Lung cancer constitutes 11.4% of all newly diagnosed
cases of cancer, and is the leading cause of cancer-associated
mortality worldwide (1). Lung
cancer is a heterogenous disease that is divided into two major
categories based on histopathology: Small cell lung cancer (SCLC)
and non-SCLC (NSCLC). NSCLC accounts for 85% of lung cancer cases,
and has several subtypes, including lung adenocarcinoma (LUAD),
lung squamous cell carcinoma and large cell lung carcinoma
(2). SCLC is a type of
neuroendocrine tumor that is classified into two subtypes: SCLC and
combined SCLC (3,4). Importantly, in addition to different
histopathological subtypes, the heterogeneity of lung cancer also
refers to differences between patients with the same subtype, or
differences among cells in the same tumor tissue (5-10).
The heterogeneity of lung cancer affects clinical treatment, since
patients with the same pathological type may have diverse reactions
to the same treatment (11-14).
To effectively treat lung cancer, it is necessary to explore the
source of lung cancer heterogeneity, identify specific antitumor
drugs and achieve personalized treatment for patients.
Conventional two-dimensional (2D) culture and
patient-derived xenograft (PDX) models are useful tools that assist
in understanding the mechanisms underlying the occurrence,
development and heterogeneity of lung cancer. However, these tools
have certain limitations. Culture methods, passage numbers and
other unexpected factors may cause tumor cell lines to lose the
phenotype and genotype of a primary tumor in a 2D culture model
(15-17).
Moreover, as 2D culture models lack extracellular matrix, stromal
cells and immune cells, they cannot accurately simulate the tumor
microenvironment (TME) and hence the conditions affecting cancers
in vivo (18-21).
Moreover, the defects of 2D culture models frequently cause
antitumor drugs to show efficacies and toxicities in vivo
that are different from those obtained in vitro during drug
screening, which causes rapid drug screening to be challenging
(22,23). Compared with 2D culture models, PDX
models in which researchers transplant surgically resected tumor
tissues into immunodeficient mice, are more accurate in
representing the phenotype, genotype and TME of the parental tumor
(24-28).
However, the following issues can be observed: i) The proportion of
transplantations that are successful in the establishment of PDX
models is sometimes too low (29-31);
ii) the process of successfully developing PDX models for drug
screening is time consuming (32,33),
and the condition of the patient often deteriorates during this
period; iii) murine stromal cells gradually replace patient stromal
cells, which changes the TME of the PDX model (34,35);
and iv) the mechanism of interaction between tumor cells and immune
cells in a PDX model is challenging to investigate because general
PDX models lack immune cells (36,37).
The narrow limitations of conventional tumor cell
lines and PDX models have driven researchers to investigate
improved preclinical tumor models that preserve the characteristics
of primary tumors to the largest extent. Such models are designed
to be built in a short amount of time for rapid antitumor drug
screening and expanded for the long-term investigation of cancer
mechanisms and modification of treatment plans. Tumor organoids,
also known as tumoroids (38),
have thus emerged. Through self-organization, stem cells generate
organoids that retain almost all the features of parental tissues
(39,40). Numerous studies have demonstrated
that tumor organoids have broad applications in different types of
cancer (41-47).
Researchers are using organoids to investigate lung cancer, with
different studies describing various methods for the establishment
of lung cancer organoids (LCOs) (48-80).
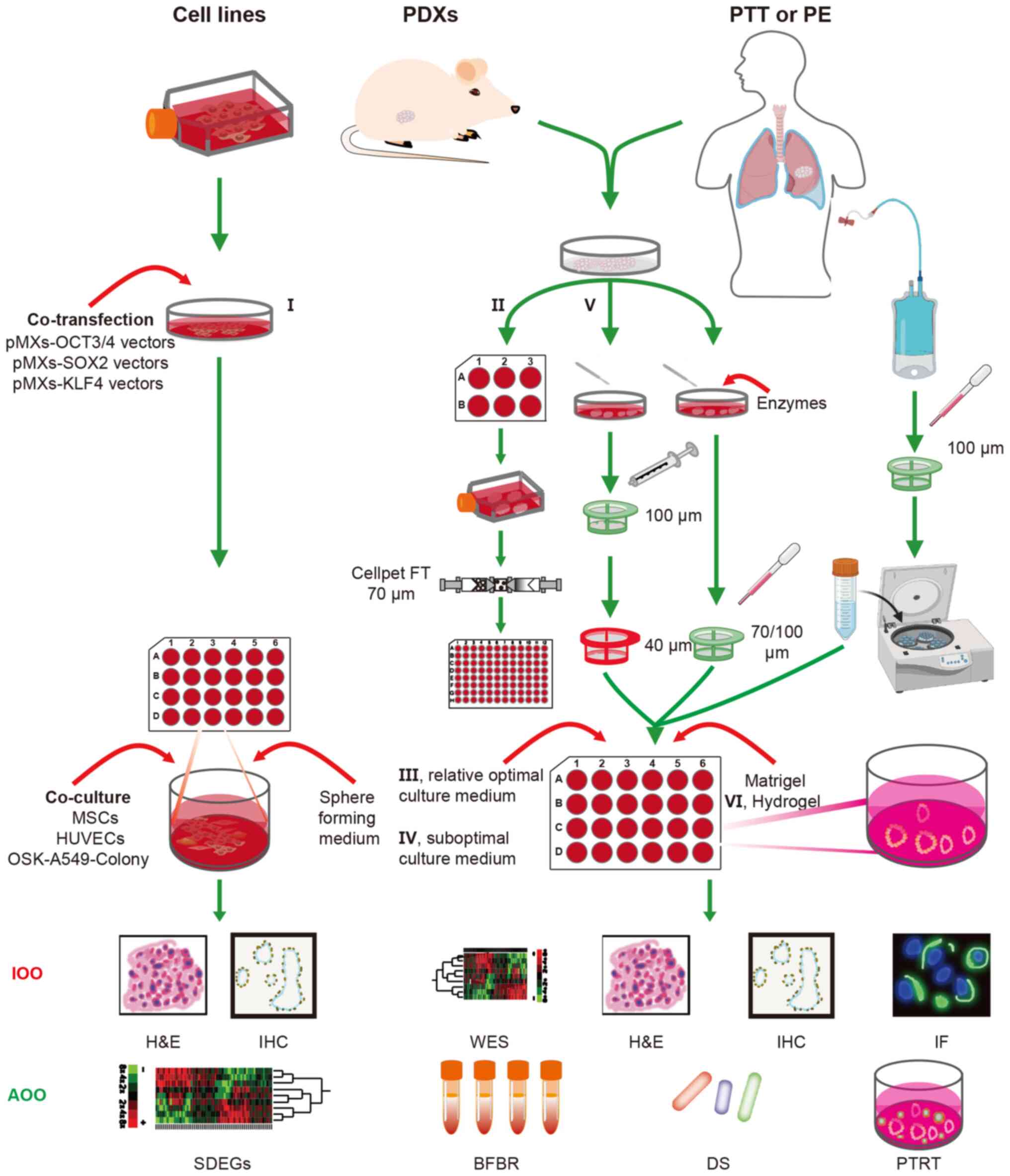
These methods can be divided into three broad categories based on
the source of cells used, namely cancer cell line-based LCOs,
PDX-derived LCOs and patient-derived LCOs (Fig. 1). At present, 17 different methods
for building LCOs have been described in the literature (Tables I and SI). These methods can be merged into six
categories based on the source of cells, the pre-treatment method
used, the composition of the medium and/or the culture scaffold: i)
Cancer stem cells (CSCs) induced by defined transcription factors
(48); ii) suspension culture
method (52,66); iii) relative optimal culture medium
with serum-free additive, amino acids, growth factor,
stemness-related signaling pathway activators and apoptosis
signaling pathway inhibitors (49-51,53,55,57,68,76,79,80);
iv) suboptimal culture medium (relative optimal culture medium
without stemness-associated signaling pathway activators) (71); v) mechanical digestion and
suboptimal culture medium (70);
and vi) hydrogel scaffold (69,78)
(Fig. 1; Tables I, II, SI
and SII). The current review
presents the advantages and drawbacks of these methods, and cites
references for relevant studies. The three broad categories of LCOs
are discussed, along with the different methods used to establish
LCOs.
Despite their disadvantages, cancer cell line-based
LCOs have certain practical utility. There are, however, some
aspects of such LCOs that require further investigation: i) Whether
lung cancer cells can be transformed into lung CSCs by transfection
with other combinations of stem cell transcription factors such as
OCT3/4, SOX2, KLF4, cellular myelocytomatosis oncogene and/or Nanog
homeobox; ii) differences among stem cells induced by transfection
with different combinations of stem cell transcription factors;
iii) whether other lung cancer cell lines, especially SCLC cell
lines, can also be transformed into lung CSCs and used to
successfully establish tumoroids using this method,; and iv)
whether this approach is suitable for use in drug screening. In
addition, as some studies have reported that pyroptosis plays an
important role in the occurrence and development of lung cancer
(88-90),
it is not yet known if cancer cell line-based LCOs can be used for
researching the mechanism of drug-induced lung cancer cell
pyroptosis. To broaden the application scope of cancer cell
line-based LCOs, these unknown factors require elucidation.
As PDXs effectively maintain the characteristics of
primary tumors, including the phenotype, genetic profile and TME,
they are widely used as preclinical models to explore the
mechanisms of tumorigenesis and development, screen antitumor drugs
and discover novel therapeutic methods (91-94).
However, the high cost and time-consuming process of developing
PDXs limit their usage (95,96).
To use the full advantages of PDXs while avoiding their
disadvantages, investigators have developed PDX-derived organoid
models that have already been applied to multiple tumor types, and
shown to preserve the genomic and transcriptomic profiles, protein
markers and drug response of primary PDXs (94,97-99).
These models have been used to study the pathogenesis of lung
cancer and screen antitumor drugs (Fig. 1). A total of four methods to create
PDX-derived LCOs are presented in the current review (Table I) (55,78-80).
Only one of these was used to establish PDX-derived organoids of
NSCLC (55), while the other three
were used to generate PDX-derived organoids of SCLC (78-80).
Delayed diagnosis, high aggressiveness,
susceptibility to relapse and poor prognosis are the basic
characteristics of SCLC (4,100-102).
Although SCLC is divided into four subtypes based on expression of
the transcription factors achaete-scute homolog 1, neuronal
differentiation 1, POU class 2 homeobox 3 and Yes1 associated
transcriptional regulator (103),
there is no specific and effective treatment for each subtype
(104). Traditional preclinical
models perform poorly in the exploration of novel markers and
treatment methods for SCLC (105). Therefore, it is urgently
necessary to develop more efficient preclinical models. Tumor
organoids have unique advantages in that regard, and researchers
have been attempting to establish SCLC organoids (71,72,77-80).
Gmeiner et al (78), Chen
et al (79) and Redin et
al (80) have each reported
methods for the culture of SCLC PDX-derived organoids to explore
drug resistance mechanisms and screen specific antitumor drugs. The
study by Gmeiner et al (78) indicated that overexpression of the
transcription factor E2 promoter binding factor 1-3 caused the
upregulation of thymidylate synthase and the increased malignancy
of SCLC. To test the efficacy of thymidylate synthase inhibitors in
the treatment of SCLC, the authors created SCLC PDX-derived
organoid models and used them to demonstrate that SCLC is sensitive
to CF10, a novel fluoropyrimidine polymer. Retinoic acid
receptor-related orphan receptors (RORs), including RORα, RORβ and
RORγ, participate in a variety of physiological and pathological
reactions through ligand-dependent interactions with co-regulators
(106). Chen et al
(79) found that the high
expression of RORγ improved SCLC cell growth and inhibited
apoptosis, while the RORγ antagonists XY018 and GSK805 eliminated
this effect in H446 and H1048 cells. These results were verified in
SCLC PDX-derived organoids. Chemical library screening, and
cellular thermal shift and surface plasmon resonance assays were
used to identify N-hydroxyapiosporamide (N-hydap), as a potent and
selective RORγ antagonist. N-hydap was more efficient at
suppressing the growth and survival of cancer cells than GSK805 and
XY018, which was confirmed using SCLC PDX-derived organoid models.
The study by Chen et al (79) provides a new approach for the
screening of targeted antitumor drugs. Redin et al (80) described another method for the
generation of SCLC PDX-derived organoids, which they used to verify
the curative effect of the YES1-specific inhibitor CH6953755 on
SCLC. All three methods involve the use of newly established, not
passaged, PDX-derived organoids to explore the mechanism of lung
cancer development and screen antitumor drugs. The study findings
indicate that short-term culture PDX-derived LCOs are reliable in
the preclinical research of lung cancer.
The aforementioned four methods can be divided into
two categories, those using relative optimal culture medium and
those using a hydrogel scaffold. The methods developed by Shi et
al (55), Chen et al
(79) and Redin et al
(80) are in the former category,
while that developed by Gmeiner et al (78) is in the latter category. The
details of these methods are presented in Table II.
These studies used four different digestion methods
to dissociate tissues into single cells. Digestive strategies using
the combination of Liberase TM, which comprises a combination of
collagenase I, II and thermolysin, with TrypLE involve lower
concentrations of enzymes with higher digestive efficiency compared
with those that use a combination of collagenase II and TrypLE.
Digestive methods using collagenase IV alone also have a higher
digestive efficiency compared with those that using a combination
of collagenase II and TrypLE. This may be due to collagenases I and
IV having higher activity than collagenase II. Notably, treatment
with TrypLE, a recombinant trypsin-like protease (107) used in two different digestive
enzyme combinations, has been shown to result in a significantly
higher cell viability compared with trypsin (108). Any pure collagenase is not able
to effectively dissociate tissues into single cells (109). The combination of any collagenase
and TrypLE may have a greater ability to generate single cells from
tissues than either enzyme used alone.
Cell-Titer Glo reagents manufactured by Promega
Corporation were used to measure the viability of organoids in the
method reported by Chen et al (79). To avoid interference with the
detection of fluorescence, phenol red-free Dulbecco's modified
Eagle's medium/Ham's F 12 nutrient medium (DMEM/F12) was used.
PDX-derived LCOs have certain advantages compared
with cancer cell line-based or patient-derived LCOs: i) The
phenotype, genetic profiles and heterogeneity of parental tumors
are more effectively preserved in PDX-derived LCOs than in lung
cancer cell line-based LCOs; ii) compared with patient samples,
PDXs are easier to obtain and can undergo long-term expansion by
passaging, which ensures the repeatability and sustainability of
experiments; iii) the establishment of PDX-derived LCOs has lower
requirements for ethical approval compared with patient-derived
LCOs; and iv) PDX-derived LCOs are easier to develop than
patient-derived LCOs (55).
However, there is a clear disadvantage of PDX-derived LCOs, in
addition to the defects common to all LCOs: PDX-derived LCOs may be
contaminated by mouse cells during short-term culture, which is
likely to affect the phenotypic identification of tumors, genotype
analysis or antitumor drug screening. These factors may restrict
their application.
Patient-derived LCOs are ideal for researching the
mechanism of initiation, development and drug resistance of lung
cancer, and for exploring new biomarkers, antitumor drugs and
treatment protocols. The samples used to create patient-derived
LCOs mainly originate from cancer tissue, including that obtained
during surgery or biopsy, or from the exfoliative tumor cells
present in pleural effusion (PE) (Fig.
1). In the current review, 13 methods used to generate LCOs
from patient samples are presented (Tables I, II, SI
and SII) (49-53,55,57,66,68-71,76).
These involve all six categories of methods used to establish
organoids. The success rates of organoid and pure LCO establishment
using these methods are variable (57,60),
which might influence subsequent mechanistic research and drug
screening. Therefore, it is necessary to compare and analyze the
details of the methods investigated in these studies to select the
ideal method for the culture of LCOs.
The relative optimal culture medium method is the
most popular, and the most representative method among all those
reported is that described by Sachs et al (57). Although different laboratories
generated LCOs via similar methods, the success rate of organoid
establishment ranged from 41 to 88%. The success rate range of pure
LCO establishment was also diverse, ranging from 7 to 92.7%
(60,63). Dijkstra et al (60) reported lower success rates of
organoid establishment (41%) and LCO establishment (17%) compared
with other studies (57,63). Patients with stage IV
adenocarcinoma accounted for 78% of all patients in the study by
Dijkstra et al suggesting that the low success rates might
be due to the degree of tumor malignancy. Kim et al
(63) used 77 malignant effusion
samples, three brain metastasis samples, a single bone metastasis
sample and two primary lung tumor samples from patients with
advanced LUAD to successfully generate LCOs, and the success rates
of organoid and pure LCO establishment were 83 and 92.7%,
respectively. According to these results, it can be concluded that
samples from malignant effusions or metastatic foci easily form
pure cancer organoids. This may be due to the airway organoid (AO)
culture medium being more suitable for normal airway epithelial
cell growth and samples from malignant effusions or metastasis foci
being less easily contaminated by normal epithelial cells than
those from primary lung tissues. If normal epithelial cells are not
removed during the pretreatment process, cancer cells are rapidly
overtaken by normal epithelial cells during organoid culture,
leading to failure of the cancer organoid culture (135,136). However, surgically resected tumor
tissues are a prominent source of material for LCO culture. To make
full use of these tissues, a number of researchers have sought to
devise improved culture methods. Kim et al (71) developed a new method using LCO
suboptimal medium free of Wnt3a, Noggin and A83-01 to culture lung
tumoroids, which improved the growth of cancer organoids and
inhibited that of normal epithelial organoids. When surgically
resected tumor tissues were used to establish organoids, the
success rates were 58-87% (71,73),
which were comparable with the 41-88% success rates of the method
described by Sachs et al (57). Moreover, a success rate of pure LCO
establishment of 71% was observed (73), which is higher than the 17%
reported by Dijkstra et al (60). Therefore, it is speculated that LCO
suboptimal medium may be superior to AO medium in cancer organoid
culture. It is noteworthy that that the pretreatment method used by
Kim et al (71) differed
from that used by Sachs et al (57). Sachs et al (57) used only collagenase to digest
tissue. By contrast, Kim et al (71) used DNase and collagenase/dispase to
isolate single lung cancer cells. Gohi et al (137) reported that digestion using a
combination of collagenase and DNase is conducive to the
maintenance of cell surface antigen integrity and cell activity.
These findings are meaningful for subsequent cancer organoid
culture. Therefore, the pretreatment method reported by Kim et
al (71) may partially
contribute to the high success rates of organoid establishment and
pure LCO establishment that were obtained. The importance of
pretreatment methods was supported by Hu et al (70), who found mechanical processing to
be more beneficial for tumor organoid formation than enzymatic
digestion, with the latter being beneficial for normal organoid
formation. Moreover, their study revealed that a medium without
R-spondin and Noggin is conducive to the establishment of pure
LCOs. Overall, it may be easier to generate higher purity LCOs with
an acceptable success rate by the use of mechanical digestion and
the suboptimal culture medium method (Tables I; SI).
In addition to the success rates of organoid
establishment and pure LCO establishment, researchers have
evaluated the sustainability of LCOs, which includes the expansion
and efficient reconstitution of cryopreserved LCOs. Short-term
organoid culture is sufficient to perform drug screening for
patients whose cancer tissue has been used to generate LCOs.
However, the long-term expansion of tumor organoids and efficient
reconstitution of cryopreserved organoids are necessary to provide
sufficient tumor organoids for the establishment of LCO biobanks
for use in long-term studies, such as those for antitumor drug
discovery, the elucidation of drug resistance mechanisms and
improvement of treatment protocols. Four methods of LCO long-term
culture are covered in the present review (55,57,71,76).
In terms of time, different definitions of tumoroid long-term
culture have been proposed. However, as regards passage number, the
definitions of tumoroid long-term culture are similar (>10
passages). The results of the study by Yokota et al
(65) showed that the AO medium is
a more robust tumor organoid culture medium than the media used by
Kim et al (71) and Shi
et al (55). While AO
medium is suitable for the growth of all lung epithelial cells,
some LCOs with particular mutations may also be long-term expanded
in AO medium (65). The study of
Yokota et al (65) revealed
that activation of the Wnt/β-catenin pathway is a prerequisite for
the maintenance of certain LCOs (TPM3-ROS1;
TP53K120Sfs*3) in long-term culture. This was
verified by Choi et al (76), who found that Wnt3A and R-spondin1
do not promote SCLC tumoroid formation but play important roles in
the long-term culture of SCLC tumoroids. Therefore, if the specific
mutations of lung cancer tissues are unknown, AO medium appears to
be a suitable choice for lung tumoroid long-term culture.
Nevertheless, the removal of activators of the Wnt/β-catenin
pathway did not influence other lung tumoroids in long-term
culture, for example LCOs (BRAFG469A;
TP53T155P) (65).
These results are consistent with those of Kim et al
(71), who found that most LCOs
could be long-term expanded in LCO suboptimal medium. In general,
the efficient reconstitution of cryopreserved LCOs is essential for
the establishment of LCO biobanks. The cell viability of LCOs
before cryopreservation and the methods of cryopreservation used
determine the success or failure of recovery. Although LCO
suboptimal medium contains fewer reagents than relative optimal
culture medium, Kim et al (71) reported that tumoroids cultured in
the former medium had a high recovery success rate (70%) after
freezing. This indicates that LCO suboptimal medium can effectively
sustain cell viability and provide high recovery success rates. In
a study by Shi et al (55),
long-term culture was achieved for 15% of NSCLC organoids, and
these organoids had good cell viability, with all of them being
recoverable after >1 year of cryopreservation and continued
passaging. Hu et al (70)
chose tumoroids with fast growth rates and short generation
intervals for cryopreservation. Of the five lung tumoroids used,
four were successfully cryopreserved and thawed. These results
showed that cell viability is a critical factor in the efficient
reconstitution of cryopreserved LCOs. Therefore, it is necessary to
screen for high-viability LCOs before cryopreservation.
In general, patient-derived LCOs are improved
preclinical models is comparison with other traditional models for
the following reasons: i) Patient-derived LCOs have excellent
fidelity because they are directly structured with patient tumor
tissue or PE; ii) patient-derived LCOs can be generated in weeks or
even days, and short-term cultured organoids are able to predict
the responses of patients to antitumor drugs. The integrated
superhydrophobic microwell array chip (InSMAR-chip) shortens the
time for drug screening to 1 week, thereby saving precious time for
patients requiring treatment (70); and iii) the combination of
patient-derived LCOs and microfluidic devices can standardize drug
screening to help clinicians in the formulation of appropriate
medication plans. Patient-derived LCOs also have certain
limitations: i) As samples originate from patients with lung
cancer, they are of high value and if the establishment of
patient-derived LCO fails, it is challenging to compensate for the
loss; ii) although patient-derived LCOs can be long-term expanded,
immortality of patient-derived LCOs in vitro has not yet
been achieved.
A number of issues remain to be resolved including:
i) How the optimization of patient-derived LCO culture methods can
be achieved to eliminate normal organic contamination and maintain
long-term lung tumoroid culture and even achieve immortality; ii)
how the standardization of patient-derived LCO culture methods can
be accomplished; iii) how the creation of a co-culture system of
patient-derived LCOs and the microenvironment can be realized; and
iv) how the success rate of LCO establishment from patient biopsy
samples can be enhanced.
LCOs derived from three major resources greatly
promote preclinical research, with applications including the
exploration of cancer mechanisms, searching for novel tumor
biomarkers, screening of antitumor drugs and improving treatment
plans. The three major types of lung tumoroids, which are cancer
cell line-based LCOs, PDX-derived LCOs and patient-derived LCOs,
are complementary, and the results of studies on the three major
types of lung tumoroids have provided a more comprehensive
understanding of the pathogenesis of lung cancer (48,50,52).
Furthermore, there are six different categories of methods that can
be used to establish LCOs. Appropriate models or methods can be
selected based on the requirements of researchers. However, the
present review puts forth several suggestions: i) AO medium is a
more robust tumor organoid culture medium than other media used for
the long-term culture of LCOs; ii) the mechanical digestion and
suboptimal culture medium method is more conducive to the
establishment of pure LCOs, while the mechanical dissociation
method is beneficial for the passage of organoids; iii) malignant
PE samples appear to have a high tendency to establish pure LCOs;
and iv) the use of CellPet FT to process organoids is beneficial
for standardization in high-throughput screening. The achievement
of standardization is important in lung tumoroid culture. The
combination of bioengineering technology, including microfluidic
devices and the InSMAR-chip, and lung tumoroid culture has
accelerated the standardization of lung tumoroid applications to
some degree. It is hypothesized that with technical progress, lung
tumoroids will have broader application prospects in the
future.
Not applicable.
Funding: No funding was received.
Not applicable.
QZ wrote the manuscript and prepared the figures. MZ
conceived and designed the study. MZ revised the manuscript
critically for important intellectual content. Both authors read
and approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang M, Herbst RS and Boshoff C: Toward
personalized treatment approaches for non-small-cell lung cancer.
Nat Med. 27:1345–1356. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 World Health Organization
Classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Rudin CM, Brambilla E, Faivre-Finn C and
Sage J: Small-cell lung cancer. Nat Rev Dis Primers.
7(3)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Prabavathy D and Ramadoss N: Heterogeneity
of small cell lung cancer stem cells. Adv Exp Med Biol. 1139:41–57.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang Y, Chang L, Yang Y, Fang W, Guan Y,
Wu A, Hong S, Zhou H, Chen G, Chen X, et al: Intratumor
heterogeneity comparison among different subtypes of non-small-cell
lung cancer through multi-region tissue and matched ctDNA
sequencing. Mol Cancer. 18(7)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
de Sousa VML and Carvalho L: Heterogeneity
in lung cancer. Pathobiology. 85:96–107. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Krohn A, Ahrens T, Yalcin A, Plönes T,
Wehrle J, Taromi S, Wollner S, Follo M, Brabletz T, Mani SA, et al:
Tumor cell heterogeneity in small cell lung cancer (SCLC):
Phenotypical and functional differences associated with
Epithelial-Mesenchymal Transition (EMT) and DNA methylation
changes. PLoS One. 9(e100249)2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liao H, Luo X, Huang Y, Yang X, Zheng Y,
Qin X, Tan J, Shen P, Tian R, Cai W, et al: Mining the prognostic
role of DNA methylation heterogeneity in lung adenocarcinoma. Dis
Markers. 2022(9389372)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Arora L, Kalia M, Dasgupta S, Singh N,
Verma AK and Pal D: Development of a Multicellular 3D tumor model
to study cellular heterogeneity and plasticity in NSCLC tumor
microenvironment. Front Oncol. 12(881207)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang Q, Li M, Yang M, Yang Y, Song F,
Zhang W, Li X and Chen K: Analysis of immune-related signatures of
lung adenocarcinoma identified two distinct subtypes: Implications
for immune checkpoint blockade therapy. Aging (Albany NY).
12:3312–3339. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu LP, Lu L, Zhao QQ, Kou QJ, Jiang ZZ,
Gui R, Luo YW and Zhao QY: Identification and validation of the
pyroptosis-related molecular subtypes of lung adenocarcinoma by
bioinformatics and machine learning. Front Cell Dev Biol.
9(756340)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kogure Y, Iwasawa S, Saka H, Hamamoto Y,
Kada A, Hashimoto H, Atagi S, Takiguchi Y, Ebi N, Inoue A, et al:
Efficacy and safety of carboplatin with nab-paclitaxel versus
docetaxel in older patients with squamous non-small-cell lung
cancer (CAPITAL): A randomised, multicentre, open-label, phase 3
trial. Lancet Healthy Longev. 2:e791–e800. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Spigel DR, Vicente D, Ciuleanu TE,
Gettinger S, Peters S, Horn L, Audigier-Valette C, Pardo Aranda N,
Juan-Vidal O, Cheng Y, et al: Second-line nivolumab in relapsed
small-cell lung cancer: CheckMate 331(☆). Ann Oncol. 32:631–641.
2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Muff R, Rath P, Ram Kumar RM, Husmann K,
Born W, Baudis M and Fuchs B: Genomic instability of osteosarcoma
cell lines in culture: Impact on the prediction of metastasis
relevant genes. PLoS One. 10(e0125611)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kasai F, Hirayama N, Ozawa M, Iemura M and
Kohara A: Changes of heterogeneous cell populations in the Ishikawa
cell line during long-term culture: Proposal for an in vitro clonal
evolution model of tumor cells. Genomics. 107:259–266.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bahcecioglu G, Basara G, Ellis BW, Ren X
and Zorlutuna P: Breast cancer models: Engineering the tumor
microenvironment. Acta Biomater. 106:1–21. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nolan JC, Frawley T, Tighe J, Soh H,
Curtin C and Piskareva O: Preclinical models for neuroblastoma:
Advances and challenges. Cancer Lett. 474:53–62. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lee SW, Kwak HS, Kang MH, Park YY and
Jeong GS: Fibroblast-associated tumour microenvironment induces
vascular structure-networked tumouroid. Sci Rep.
8(2365)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Salinas-Vera YM, Valdés J, Hidalgo-Miranda
A, Cisneros-Villanueva M, Marchat LA, Nuñez-Olvera SI, Ramos-Payán
R, Pérez-Plasencia C, Arriaga-Pizano LA, Prieto-Chávez JL, et al:
Three-dimensional organotypic cultures reshape the microRNAs
transcriptional program in breast cancer cells. Cancers (Basel).
14(2490)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jo Y, Choi N, Kim K, Koo HJ, Choi J and
Kim HN: Chemoresistance of cancer cells: Requirements of tumor
microenvironment-mimicking in vitro models in anti-cancer drug
development. Theranostics. 8:5259–5275. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shang M, Soon RH, Lim CT, Khoo BL and Han
J: Microfluidic modelling of the tumor microenvironment for
anti-cancer drug development. Lab Chip. 19:369–386. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Siolas D and Hannon GJ: Patient-derived
tumor xenografts: Transforming clinical samples into mouse models.
Cancer Res. 73:5315–5319. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lin D, Wyatt AW, Xue H, Wang Y, Dong X,
Haegert A, Wu R, Brahmbhatt S, Mo F, Jong L, et al: High fidelity
patient-derived xenografts for accelerating prostate cancer
discovery and drug development. Cancer Res. 74:1272–1283.
2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xiao T, Li W, Wang X, Xu H, Yang J, Wu Q,
Huang Y, Geradts J, Jiang P, Fei T, et al: Estrogen-regulated
feedback loop limits the efficacy of estrogen receptor-targeted
breast cancer therapy. Proc Natl Acad Sci USA. 115:7869–7878.
2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yoshida GJ: Applications of
patient-derived tumor xenograft models and tumor organoids. J
Hematol Oncol. 13(4)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li Z, Zheng W, Wang H, Cheng Y, Fang Y, Wu
F, Sun G, Sun G, Lv C and Hui B: Application of animal models in
cancer research: Recent progress and future prospects. Cancer Manag
Res. 13:2455–2475. 2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kuwata T, Yanagihara K, Iino Y, Komatsu T,
Ochiai A, Sekine S, Taniguchi H, Katai H, Kinoshita T and Ohtsu A:
Establishment of novel gastric cancer patient-derived xenografts
and cell lines: Pathological comparison between primary tumor,
patient-derived, and cell-line derived xenografts. Cells.
8(585)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Recondo G, Mahjoubi L, Maillard A, Loriot
Y, Bigot L, Facchinetti F, Bahleda R, Gazzah A, Hollebecque A,
Mezquita L, et al: Feasibility and first reports of the MATCH-R
repeated biopsy trial at Gustave Roussy. NPJ Precis Oncol.
4(27)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Heo EJ, Cho YJ, Cho WC, Hong JE, Jeon HK,
Oh DY, Choi YL, Song SY, Choi JJ, Bae DS, et al: Patient-derived
xenograft models of epithelial ovarian cancer for preclinical
studies. Cancer Res Treat. 49:915–926. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tucker ER, George S, Angelini P, Bruna A
and Chesler L: The promise of Patient-derived preclinical models to
accelerate the implementation of personalised medicine for children
with neuroblastoma. J Pers Med. 11(248)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhuang Y, Grainger JM, Vedell PT, Yu J,
Moyer AM, Gao H, Fan XY, Qin S, Liu D, Kalari KR, et al:
Establishment and characterization of immortalized human breast
cancer cell lines from breast cancer patient-derived xenografts
(PDX). NPJ Breast Cancer. 7(79)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Martinez-Garcia R, Juan D, Rausell A,
Muñoz M, Baños N, Menéndez C, Lopez-Casas PP, Rico D, Valencia A
and Hidalgo M: Transcriptional dissection of pancreatic tumors
engrafted in mice. Genome Med. 6(27)2014.PubMed/NCBI View
Article : Google Scholar
|
|
35
|
Hakuno SK, Michiels E, Kuhlemaijer EB,
Rooman I, Hawinkels L and Slingerland M: Multicellular modelling of
Difficult-to-Treat gastrointestinal cancers: Current possibilities
and challenges. Int J Mol Sci. 23(3147)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jung J, Seol HS and Chang S: The
generation and application of Patient-derived xenograft model for
cancer research. Cancer Res Treat. 50:1–10. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Meraz IM, Majidi M, Meng F, Shao R, Ha MJ,
Neri S, Fang B, Lin SH, Tinkey PT, Shpall EJ, et al: An Improved
Patient-derived xenograft humanized mouse model for evaluation of
lung cancer immune responses. Cancer Immunol Res. 7:1267–1279.
2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ganesh K, Wu C, O'Rourke KP, Szeglin BC,
Zheng Y, Sauvé CG, Adileh M, Wasserman I, Marco MR, Kim AS, et al:
A rectal cancer organoid platform to study individual responses to
chemoradiation. Nat Med. 25:1607–1614. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xia X, Li F, He J, Aji R and Gao D:
Organoid technology in cancer precision medicine. Cancer Lett.
457:20–27. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Brassard JA and Lutolf MP: Engineering
stem cell Self-organization to Build better organoids. Cell Stem
Cell. 24:860–876. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L,
Xia F, Fu G, Deng Y, Pan M, et al: Patient-derived organoids
predict chemoradiation responses of locally advanced rectal cancer.
Cell Stem Cell. 26:17–26.e6. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lõhmussaar K, Oka R, Espejo Valle-Inclan
J, Smits MHH, Wardak H, Korving J, Begthel H, Proost N, van de Ven
M, Kranenburg OW, et al: Patient-derived organoids model cervical
tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem
Cell. 28:1380–1396.e6. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lee SH, Hu W, Matulay JT, Silva MV,
Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, et
al: Tumor evolution and drug response in patient-derived organoid
models of bladder cancer. Cell. 173:515–528.e17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Servant R, Garioni M, Vlajnic T, Blind M,
Pueschel H, Müller DC, Zellweger T, Templeton AJ, Garofoli A,
Maletti S, et al: Prostate cancer patient-derived organoids:
Detailed outcome from a prospective cohort of 81 clinical
specimens. J Pathol. 254:543–555. 2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen P, Zhang X, Ding R, Yang L, Lyu X,
Zeng J, Lei JH, Wang L, Bi J, Shao N, et al: Patient-derived
organoids can guide personalized-therapies for patients with
advanced breast cancer. Adv Sci (Weinh). 8(e2101176)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Seidlitz T and Stange DE: Gastrointestinal
cancer organoids-applications in basic and translational cancer
research. Exp Mol Med. 53:1459–1470. 2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Broutier L, Mastrogiovanni G, Verstegen
MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R,
Sidorova O, Gaspersz MP, et al: Human primary liver cancer-derived
organoid cultures for disease modeling and drug screening. Nat Med.
23:1424–1435. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ogawa H, Koyanagi-Aoi M, Otani K, Zen Y,
Maniwa Y and Aoi T: Interleukin-6 blockade attenuates lung cancer
tissue construction integrated by cancer stem cells. Sci Rep.
7(12317)2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li YF, Gao Y, Liang BW, Cao XQ, Sun ZJ, Yu
JH, Liu ZD and Han Y: Patient-derived organoids of non-small cells
lung cancer and their application for drug screening. Neoplasma.
67:430–437. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Han Y, Lee T, He Y, Raman R, Irizarry A,
Martin ML and Giaccone G: The regulation of CD73 in non-small cell
lung cancer. Eur J Cancer. 170:91–102. 2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wang Y, Jiang T, Qin Z, Jiang J, Wang Q,
Yang S, Rivard C, Gao G, Ng TL, Tu MM, et al: HER2 exon 20
insertions in non-small-cell lung cancer are sensitive to the
irreversible pan-HER receptor tyrosine kinase inhibitor pyrotinib.
Ann Oncol. 30:447–455. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhang P, He B, Cai Q, Tu G, Peng X, Zhao
Z, Peng W, Yu F, Wang M, Tao Y, et al: Decreased IL-6 and NK cells
in Early-stage lung adenocarcinoma presenting as ground-glass
opacity. Front Oncol. 11(705888)2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li Z, Qian Y, Li W, Liu L, Yu L, Liu X, Wu
G, Wang Y, Luo W, Fang F, et al: Human Lung adenocarcinoma-derived
organoid models for drug screening. iScience.
23(101411)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Li Z, Yu L, Chen D, Meng Z, Chen W and
Huang W: Protocol for generation of lung adenocarcinoma organoids
from clinical samples. STAR Protoc. 2(100239)2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Shi R, Radulovich N, Ng C, Liu N, Notsuda
H, Cabanero M, Martins-Filho SN, Raghavan V, Li Q, Mer AS, et al:
Organoid cultures as preclinical models of Non-small cell lung
cancer. Clin Cancer Res. 26:1162–1174. 2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Liu T, Guo W, Luo K, Li L, Dong J, Liu M,
Shi X, Wang Z, Zhang J, Yin J, et al: Smoke-induced SAV1 gene
promoter hypermethylation disrupts YAP negative feedback and
promotes malignant progression of non-small cell lung cancer. Int J
Biol Sci. 18:4497–4512. 2022.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Sachs N, Papaspyropoulos A, Zomer-van
Ommen DD, Heo I, Böttinger L, Klay D, Weeber F, Huelsz-Prince G,
Iakobachvili N, Amatngalim GD, et al: Long-term expanding human
airway organoids for disease modeling. EMBO J.
38(e100300)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dijkstra KK, Cattaneo CM, Weeber F,
Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL,
Kaing S, Kelderman S, et al: Generation of Tumor-Reactive T cells
by Co-culture of peripheral blood lymphocytes and tumor organoids.
Cell. 174:1586–1598.e12. 2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Cattaneo CM, Dijkstra KK, Fanchi LF,
Kelderman S, Kaing S, van Rooij N, van den Brink S, Schumacher TN
and Voest EE: Tumor organoid-T-cell coculture systems. Nat Protoc.
15:15–39. 2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Dijkstra KK, Monkhorst K, Schipper LJ,
Hartemink KJ, Smit EF, Kaing S, de Groot R, Wolkers MC, Clevers H,
Cuppen E, et al: Challenges in establishing pure lung cancer
organoids limit their utility for personalized medicine. Cell Rep.
31(107588)2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Bie Y, Wang J, Xiong L, Wang D, Liao J,
Zhang Y and Lin H: Lung adenocarcinoma organoids harboring EGFR
19Del and L643V double mutations respond to osimertinib and
gefitinib: A case report. Medicine (Baltimore).
100(e24793)2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Sándor GO, Soós A, Lörincz P, Rojkó L,
Harkó T, Bogyó L, Tölgyes T, Bursics A, Buzás EI, Moldvay J, et al:
Wnt activity and cell proliferation are coupled to extracellular
vesicle release in multiple organoid models. Front Cell Dev Biol.
9(670825)2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kim SY, Kim SM, Lim S, Lee JY, Choi SJ,
Yang SD, Yun MR, Kim CG, Gu SR, Park C, et al: Modeling clinical
responses to targeted therapies by patient-derived organoids of
advanced lung adenocarcinoma. Clin Cancer Res. 27:4397–4409.
2021.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Padmanabhan J, Saha B, Powell C, Mo Q,
Perez BA and Chellappan S: Inhibitors targeting CDK9 show high
efficacy against osimertinib and AMG510 resistant lung
adenocarcinoma cells. Cancers (Basel). 13(3909)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Yokota E, Iwai M, Yukawa T, Yoshida M,
Naomoto Y, Haisa M, Monobe Y, Takigawa N, Guo M, Maeda Y, et al:
Clinical application of a lung cancer organoid (tumoroid) culture
system. NPJ Precis Oncol. 5(29)2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Tamura H, Higa A, Hoshi H, Hiyama G,
Takahashi N, Ryufuku M, Morisawa G, Yanagisawa Y, Ito E, Imai JI,
et al: Evaluation of anticancer agents using patient-derived tumor
organoids characteristically similar to source tissues. Oncol Rep.
40:635–646. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Takahashi N, Hoshi H, Higa A, Hiyama G,
Tamura H, Ogawa M, Takagi K, Goda K, Okabe N, Muto S, et al: An in
vitro system for evaluating molecular targeted drugs using lung
patient-derived tumor organoids. Cells. 8(481)2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ma X, Yang S, Jiang H, Wang Y and Xiang Z:
Transcriptomic analysis of tumor tissues and organoids reveals the
crucial genes regulating the proliferation of lung adenocarcinoma.
J Transl Med. 19(368)2021.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Mazzocchi A, Dominijanni A and Soker S:
Pleural effusion aspirate for use in 3D lung cancer modeling and
chemotherapy screening. Methods Mol Biol. 2394:471–483.
2022.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Hu Y, Sui X, Song F, Li Y, Li K, Chen Z,
Yang F, Chen X, Zhang Y, Wang X, et al: Lung cancer organoids
analyzed on microwell arrays predict drug responses of patients
within a week. Nat Commun. 12(2581)2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ,
Chun SM, Jung DJ, Shin TH, Jeong GS, Kim DK, et al: Patient-derived
lung cancer organoids as in vitro cancer models for therapeutic
screening. Nat Commun. 10(3991)2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Jung DJ, Shin TH, Kim M, Sung CO, Jang SJ
and Jeong GS: A one-stop microfluidic-based lung cancer organoid
culture platform for testing drug sensitivity. Lab Chip.
19:2854–2865. 2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Chen JH, Chu XP, Zhang JT, Nie Q, Tang WF,
Su J, Yan HH, Zheng HP, Chen ZX, Chen X, et al: Genomic
characteristics and drug screening among organoids derived from
non-small cell lung cancer patients. Thorac Cancer. 11:2279–2290.
2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Chen X, Liu Y, Wang Y, Wang C, Chen X,
Xiong Y, Liu L, Yuan X, Tang H, Shu C, et al: CYP4F2-catalyzed
metabolism of arachidonic acid promotes stromal cell-mediated
immunosuppression in non-small cell lung cancer. Cancer Res.
82:4016–4030. 2022.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Peng KC, Su JW, Xie Z, Wang HM, Fang MM,
Li WF, Chen YQ, Guan XH, Su J, Yan HH, et al: Clinical outcomes of
EGFR+/METamp+ vs. EGFR+/METamp-untreated patients with advanced
non-small cell lung cancer. Thorac Cancer. 13:1619–1630.
2022.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Choi SY, Cho YH, Kim DS, Ji W, Choi CM,
Lee JC, Rho JK and Jeong GS: Establishment and long-term expansion
of small cell lung cancer patient-derived tumor organoids. Int J
Mol Sci. 22(1349)2021.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Choi YJ, Lee H, Kim DS, Kim DH, Kang MH,
Cho YH, Choi CM, Yoo J, Lee KO, Choi EK, et al: Discovery of a
novel CDK7 inhibitor YPN-005 in small cell lung cancer. Eur J
Pharmacol. 907(174298)2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Gmeiner WH, Miller LD, Chou JW,
Dominijanni A, Mutkus L, Marini F, Ruiz J, Dotson T, Thomas KW,
Parks G, et al: Dysregulated pyrimidine biosynthesis contributes to
5-FU resistance in SCLC Patient-derived organoids but response to a
novel polymeric fluoropyrimidine, CF10. Cancers (Basel).
12(788)2020.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Chen J, Hu Y, Zhang J, Wang Q, Wu X, Huang
W, Wang Q, Cai G, Wang H, Ou T, et al: Therapeutic targeting RORγ
with natural product N-hydroxyapiosporamide for small cell lung
cancer by reprogramming neuroendocrine fate. Pharmacol Res.
178(106160)2022.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Redin E, Garrido-Martin EM, Valencia K,
Redrado M, Solorzano JL, Carias R, Echepare M, Exposito F, Serrano
D, Ferrer I, et al: YES1 is a druggable oncogenic target in Small
Cell Lung Cancer. J Thorac Oncol. 17:1387–1403. 2022.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Lancaster MA and Knoblich JA:
Organogenesis in a dish: Modeling development and disease using
organoid technologies. Science. 345(1247125)2014.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Suzuka J, Tsuda M, Wang L, Kohsaka S,
Kishida K, Semba S, Sugino H, Aburatani S, Frauenlob M, Kurokawa T,
et al: Rapid reprogramming of tumour cells into cancer stem cells
on double-network hydrogels. Nat Biomed Eng. 5:914–925.
2021.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Xu Z, Jia Y, Huang X, Feng N and Li Y:
Rapid induction of pancreatic cancer cells to cancer stem cells via
heterochromatin modulation. Cell Cycle. 17:1487–1495.
2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Ishiguro T, Ohata H, Sato A, Yamawaki K,
Enomoto T and Okamoto K: Tumor-derived spheroids: Relevance to
cancer stem cells and clinical applications. Cancer Sci.
108:283–289. 2017.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Weiswald LB, Bellet D and Dangles-Marie V:
Spherical cancer models in tumor biology. Neoplasia. 17:1–15.
2015.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Oshima N, Yamada Y, Nagayama S, Kawada K,
Hasegawa S, Okabe H, Sakai Y and Aoi T: Induction of cancer stem
cell properties in colon cancer cells by defined factors. PLoS One.
9(e101735)2014.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega
VB, Wong E, Orlov YL, Zhang W, Jiang J, et al: Integration of
external signaling pathways with the core transcriptional network
in embryonic stem cells. Cell. 133:1106–1117. 2008.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Zhang CC, Li CG, Wang YF, Xu LH, He XH,
Zeng QZ, Zeng CY, Mai FY, Hu B and Ouyang DY: Chemotherapeutic
paclitaxel and cisplatin differentially induce pyroptosis in A549
lung cancer cells via caspase-3/GSDME activation. Apoptosis.
24:312–325. 2019.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Long K, Gu L, Li L, Zhang Z, Li E, Zhang
Y, He L, Pan F, Guo Z and Hu Z: Small-molecule inhibition of APE1
induces apoptosis, pyroptosis, and necroptosis in non-small cell
lung cancer. Cell Death Dis. 12(503)2021.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Song J, Sun Y, Cao H, Liu Z, Xi L, Dong C,
Yang R and Shi Y: A novel pyroptosis-related lncRNA signature for
prognostic prediction in patients with lung adenocarcinoma.
Bioengineered. 12:5932–5949. 2021.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Coleman CN, Higgins GS, Brown JM, Baumann
M, Kirsch DG, Willers H, Prasanna PG, Dewhirst MW, Bernhard EJ and
Ahmed MM: Improving the predictive value of preclinical studies in
support of radiotherapy clinical trials. Clin Cancer Res.
22:3138–3147. 2016.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Sereti E, Karagianellou T, Kotsoni I,
Magouliotis D, Kamposioras K, Ulukaya E, Sakellaridis N,
Zacharoulis D and Dimas K: Patient derived xenografts (PDX) for
personalized treatment of pancreatic cancer: Emerging allies in the
war on a devastating cancer? J Proteomics. 188:107–118.
2018.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Invrea F, Rovito R, Torchiaro E, Petti C,
Isella C and Medico E: Patient-derived xenografts (PDXs) as model
systems for human cancer. Curr Opin Biotechnol. 63:151–156.
2020.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Beshiri ML, Tice CM, Tran C, Nguyen HM,
Sowalsky AG, Agarwal S, Jansson KH, Yang Q, McGowen KM, Yin J, et
al: A PDX/Organoid biobank of advanced prostate cancers captures
genomic and phenotypic heterogeneity for disease modeling and
therapeutic screening. Clin Cancer Res. 24:4332–4345.
2018.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Fujii E, Kato A and Suzuki M:
Patient-derived xenograft (PDX) models: Characteristics and points
to consider for the process of establishment. J Toxicol Pathol.
33:153–160. 2020.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Abdolahi S, Ghazvinian Z, Muhammadnejad S,
Saleh M, Asadzadeh Aghdaei H and Baghaei K: Patient-derived
xenograft (PDX) models, applications and challenges in cancer
research. J Transl Med. 20(206)2022.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Fong ELS, Toh TB, Lin QXX, Liu Z, Hooi L,
Mohd Abdul Rashid MB, Benoukraf T, Chow EK, Huynh TH and Yu H:
Generation of matched patient-derived xenograft in vitro-in vivo
models using 3D macroporous hydrogels for the study of liver
cancer. Biomaterials. 159:229–240. 2018.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Nelson SR, Zhang C, Roche S, O'Neill F,
Swan N, Luo Y, Larkin A, Crown J and Walsh N: Modelling of
pancreatic cancer biology: Transcriptomic signature for 3D
PDX-derived organoids and primary cell line organoid development.
Sci Rep. 10(2778)2020.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Romero-Calvo I, Weber CR, Ray M, Brown M,
Kirby K, Nandi RK, Long TM, Sparrow SM, Ugolkov A, Qiang W, et al:
Human organoids share structural and genetic features with primary
pancreatic adenocarcinoma tumors. Mol Cancer Res. 17:70–83.
2019.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Chauhan AF and Liu SV: Small cell lung
cancer: Advances in diagnosis and management. Semin Respir Crit
Care Med. 41:435–446. 2020.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Wang Y, Zou S, Zhao Z, Liu P, Ke C and Xu
S: New insights into small-cell lung cancer development and
therapy. Cell Biol Int. 44:1564–1576. 2020.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Wang WZ, Shulman A, Amann JM, Carbone DP
and Tsichlis PN: Small cell lung cancer: Subtypes and therapeutic
implications. Semin Cancer Biol. 86:543–554. 2022.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Ireland AS, Micinski AM, Kastner DW, Guo
B, Wait SJ, Spainhower KB, Conley CC, Chen OS, Guthrie MR, Soltero
D, et al: MYC drives temporal evolution of small cell lung cancer
subtypes by reprogramming neuroendocrine fate. Cancer Cell.
38:60–78.e12. 2020.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Kalemkerian GP, Loo BW, Akerley W, Attia
A, Bassetti M, Boumber Y, Decker R, Dobelbower MC, Dowlati A,
Downey RJ, et al: NCCN Guidelines Insights: Small cell lung cancer,
version 2.2018. J Natl Compr Canc Netw. 16:1171–1182.
2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Drapkin BJ and Rudin CM: Advances in
small-cell lung cancer (SCLC) translational research. Cold Spring
Harb Perspect Med. 11(a038240)2021.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Fan J, Lv Z, Yang G, Liao TT, Xu J, Wu F,
Huang Q, Guo M, Hu G, Zhou M, et al: Retinoic acid receptor-related
orphan receptors: Critical roles in tumorigenesis. Front Immunol.
9(1187)2018.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Hogan S, O'Gara JP and O'Neill E: Novel
treatment of staphylococcus aureus Device-related infections using
fibrinolytic agents. Antimicrob Agents Chemother. 62:e02008–17.
2018.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Gobin CM, Menefee JN, Lattimore CC, Doty A
and Fredenburg KM: Cell Dissociation enzymes affect Annexin
V/Flow-cytometric apoptotic assay outcomes After miRNA-based
transient transfection. Anticancer Res. 42:2819–2825.
2022.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Maruyama I, Yoshida C, Kobayashi M,
Oyamada H and Momose K: Preparation of single smooth muscle cells
from guinea pig taenia coli by combinations of purified collagenase
and papain. J Pharmacol Methods. 18:151–161. 1987.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Wise DR and Thompson CB: Glutamine
addiction: A new therapeutic target in cancer. Trends Biochem Sci.
35:427–433. 2010.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Richards NG and Schuster SM: Mechanistic
issues in asparagine synthetase catalysis. Adv Enzymol Relat Areas
Mol Biol. 72:145–198. 1998.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Wellen KE, Lu C, Mancuso A, Lemons JM,
Ryczko M, Dennis JW, Rabinowitz JD, Coller HA and Thompson CB: The
hexosamine biosynthetic pathway couples growth factor-induced
glutamine uptake to glucose metabolism. Genes Dev. 24:2784–2799.
2010.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Zhang J, Pavlova NN and Thompson CB:
Cancer cell metabolism: The essential role of the nonessential
amino acid, glutamine. EMBO J. 36:1302–1315. 2017.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Heeneman S, Deutz NE and Buurman WA: The
concentrations of glutamine and ammonia in commercially available
cell culture media. J Immunol Methods. 166:85–91. 1993.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Schneider M, Marison IW and von Stockar U:
The importance of ammonia in mammalian cell culture. J Biotechnol.
46:161–185. 1996.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Imamoto Y, Tanaka H, Takahashi K, Konno Y
and Suzawa T: Advantages of AlaGln as an additive to cell culture
medium: Use with anti-CD20 chimeric antibody-producing POTELLIGENT™
CHO cell lines. Cytotechnology. 65:135–143. 2013.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Yoshida Y, Soma T, Matsuzaki T and
Kishimoto J: Wnt activator CHIR99021-stimulated human dermal
papilla spheroids contribute to hair follicle formation and
production of reconstituted follicle-enriched human skin. Biochem
Biophys Res Commun. 516:599–605. 2019.PubMed/NCBI View Article : Google Scholar
|
|
118
|
An WF, Germain AR, Bishop JA, Nag PP,
Metkar S, Ketterman J, Walk M, Weiwer M, Liu X, Patnaik D, et al:
Discovery of potent and highly selective inhibitors of GSK3b. In:
Probe Reports from the NIH Molecular Libraries Program. National
Center for Biotechnology Information (US), Bethesda (MD), 2010.
|
|
119
|
Takahashi T and Shiraishi A: Stem cell
signaling pathways in the small intestine. Int J Mol Sci.
21(2032)2020.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Vincan E, Schwab RHM, Flanagan DJ, Moselen
JM, Tran BM, Barker N and Phesse TJ: The Central role of wnt
signaling and organoid technology in personalizing anticancer
therapy. Prog Mol Biol Transl Sci. 153:299–319. 2018.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Yoshida T, Singh AK, Bishai WR, McConkey
DJ and Bivalacqua TJ: Organoid culture of bladder cancer cells.
Investig Clin Urol. 59:149–151. 2018.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Djomehri SI, Burman B, Gonzalez ME,
Takayama S and Kleer CG: A reproducible scaffold-free 3D organoid
model to study neoplastic progression in breast cancer. J Cell
Commun Signal. 13:129–143. 2019.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Ahn Y, An JH, Yang HJ, Lee DG, Kim J, Koh
H, Park YH, Song BS, Sim BW, Lee HJ, et al: Human blood vessel
organoids penetrate human cerebral organoids and form a Vessel-like
system. Cells. 10(2036)2021.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Li Y, Wang R, Huang D, Ma X, Mo S, Guo Q,
Fu G, Li Y, Xu X, Hu X, et al: A novel human colon signet-ring cell
carcinoma organoid line: Establishment, characterization and
application. Carcinogenesis. 41:993–1004. 2020.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Ma HC, Zhu YJ, Zhou R, Yu YY, Xiao ZZ and
Zhang HB: Lung cancer organoids, a promising model still with long
way to go. Crit Rev Oncol Hematol. 171(103610)2022.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Maddalo G, Spolverato Y, Rugge M and
Farinati F: Gastrin: From pathophysiology to cancer prevention and
treatment. Eur J Cancer Prev. 23:258–263. 2014.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Zheng B, Ko KP, Fang X, Wang X, Zhang J,
Jun S, Kim BJ, Luo W, Kim MJ, Jung YS, et al: A new murine
esophageal organoid culture method and organoid-based model of
esophageal squamous cell neoplasia. iScience.
24(103440)2021.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Tsai S, McOlash L, Palen K, Johnson B,
Duris C, Yang Q, Dwinell MB, Hunt B, Evans DB, Gershan J, et al:
Development of primary human pancreatic cancer organoids, matched
stromal and immune cells and 3D tumor microenvironment models. BMC
Cancer. 18(335)2018.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Kawasaki K, Toshimitsu K, Matano M, Fujita
M, Fujii M, Togasaki K, Ebisudani T, Shimokawa M, Takano A,
Takahashi S, et al: An organoid biobank of neuroendocrine neoplasms
enables genotype-phenotype mapping. Cell. 183:1420–1435.e21.
2020.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Shiota J, Samuelson LC and Razumilava N:
Hepatobiliary organoids and their applications for studies of liver
health and disease: Are We There Yet? Hepatology. 74:2251–2263.
2021.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Dong R, Zhang B and Zhang X: Liver
organoids: An in vitro 3D model for liver cancer study. Cell
Biosci. 12(152)2022.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Sato T, Stange DE, Ferrante M, Vries RG,
Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J,
Siersema PD, et al: Long-term expansion of epithelial organoids
from human colon, adenoma, adenocarcinoma, and Barrett's
epithelium. Gastroenterology. 141:1762–1772. 2011.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Fatehullah A, Tan SH and Barker N:
Organoids as an in vitro model of human development and disease.
Nat Cell Biol. 18:246–254. 2016.PubMed/NCBI View Article : Google Scholar
|
|
134
|
van de Wetering M, Francies HE, Francis
JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J,
Taylor-Weiner A, Kester L, et al: Prospective derivation of a
living organoid biobank of colorectal cancer patients. Cell.
161:933–945. 2015.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Karthaus WR, Iaquinta PJ, Drost J,
Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel
H, Sachs N, et al: Identification of multipotent luminal progenitor
cells in human prostate organoid cultures. Cell. 159:163–175.
2014.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Verissimo CS, Overmeer RM, Ponsioen B,
Drost J, Mertens S, Verlaan-Klink I, Gerwen BV, van der Ven M,
Wetering MV, Egan DA, et al: Targeting mutant RAS in
patient-derived colorectal cancer organoids by combinatorial drug
screening. Elife. 5(e18489)2016.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Gohi B, Liu XY, Zeng HY, Xu S, Ake KMH,
Cao XJ, Zou KM and Namulondo S: Enhanced efficiency in isolation
and expansion of hAMSCs via dual enzyme digestion and
micro-carrier. Cell Biosci. 10(2)2020.PubMed/NCBI View Article : Google Scholar
|