.0 (https://rna.sysu.edu.cn/rmbase/) and PrimerBank
databases (https://pga.mgh.harvard.edu/primerbank/index.html)
were used to create primers, and all sequences are displayed in
Table I. An RT kit (cat. no.
RR047A; Takara Bio, Inc.) was used to reverse transcribe mRNA into
cDNA according to the manufacturer's instructions, and Novostart
SYBR qPCR SuperMix Plus (cat. no. E096-01B; Novoprotein Scientific,
Inc.) was used on the prepared cDNA. The thermocycling conditions
were as follows: Initial denaturation at 95˚C for 1 min, followed
by 40 cycles of denaturation at 95˚C for 20 sec, and annealing and
extension at 60˚C for 1 min. The relative expression levels of
METLL14 were calculated using the 2-ΔΔCq method
(24).
 | Table IQuantitative PCR primers used in the
present study. |
Table I
Quantitative PCR primers used in the
present study.
| Gene | Length, bp | Forward primer,
5'-3' | Reverse primer,
5'-3' |
|---|
| METTL14 | 169 |
CGGGGACTTCATTCATGCTA |
CCAGCCTGGTCGAATTGTA |
| CBLL1 | 76 |
GATCCTTGGGTGGTCTTGAT |
GGTTTCGCTTTGTTTGCTTG |
| RBM15 | 122 |
CTCCGACGACCCGCAACAAT |
CCACCAGAGCCCCCTAACTT |
| KIAA1429 | 203 |
GAGTAAGAGCCCATAGCAGT |
TAGCACCAGACCATCAGTATTCAC |
| ERK1 | 145 |
TTTTCCCCAAGTCAGACTCC |
GACTGGCCCACCTCATC |
| ERK2 | 86 |
AACTTGTGTTAGGGCTGTGA |
AAGGTCTGAAGAACCACCTG |
| p38 | 169 |
CTCATTAACAGGATGCCAAGC |
CTTGGGCCGCTGTAATTCTC |
| JNK | 146 |
TGTGTGGAATCAAGCACCTTC |
AGGCGTCATCATAAAACTCGTTC |
| β-actin | 96 |
CCCTGGAGAAGAGCTACGAG |
GGAAGGAAGGCTGGAAGAGT |
Western blot analysis
Synovial tissue samples were collected and lysed
using 600 µl RIPA cell lysis buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology). Total protein was extracted from
tissue via centrifugation for 15 min at 4˚C and 12,000 x g. The
protein concentration was determined by an ultra micro
spectrophotometer (OD1000+; Nanjing Wuyi Technology Co., Ltd.).
Proteins (50 µg/lane) were subsequently separated on 10%
polyacrylamide gels using SDS-PAGE and transferred onto PVDF
membranes (cat. no. IPVH00010; MilliporeSigma). Membranes were
blocked with 5% skimmed milk powder at room temperature for 2-4 h,
and then incubated with anti-METTL14 (1:1,000; cat. no. ab220030;
Abcam) and anti-GAPDH (1:2,000; cat. no. TA-08; ZSGB-Bio, Inc.) at
4˚C overnight. Subsequently, membranes were incubated with a
HRP-conjugated goat anti-mouse IgG secondary antibody (1:20,000;
cat. no. ZB-2305; OriGene Technologies, Inc.) at room temperature
for 2 h. Super-sensitive ECL chemiluminescent substrate (cat. no.
BL520A; Biosharp, Inc.) was used to develop the membrane and a Gel
Imaging System (Shanghai Peiqing Technology Co., Ltd.) was used to
capture, and protein expression was semi-quantified using using
ImageJ software (version 1.80; National Institutes of Health).
Detection of total RNA m6A
modification levels
TRIzol reagent was used to isolate total RNA from
peripheral blood samples, and a NanoDrop 3000 (NanoDrop; Thermo
Fisher Scientific, Inc.) was used to assess the quality of the
extracted RNA. The EpiQuik m6A RNA Methylation Quantification kit
(cat. no. P-9005-48; EpiGentek) was used to quantify the m6A
content according to the manufacturer's protocols.
Flow cytometry
Peripheral blood was extracted from patients and
combined with 10 µl anti-FITC-CD68 (cat. no. MA1-82715; Thermo
Fisher Scientific, Inc.), anti-PE-CD86 (cat. no. MHCD8604; Thermo
Fisher Scientific, Inc.) or anti-PE-CD206 (cat. no. MA5-23594;
Thermo Fisher Scientific, Inc.). Samples were incubated for 30 min
at 4˚C in the dark. Subsequently, samples were centrifuged at 1,500
x g for 5 min at room temperature, and the supernatant was
discarded. Samples were washed twice with saline, and resuspended
in 500 µl saline for detection. Flow cytometry was conducted using
a CytoFLEX flow cytometer (Beckman Coulter, Inc.), and FlowJo
analysis software (version 10.8.1; Beckman Coulter, Inc.) was used
to assess the levels of CD68/CD86/CD206 in cells.
ELISA
Peripheral blood samples were separated via
centrifugation at 600 x g for 10 min at room temperature, and were
subsequently stored at -80˚C. The levels of inflammatory
components, TNF-α (cat. no. E-EL-H0109c; Elabscience Biotechnology,
Inc.) and IL-10 (cat. no. E-EL-H6154; Elabscience Biotechnology,
Inc.) were determined using ELISA kits, following the
manufacturer's protocols.
Kyoto encyclopedia of genes and
genomes (KEGG) and gene ontology (GO) enrichment analyses
The linear models for microarray data package
(https://bioconductor.org/packages/release/bioc/html/limma.html)
in the R programming language was used to analyze the GEO dataset.
Genes associated with METTL14 (P<0.01 and |R|>0.6) were
subjected to functional term enrichment analysis using the GO
database and signaling pathway enrichment analysis using the KEGG
database. The DAVID database (https://david-d.ncifcrf.gov/) was used to perform
these enrichment analyses. GO signaling pathway enrichment analysis
included three main components; namely, biological process (BP),
cellular component (CC) and molecular function (MF).
Statistical analysis
Quantitative data conforming to a normal
distribution are presented as the mean ± SD. The Mann-Whitney U
test was used for data that did not follow a normal distribution.
Comparisons between multiple groups were made using one-way ANOVA
followed by the least significant difference post hoc test.
Differences between two groups were analyzed using unpaired
Student's t-test. Spearman's rank correlation coefficient tests
were used to assess correlations. The χ2 test was used
for statistical analysis of sex comparisons among groups. Receiver
operating characteristic (ROC) curves were used to evaluate the
accuracy of METTL14 in predicting pain indicators (VAS). P<0.05
was considered to indicate a statistically significant
difference.
Results
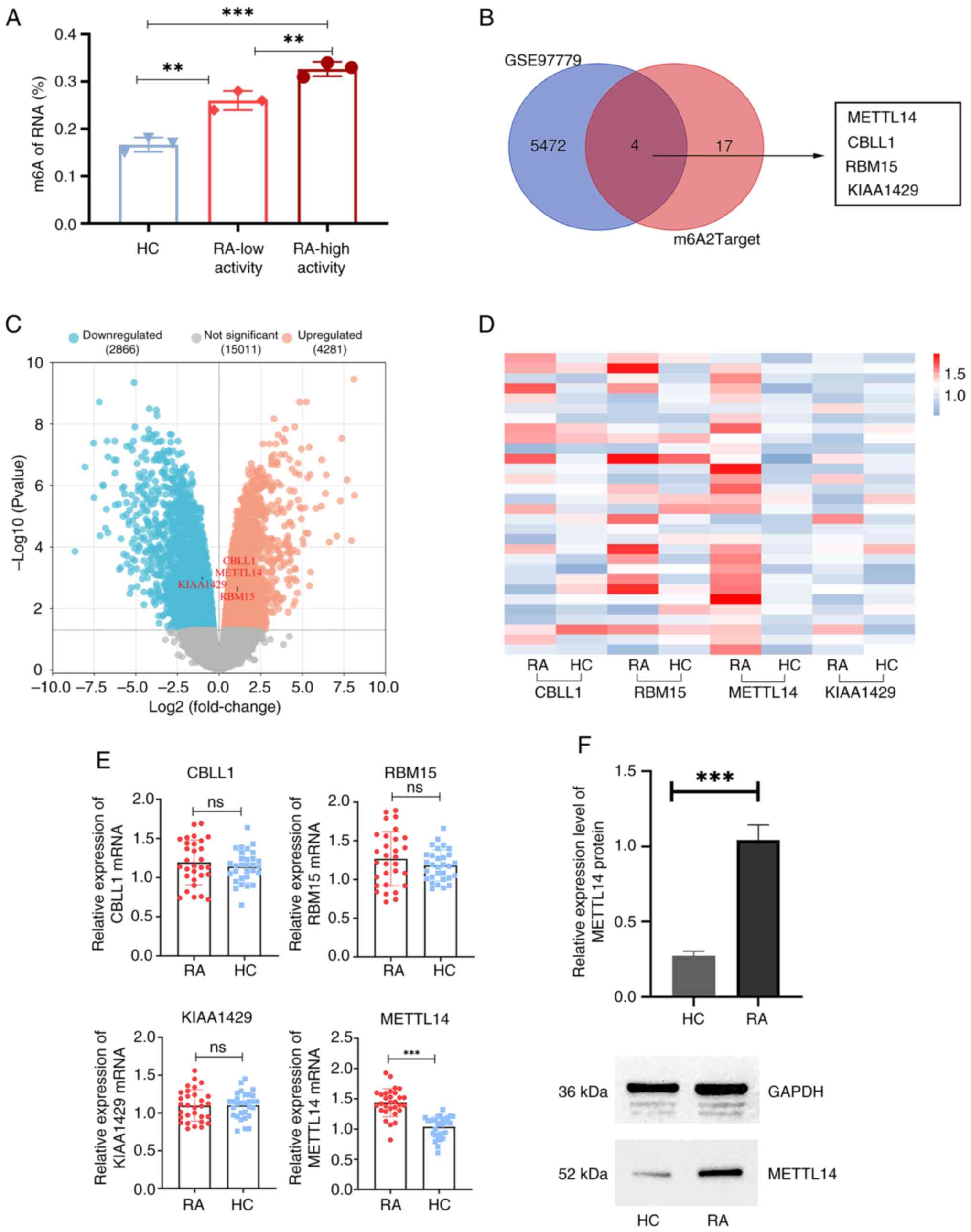
Expression levels of m6A methylation
modifier enzyme genes are associated with differential expression
in RA macrophages
RA disease activity was distinguished according to
the Disease Activity Score-28 (DAS28) of the patients. Through the
determination of m6A levels in the peripheral blood, the results of
the present study demonstrated that m6A expression levels in the
low-activity RA group were significantly increased compared with
those in the HC group (Fig. 1A).
In addition, m6A levels in the high-activity RA group were
significantly increased compared with those in the low-activity RA
group and the HC group (Fig. 1A).
Using the GEO2R function, 5,476 differentially expressed genes were
identified in RA macrophages in the GSE97779 dataset. Subsequently,
m6A methylation modifier enzyme genes were intersected with the
differentially expressed genes in RA macrophages. The results of
the present study demonstrated that four genes (METTL14, CBLL1,
RBM15 and KIAA1429) were methyltransferases (Fig. 1B and C). Therefore, it was hypothesized that
the polarization process of RA macrophages may involve these four
genes. Subsequently, the results of the present study demonstrated
that the mRNA expression levels of METTL14 were significantly
higher in the RA group than those in the HC group (Fig. 1D and E). There was no significant difference
between RA and HC groups regarding the other three
methyltransferases. In addition, the results of western blot
analysis demonstrated that the protein expression levels of METTL14
were significantly increased in the synovial tissue of patients
with RA, compared with those in the HC group (Fig. 1F).
METTL14 is associated with markers of
immunological inflammation and clinical manifestation in patients
with RA
The results of the present study demonstrated that
METTL14 was differentially expressed in patients with RA, compared
with in HC individuals. Thus, the potential association of METTL14
with immunological inflammation was determined in patients with RA,
and the results demonstrated that METTL14 was positively associated
with CRP and RF (Fig. 2A).
Subsequently, the mRNA expression levels of METTL14 were compared
between the subgroups of patients with RA, who were classified
according to the presence of joint tenderness, morning stiffness,
joint swelling, joint mobility problems and other clinical
symptoms. The results demonstrated that patients in the RA group
experienced joint tenderness, and these patients exhibited
significantly higher METTL14 expression levels compared with those
in the group without joint pain (Fig.
2B). In addition, METTL14 has a high accuracy in predicting
pain indicators of VAS, according to the results of the ROC curve
analysis (Fig. 2C).
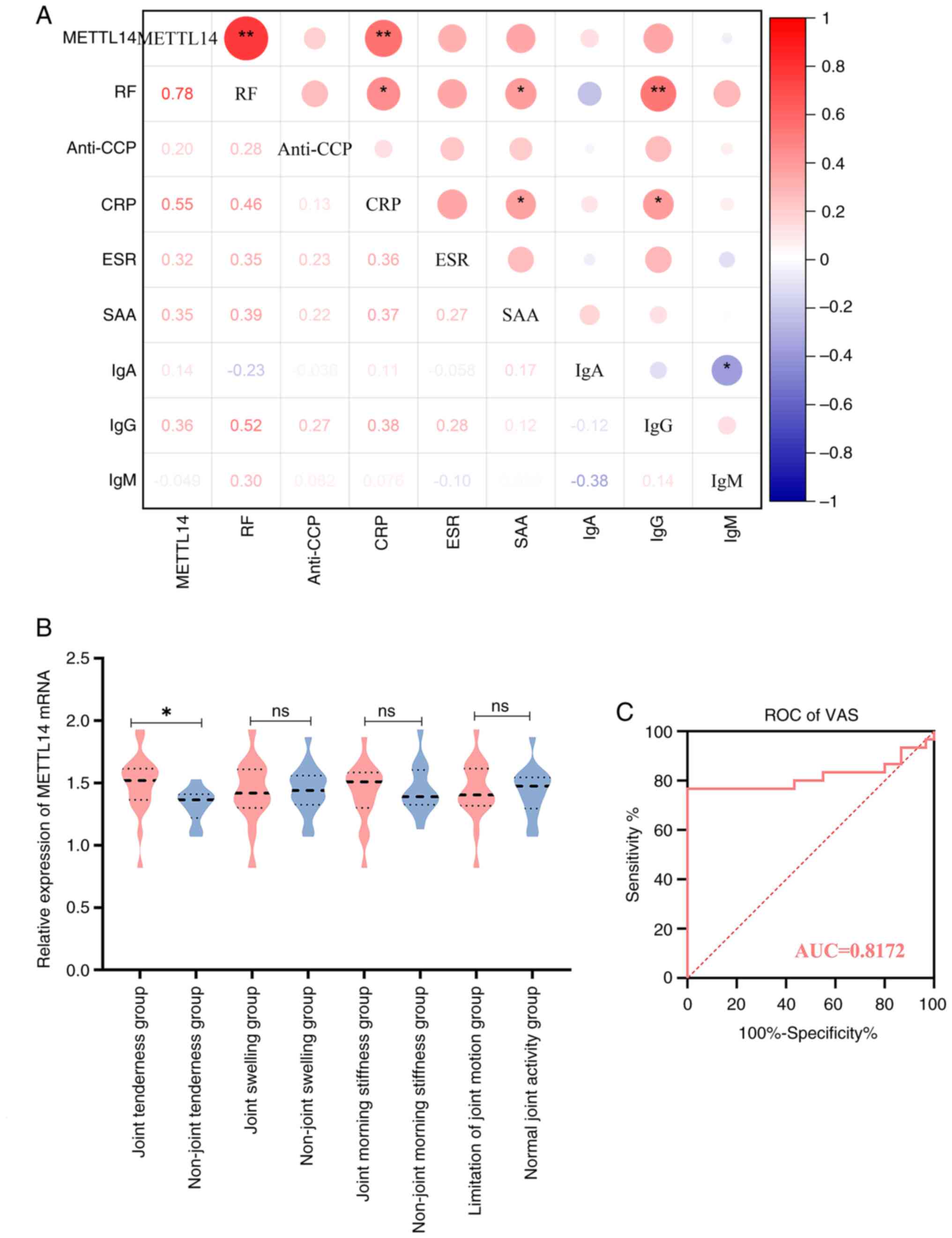 | Figure 2Association of immune-inflammatory
markers, clinical symptoms and METTL14 in patients with RA. (A)
Correlation analysis of METTL14 and immune-inflammatory markers in
patients with RA. Spearman correlation analysis was used to analyze
the data. *P<0.05, **P<0.01. (B)
Analysis of METTL14 and clinical symptoms in patients with RA. Data
are presented as median ± IQR, and data were statistically analyzed
using the Mann-Whitney U test. *P<0.05. (C) ROC
curve. RA, rheumatoid arthritis; METTL14, methyltransferase 14; RF,
rheumatoid factor; CRP, C-reactive protein; ROC, receiver operative
characteristic; VAS, Visual Analogue Scale; AUC, area under the
curve; ns, not significant. |
METTL14 is associated with cytokines
and markers of macrophage polarization in patients with RA
The levels of the M1 macrophage marker
CD68+CD86+ and the M2 macrophage marker
CD68+CD206+ were determined. The results of
the present study demonstrated that the levels of the M1 macrophage
marker CD68+CD86+ and the pro-inflammatory
factor TNF-α were significantly increased in the RA group, compared
with those in the HC group (Fig.
3A). By contrast, the levels of the M2 macrophage marker
CD68+CD206+ and the anti-inflammatory factor
IL-10 were significantly lower in the RA group, compared with those
in the HC group (Fig. 3A). The
results also demonstrated that CD68+CD86+ was
positively correlated with TNF-α, and
CD68+CD206+ was positively correlated with
IL-10 (Fig. 3B). Collectively,
these results highlighted that RA may be associated with macrophage
polarization and an unbalanced inflammatory factor profile. In
addition, the results revealed a positive correlation between
METTL14, TNF-α and CD68+CD86+ (Fig. 3C). Based on the median METTL14
expression level, patients with RA were divided into two groups;
namely, METTL14 low-expression and METTL14 high-expression groups.
While there were no significant differences in the levels of
CD68+CD206+ and IL-10 between these groups,
the METTL14 high-expression group exhibited significantly higher
levels of TNF-α and CD68+CD86+ than those in
the low-expression group (Fig.
3D). These results suggested that high METTL14 expression may
be closely associated with M1 macrophage markers and
pro-inflammatory factors. The flow cytometry plots of macrophage
polarization markers are shown in Fig. S1.
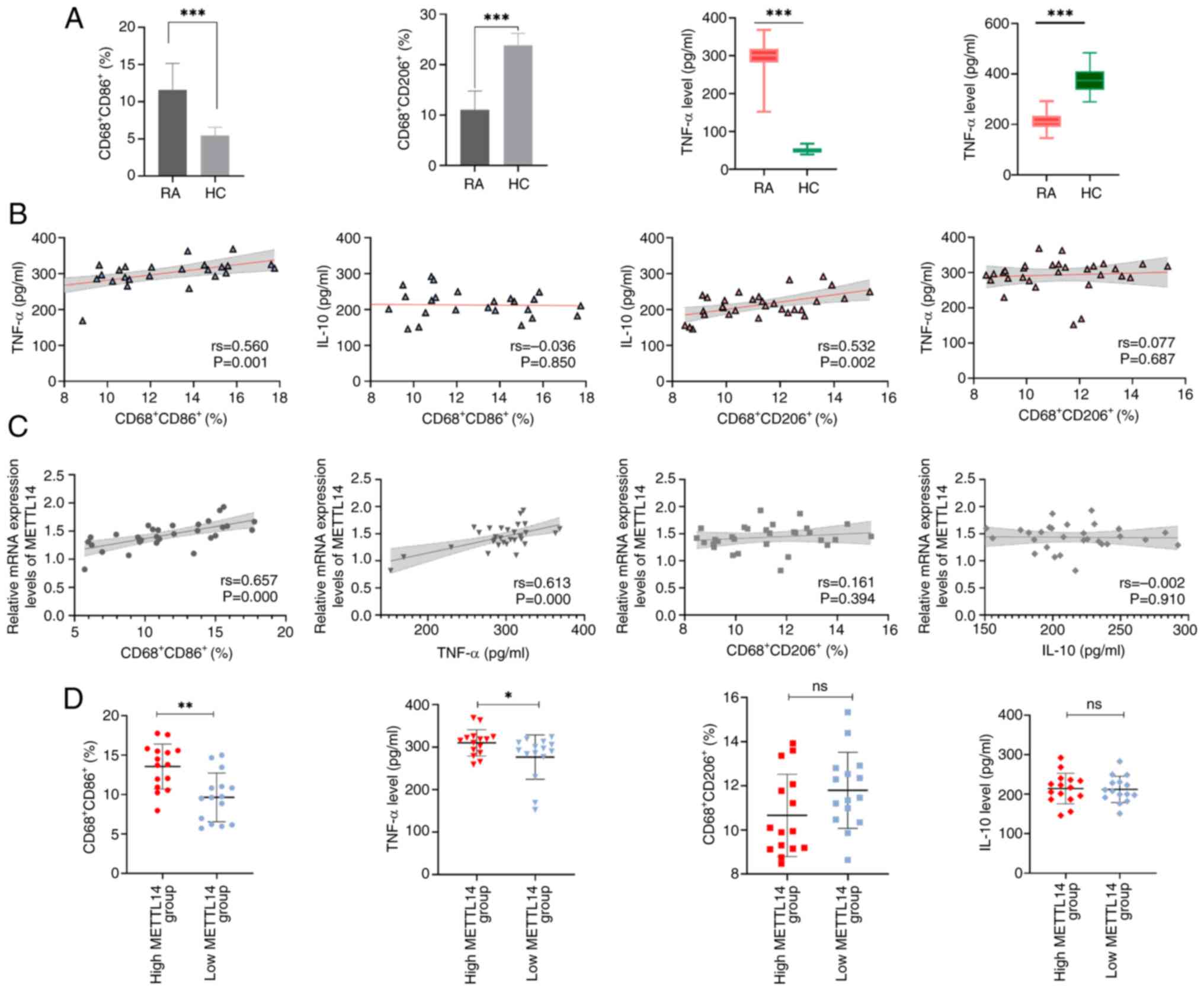 | Figure 3METTL14 expression is associated with
cytokines and indicators of macrophage polarization. (A) Levels of
inflammatory factors, TNF-α and IL-10, and macrophage markers,
CD68+CD86+ and
CD68+CD206+ (n=30). Data are presented as the
mean ± SD and were statistically analyzed using the unpaired
two-independent sample t-test. ***P<0.001. (B)
Correlation between TNF-α, IL-10, CD68+CD86+
and CD68+CD206+. Spearman correlation
analysis was used to analyze the data. (C) Correlation between
METTL14, TNF-α, IL-10, CD68+CD86+ and
CD68+CD206+. Spearman correlation analysis
was used to analyze the data. (D) CD68+CD86+
and CD68+CD206+, TNF-α and IL-10 expression
levels in the METTL14 low-expression and METTL14 high-expression
groups. Data are presented as the median ± IQR, and data were
statistically analyzed using the Mann-Whitney U test.
*P<0.05, **P<0.01. RA, rheumatoid
arthritis; HC, healthy control; METTL14, methyltransferase 14; ns,
not significant. |
KEGG and GO enrichment analyses
The linear models for microarray data package in the
R programming language was used to analyze the GSE97779 dataset and
produce the differentially expressed gene expression matrix.
Correlation analysis was used to determine genes correlated with
METTL14, using P<0.01 and |R|>0.6. For enrichment analysis,
genes that were highly correlated with METTL14 were submitted to
the DAVID database. According to the GO analysis, the enriched BP
terms were associated with an ‘inflammatory response’, an ‘intimate
immune response’, ‘IL-6 positive regulation’ and ‘cytokine-mediated
signaling’ (Fig. 4B). In addition,
the enriched CC and MF terms were associated with ‘peroxisome and
peroxisomal membrane processes’ (Fig.
4C and D). The results of the
KEGG analysis demonstrated that METTL14 was highly associated with
the ‘MAPK signaling pathway’ (Fig.
4A). The MAPK pathway was selected for subsequent analysis,
based on the P-value and count value of the result as the most
enriched pathway relevant to the topic of this study.
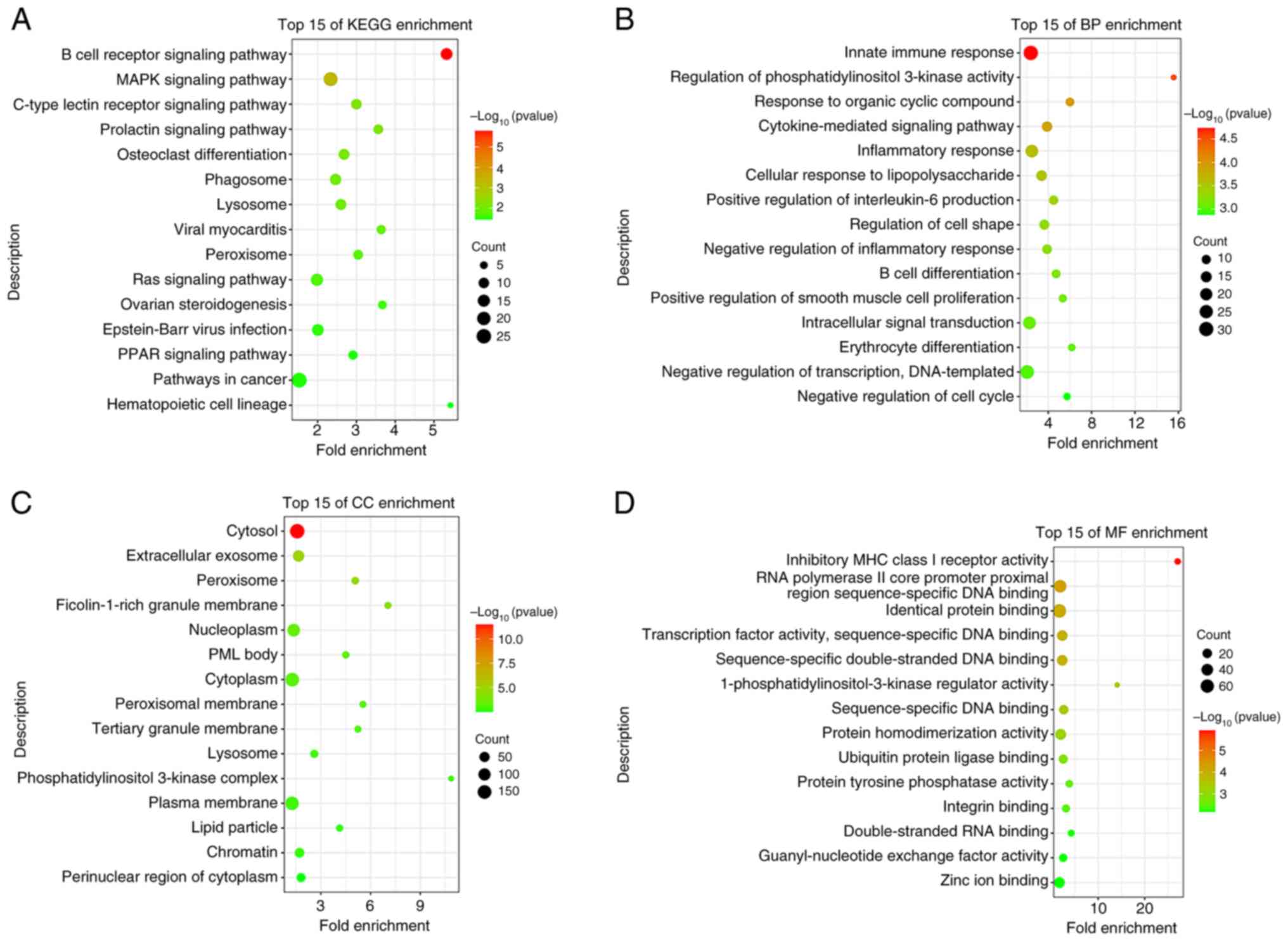 | Figure 4KEGG and GO enrichment analyses.
Enrichment of METTL14-associated genes with (A) KEGG signaling
pathways, (B) GO BP terms, (C) GO CC terms and (D) GO MF terms. The
closer the color of the dot is to red, the smaller the P-value, and
the larger the area of the dot, the more genes are enriched in this
order. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene
Ontology; BP, biological process; CC, cellular component; MF,
molecular function; METTL14, methyltransferase 14. |
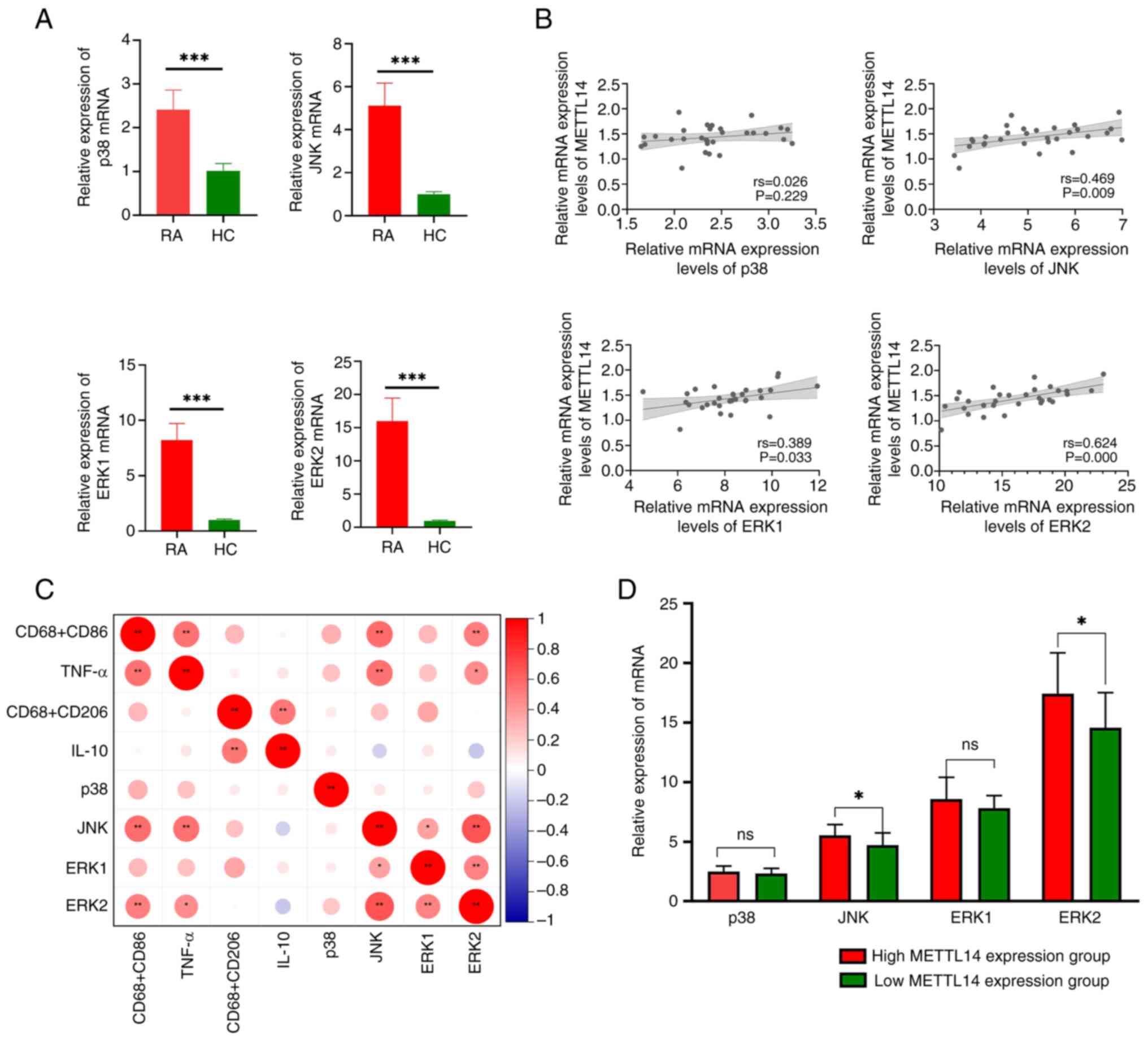
METTL14 is associated with the MAPK
signaling pathway
The results of the present study demonstrated that
the expression levels of JNK, ERK2, p38 and ERK1 were markedly
increased in the RA group, compared with those in the HC group
(Fig. 5A). Notably, the results of
the correlation analysis revealed that there was no statistically
significant association between METTL14 and p38; however, there was
a statistically significant correlation between METTL14 and JNK,
ERK1 and ERK2 (Fig. 5B). In
addition, there was a significant correlation between JNK, ERK2,
TNF-α and CD68+CD86+ (Fig. 5C). Based on the median METTL14
expression level, patients with RA were divided into two groups;
namely, METTL14 low-expression and METTL14 high-expression groups.
It was revealed that the METTL14 high-expression group exhibited
increased JNK and ERK2 expression levels compared with those in the
METTL14 low-expression group; however, no statistically significant
differences were observed in the expression levels of p38 and ERK1
between the groups (Fig. 5D).
These findings indicated that METTL14 may increase downstream
inflammatory responses via the MAPK signaling pathway.
Discussion
Antitumor immunity and healthy immunological
responses are impacted by alterations in m6A methylation (14). The results of previous studies have
indicated that m6A methylation alterations control the immune
response mechanism in innate immune cells, including natural killer
cells, dendritic cells and macrophages (25,26).
Furthermore, it has been suggested that m6A
methyltransferase-activated cells, such as synovial macrophages,
may serve a role in the pathophysiology of RA and ultimately
promote joint destruction (27,28).
Through the determination of m6A levels in the peripheral blood of
patients, the present study demonstrated that m6A levels in the
low-activity RA group were significantly increased compared with
those in the HC group, and the m6A levels in the high-activity RA
group were significantly increased compared with those in the
low-activity RA group.
Using bioinformatics analysis, differentially
expressed m6A methylation modifier enzyme genes in macrophages were
predicted. In total, it was suggested that four genes (METTL14,
CBLL1, RBM15 and KIAA1429) that alter methylation may be implicated
in the process of differential expression in RA macrophages. The
present study demonstrated that the peripheral blood and synovial
tissue of patients with RA exhibited higher expression levels of
METTL14 compared with those of HC individuals; however, there was
no significant difference in the expression levels of KIAA1429,
RBM15 or CBLL1. The methyltransferase complex that stabilizes
METTL3 and identifies target RNAs includes METTL14 as a crucial
component (29,30). Li et al (31) previously demonstrated that the
synovial tissues of rats with RA exhibited considerably higher
levels of METTL14 expression, compared with HCs. Moreover, the
potential association between METTL14 and clinical symptoms was
determined in the present study. The results of the correlation
analysis revealed a positive correlation between METTL14 and both
RF and CRP. These two indicators are markers of RA disease
activity. The present study also revealed that patients with RA
experiencing joint pain exhibited higher levels of METTL14
expression than those without joint pain. In addition, METTL14
exhibited a high sensitivity in predicting VAS, highlighting that
METTL14 may be strongly associated with clinical inflammatory
markers in RA. Thus, METTL14 may exhibit potential as a molecular
target for the treatment of RA.
Macrophage polarization includes the imbalance
between pro-inflammatory M1 and anti-inflammatory M2 macrophage
activity (32,33). Macrophage polarization causes the
breakdown of articular bone and cartilage, and induces synovial
inflammation, autoimmunity and joint injury (34,35).
Flow cytometry was used to determine the levels of M1 and M2
macrophages in the peripheral blood samples of patients, to further
explore the potential association between METTL14 expression,
macrophage polarization and inflammatory response. The results of
the present study revealed a positive correlation between the
pro-inflammatory cytokine TNF-α, the M1 macrophage marker
CD68+CD86+ and METTL14. The METTL14
high-expression group exhibited markedly higher levels of TNF-α and
CD68+CD86+ compared with those in the METTL14
low-expression group. The results of a previous study demonstrated
that METTL14 may suppress TICAM2 and block the macrophage Toll-like
receptor 4 (TLR4) pathway. In addition, macrophage polarization
towards the M1 type was revealed to be promoted by a combination of
TLR4 and METTL14 agonists (36).
The results of another previous study also revealed that METTL14
knockdown can facilitate macrophage M2 polarization (37). In a rat model of RA, METTL14
knockdown was shown to significantly reduce fibroblast-like
synovial cell activation and the generation of the inflammatory
markers, IL-6, IL-18 and CXCL10(31). Thus, METTL14 may facilitate
macrophage conversion to the M1 type through upregulation of its
expression. This, in turn, triggers a large-scale release of
pro-inflammatory factors, which causes an imbalance in joint
inflammation.
METTL14 expression is associated with numerous
clinical characteristics and carries out a crucial additive
methylation function during m6A alteration (38). Thus, METTL14 may serve a role in
modulating signaling pathways and the expression of genes involved
in RA. GO enrichment analysis revealed that METTL14 was enriched in
immune-inflammatory response biological activities, including the
positive regulation of IL-6 production, innate immune response and
inflammatory response. These findings indicated that METTL14
interacts with several targets and pathways. Moreover, METTL14 has
been reported to regulate autophagy, glycolysis, ATPase activity
and immunological response (39).
Notably, among pathways associated with inflammation, KEGG
enrichment analysis demonstrated that METTL14 was associated with
the MAPK signaling pathway. ERK, p38 and JNK are members of the
MAPK family. The results of the present study demonstrated that the
expression levels of JNK, ERK2, p38 and ERK1 were higher in the RA
group compared with those in the HC group. The results of previous
studies have highlighted that the MAPK signaling pathway is crucial
for the transcriptional activation of cytokines associated with the
pathophysiology of RA (2,40). The pro-inflammatory factor TNF-α
and M1 macrophage marker CD68+CD86+ were
significantly correlated with JNK and ERK2 in the present study.
The results of a previous study have revealed that MAPK serves a
crucial role in controlling macrophage polarization (41). Thus, RA macrophage polarization may
involve activation of the MAPK signaling pathway. The results of
the present study also revealed a significant positive correlation
between METTL14 and JNK, ERK1 and ERK2. Furthermore, when compared
with the METTL14 low-expression group, JNK and ERK2 expression
levels were significantly higher in the high-expression group.
Therefore, METTL14 may mediate the transition of RA macrophages
into the M1 type and result in the increased production of
inflammatory markers, which may lead to activation of the JNK/ERK2
signaling axis through upregulated expression. The JNK signaling
pathway modifies macrophage polarization through upregulating the
expression of the downstream transcription factor, c-Myc. The JNK
signaling pathway is key in regulating macrophage polarization and
the production of inflammatory cytokines (42,43).
The results of a previous study have demonstrated that macrophage
inflammatory responses are markedly reduced following METTL14
knockdown (37). Collectively,
these results highlight the critical roles of m6A methylation and
METTL14 expression in macrophage polarization and the potential
immunomodulation of RA. The present study indicated that, via the
JNK/ERK2 signaling axis, METTL14 may mediate the polarization of RA
macrophages.
Notably, the present study has some limitations. For
example, bias may be present due to the inclusion of one
institution, from which the samples were obtained. In addition,
in vitro and in vivo experiments were not carried out
due to time constraints. Thus, future investigations should be
carried out with increased sample sizes, animals and cells, to
verify the involvement of METTL14 in RA macrophage
polarization.
In conclusion, the present study revealed that joint
pain in RA was associated with the increased expression of METTL14.
RF and CRP levels were also positively correlated with METTL14, and
METTL14 exhibited a high sensitivity in predicting VAS. The results
of the present study also demonstrated that inflammatory cytokines,
such as TNF-α, were associated with proteins in the MAPK pathway,
such as JNK and ERK2, and the M1 macrophage marker
CD68+CD86+ was positively correlated with the
expression levels of METTL14. In addition, JNK and ERK2 were
positively correlated with TNF-α and
CD68+CD86+. Compared with the METTL14
low-expression group, JNK and ERK2 expression levels were markedly
increased in the METTL14 high-expression group. Notably, via the
JNK/ERK2 signaling pathway, the m6A methyltransferase METTL14 may
promote the conversion of macrophages to the M1 type in RA;
therefore, intensifying the inflammatory response.
Supplementary Material
Flow cytometry plots of macrophage
polarization markers. (A) Expression levels of
CD68+CD86+ and
CD68+CD206+ in RA and HC groups. (B)
Expression levels of CD68+CD86+ and
CD68+CD206+ in the METTL14 low and
high-expression groups. RA, rheumatoid arthritis; HC, healthy
control; METTL14, methyltransferase 14.
Clinical details of patients with RA
and HC individuals.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the General Project
of National Natural Science Foundation of China (grant no.
822745501), the Xin'an Institute of Medicine and Chinese Medicine
Modernization ‘Jie Bang Gua Shuai’ Project (grant no.
2023CXMMTCM020) and the Anhui Province Natural Fund Project (grant
no. 2308085MH291).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LW was responsible for designing the study, and
writing, reviewing and editing the manuscript. ZZ was responsible
for data analysis, specimen and data collection, and writing the
original draft. LW and ZZ confirm the authenticity of all the raw
data. Both authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study complied with the Declaration of
Helsinki and was approved by The Institutional Review Board Ethics
Committee of The First Affiliated Hospital of Anhui University of
Chinese Medicine (ethics approval no. 2019AH-12). Written informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wan L, Liu J, Huang C, Zhu Z, Wang K, Sun
G, Zhu L and Hu Z: Comprehensive analysis and functional
characteristics of differential expression of N6-methyladenosine
methylation modification in the whole transcriptome of rheumatoid
arthritis. Mediators Inflamm. 2022(4766992)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Liu S, Ma H, Zhang H, Deng C and Xin P:
Recent advances on signaling pathways and their inhibitors in
rheumatoid arthritis. Clin Immunol. 230(108793)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Alivernini S, Firestein GS and McInnes IB:
The pathogenesis of rheumatoid arthritis. Immunity. 55:2255–2270.
2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kurowska-Stolarska M and Alivernini S:
Synovial tissue macrophages in joint homeostasis, rheumatoid
arthritis and disease remission. Nat Rev Rheumatol. 18:384–397.
2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Jang S, Kwon EJ and Lee JJ: Rheumatoid
arthritis: Pathogenic roles of diverse immune cells. Int J Mol Sci.
23(905)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jiang P and Li X: Regulatory mechanism of
lncRNAs in M1/M2 macrophages polarization in the diseases of
different etiology. Front Immunol. 13(835932)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cutolo M, Campitiello R, Gotelli E and
Soldano S: The role of M1/M2 macrophage polarization in rheumatoid
arthritis synovitis. Front Immunol. 13(867260)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hasegawa T and Ishii M: Pathological
osteoclasts and precursor macrophages in inflammatory arthritis.
Front Immunol. 13(867368)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gao L and Lu Q: The critical importance of
epigenetics in autoimmune-related skin diseases. Front Med.
17:43–57. 2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nair N, Barton A and Wilson AG:
Cell-specific epigenetic drivers of pathogenesis in rheumatoid
arthritis. Epigenomics. 13:549–560. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen Q, Li H, Liu Y and Zhao M: Epigenetic
regulation of immune and inflammatory responses in rheumatoid
arthritis. Front Immunol. 13(881191)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jain N, Lord JM and Vogel V:
Mechanoimmunology: Are inflammatory epigenetic states of
macrophages tuned by biophysical factors? APL Bioeng.
6(031502)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ghiboub M, Koster J, Craggs PD, Li Yim
AYF, Shillings A, Hutchinson S, Bingham RP, Gatfield K, Hageman IL,
Yao G, et al: Modulation of macrophage inflammatory function
through selective inhibition of the epigenetic reader protein
SP140. BMC Biol. 20(182)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu S, Li XF, Wu YY, Yin SQ, Huang C and Li
J: N6-methyladenosine and rheumatoid arthritis: A comprehensive
review. Front Immunol. 12(731842)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gan L, Zhao Y, Fu Y and Chen Q: The
potential role of m6A modifications on immune cells and
immunotherapy. Biomed Pharmacother. 160(114343)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Elsabbagh RA, Rady M, Watzl C, Abou-Aisha
K and Gad MZ: Impact of N6-methyladenosine (m6A)
modification on immunity. Cell Commun Signal.
20(140)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li C, Zhu M, Wang J, Wu H, Liu Y and Huang
D: Role of m6A modification in immune microenvironment of digestive
system tumors. Biomed Pharmacother. 164(114953)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Geng Q, Cao X, Fan D, Wang Q, Wang X,
Zhang M, Zhao L, Jiao Y, Deng T, Liu H, et al: Potential medicinal
value of N6-methyladenosine in autoimmune diseases and tumours. Br
J Pharmacol: Jun 9, 2023 (Epub ahead of print).
|
|
19
|
Wang Y, Li L, Li J, Zhao B, Huang G, Li X,
Xie Z and Zhou Z: The emerging role of m6A modification in
regulating the immune system and autoimmune diseases. Front Cell
Dev Biol. 9(755691)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ma Z, Sugimura R and Lui KO: The role of
m6A mRNA modification in normal and malignant hematopoiesis. J
Leukoc Biol. 115:100–115. 2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kang K, Park SH, Chen J, Qiao Y,
Giannopoulou E, Berg K, Hanidu A, Li J, Nabozny G, Kang K, et al:
Interferon-γ represses M2 gene expression in human macrophages by
disassembling enhancers bound by the transcription factor MAF.
Immunity. 47:235–250. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kuriya B, Schieir O, Lin D, Xiong J, Pope
J, Boire G, Haraoui B, Thorne JC, Tin D, Hitchon C, et al:
Thresholds for the 28-joint disease activity score (DAS28) using
C-reactive protein are lower compared to DAS28 using erythrocyte
sedimentation rate in early rheumatoid arthritis. Clin Exp
Rheumatol. 35:799–803. 2017.PubMed/NCBI
|
|
23
|
Castrejón I, Chua JR and Pincus T: A
RheuMetric physician checklist to quantitate levels of
inflammation, damage and distress on 0-10 visual analogue scales.
Clin Exp Rheumatol. 35 (Suppl 107):S21–S25. 2017.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Geng Q, Cao X, Fan D, Gu X, Zhang Q, Zhang
M, Wang Z, Deng T and Xiao C: Diagnostic gene signatures and
aberrant pathway activation based on m6A methylation regulators in
rheumatoid arthritis. Front Immunol. 13(1041284)2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang X, Li X, Jia H, An G and Ni J: The
m6A methyltransferase METTL3 modifies PGC-1α mRNA
promoting mitochondrial dysfunction and oxLDL-induced inflammation
in monocytes. J Biol Chem. 297(101058)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Saeki N and Imai Y: Crosstalk between
synovial macrophages and fibroblasts in rheumatoid arthritis.
Histol Histopathol. 38:1231–1238. 2023.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Boutet MA, Courties G, Nerviani A, Le Goff
B, Apparailly F, Pitzalis C and Blanchard F: Novel insights into
macrophage diversity in rheumatoid arthritis synovium. Autoimmun
Rev. 20(102758)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tong J, Flavell RA and Li HB: RNA
m6A modification and its function in diseases. Front
Med. 12:481–489. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dou X, Huang L, Xiao Y, Liu C, Li Y, Zhang
X, Yu L, Zhao R, Yang L, Chen C, et al: METTL14 is a chromatin
regulator independent of its RNA N6-methyladenosine
methyltransferase activity. Protein Cell. 14:683–697.
2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li X, Xu X, Zhang Q, Ling M, Li X and Tan
X: METTL14 promotes fibroblast-like synoviocytes activation via the
LASP1/SRC/AKT axis in rheumatoid arthritis. Am J Physiol Cell
Physiol. 324:C1089–C1100. 2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li H, Feng Y, Zheng X, Jia M, Mei Z, Wang
Y, Zhang Z, Zhou M and Li C: M2-type exosomes nanoparticles for
rheumatoid arthritis therapy via macrophage re-polarization. J
Control Release. 341:16–30. 2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Song Y, Gao N, Yang Z, Zhang L, Wang Y,
Zhang S and Fan T: Characteristics, polarization and targeted
therapy of mononuclear macrophages in rheumatoid arthritis. Am J
Transl Res. 15:2109–2121. 2023.PubMed/NCBI
|
|
34
|
Gu Q, Yang H and Shi Q: Macrophages and
bone inflammation. J Orthop Translat. 10:86–93. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li Y, Zhou Y, Wang Y, Crawford R and Xiao
Y: Synovial macrophages in cartilage destruction and
regeneration-lessons learnt from osteoarthritis and synovial
chondromatosis. Biomed Mater. 17:2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Huang X, Wang L, Guo H and Zhang W:
Macrophage membrane-coated nanovesicles for dual-targeted drug
delivery to inhibit tumor and induce macrophage polarization.
Bioact Mater. 23:69–79. 2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zheng Y, Li Y, Ran X, Wang D, Zheng X,
Zhang M, Yu B, Sun Y and Wu J: Mettl14 mediates the inflammatory
response of macrophages in atherosclerosis through the NF-κB/IL-6
signaling pathway. Cell Mol Life Sci. 79(311)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Xie Q, Li Z, Luo X, Wang D, Zhou Y, Zhao
J, Gao S, Yang Y, Fu W, Kong L and Sun T: piRNA-14633 promotes
cervical cancer cell malignancy in a METTL14-dependent m6A RNA
methylation manner. J Transl Med. 20(51)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li T, Wang T, Jing J and Sun L: Expression
pattern and clinical value of key m6A RNA modification regulators
in abdominal aortic aneurysm. J Inflamm Res. 14:4245–4258.
2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fang Q, Zhou C and Nandakumar KS:
Molecular and cellular pathways contributing to joint damage in
rheumatoid arthritis. Mediators Inflamm.
2020(3830212)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Neamatallah T: Mitogen-activated protein
kinase pathway: A critical regulator in tumor-associated macrophage
polarization. J Microsc Ultrastruct. 7:53–56. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hao J, Hu Y, Li Y, Zhou Q and Lv X:
Involvement of JNK signaling in IL4-induced M2 macrophage
polarization. Exp Cell Res. 357:155–162. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang Y, Han CC, Cui D, Li Y, Ma Y and Wei
W: Is macrophage polarization important in rheumatoid arthritis?
Int Immunopharmacol. 50:345–352. 2017.PubMed/NCBI View Article : Google Scholar
|