Introduction
The quality of life for an individual is negatively
affected by autism spectrum disorder (ASD), a neurodevelopmental
condition that affects social communications and results in
obstinate and repetitive behaviors (1). Gastrointestinal (GI) tract issues,
such as food intolerance, abdominal pain, inflammatory disease,
digestive disorders, diarrhea, or constipation are common in
autistic patients in addition to the characteristic cognitive
traits (2). Although ASD can be
identified in children as young as 3 years old, symptoms persist
throughout an the life of an individual (3). The human gut microbiome (GM) is home
to a complex ecosystem of microorganisms living in the GI tract
including viruses, eukaryotes, archaea and bacteria (4). Additionally, saprophytic commensal
flora in the gut plays a crucial role in modulating a variety of
local functions, including nutrient absorption, maintenance of the
intestinal barrier, stimulation and regulation of the host immune
system and defense against pathogenic microorganisms (4-7).
An imbalance in the gut microbiota is referred to as dysbiosis and
can interfere with a wide range of biological mechanisms.
Alterations in systemic metabolism, neuroplasticity and the
neuroimmune system, all of which have been linked to ASD (8-13),
may result from dysbiosis. Additionally, the GM interacts with
relevant host, microbiota and environmental factors that can result
in gut dysbiosis (14,15). Numerous studies have shown that
gastrointestinal dysfunction coexists with ASD (16-21).
According to literature reviews of the human gut microbiota, the
coordination between the brain and the microflora of the
gastrointestinal tract can be disrupted by changes in the
composition of the GM (22). The
ability to identify microorganisms from all domains of life is the
primary advantage of metagenomic whole genome shotgun sequencing
(mWGS) for taxonomic classification over amplicon sequencing or
marker gene approaches (23).
Given the complex enteric nervous system in the gut directly
interferes with the brain and permits the bidirectional flow of
information, microbiome research that focuses on the relationship
between ASD and gut microbiota is crucial. This enhances functions
and emotions that may be affected by gastrointestinal contents,
such as cognition and function (22). Therefore, changes in the
composition of the microbiome may result in disturbed
host-microbiota homeostasis and cause autism (24). Changes in the GM composition have
been observed in individuals with ASD (25). The immune pathway is largely
involved in regulating microbial profiles, supporting the theory of
a complex relationship between ASD, immune dysregulation, and
altered microbiome that may aid in the identification of molecular
biomarkers for the diagnosis of ASD (26). According to a recent study, the
gut-brain and gut metabolic models of ASD with GI symptoms showed
abnormalities in several cases, including neurotoxin-related
p-cresol degradation and short-chain fatty acid (SCFA)
degradation/synthesis, which are closely associated with ASD
behaviors in animal models (27).
Additionally, the GM is the primary contributing factor for
behavioral symptoms and gastrointestinal symptoms linked to ASD
(28). Through pathways connected
to the gut-brain axis, the gut microbiota and their metabolic
products, such as SCFAs, may have an effect on the metabolism of
central neurotransmitters (29).
Furthermore, a variety of factors, such as antibiotics, pH, oxygen
levels, dietary supplements including probiotics and prebiotics,
and bacterial load, can alter the composition of the gut microbiota
(30). Probiotic administration to
regulate the gut-brain axis may improve gastrointestinal symptoms,
reduce inflammation and restore behavioral symptoms associated with
ASD, as well as the gut microbiota composition and intestinal
barrier function in animal and human models (31). The gut-brain axis is altered by gut
dysbiosis, which increases the risk of developing ASD and other
disorders (32). It has been
established that the gut-brain axis links various brain functions,
including the emotional and cognitive functions of the limbic
system, prefrontal cortex, and hypothalamus (33). The central nervous system, the
hypothalamic-pituitary-adrenal axis, the enteric nervous system and
the autonomic nervous system all have bidirectional, intricately
integrated signaling pathways that together make up the gut-brain
axis (34). Through vagal
stimulation and inflammatory mediators and their metabolites, the
GM can directly affect these processes (35). Regarding somatic complaints and
symptoms, self-dysregulation, criminal behavior, and cognitive
difficulties are markedly associated with changes in taxonomic
diversity in individuals with ASD (26).
To classify and identify molecular biomarkers for
ASD, the present study aimed to evaluate the differences in the
composition of the gut microbiota between children with autism and
their healthy siblings.
Materials and methods
Sampling
Fecal samples were taken at Pediatric Clinics in
King Abdulaziz University Hospital in Jeddah, Saudi Arabia, over 1
month in March 2022. A total of eight children, aged 3-10 years
old, including four patients with ASD (three males and one female)
and four of their siblings who were healthy controls (two females
and two males), were recruited; the characteristics of the children
with ASD are presented in Tables I
and II. The Biomedical Ethics
Research Committee at King Abdulaziz University (Jeddah, Kingdom of
Saudi Arabia) approved the present study (approval no.
10-CEGMR-Bioeth-2021). To guarantee the integrity of the DNA for
deep sequencing and microbiome analysis, all samples were collected
using particular collection tubes (ISWAB microbiome collection
tube).
 | Table IDemographic characteristics of
children with ASD. |
Table I
Demographic characteristics of
children with ASD.
| Sample | Sex | Age, years | Severity of
ASD | Symptoms |
|---|
| 1 | Male | 6 | Mild/level 1 | Hyperactivity,
repetitive behaviour, lack of eye contact, walking on tiptoes and
severe constipation |
| 2 | Female | 5 | Severe/level 1 | Hyperactivity,
repetitive behaviour, lack of eye contact, delay in language
acquisition and diarrhoea |
| 3 | Male | 8 | Mild/level 1 | Hyperactivity and
attention deficit |
| 4 | Male | 5 | Severe/level 2 | Hyperactivity,
attention deficit, cognitive and emotional symptoms, lack of eye
contact and delay in language acquisition |
 | Table IIDemographic characteristics of
children as controls. |
Table II
Demographic characteristics of
children as controls.
| Sample | Sex | Age, years | Control | Symptoms |
|---|
| 5 | Male | 9 | Healthy | Did not suffer from
any symptoms |
| 6 | Female | 5 | Healthy | Did not suffer from
any symptoms |
| 7 | Male | 5 | Healthy | Did not suffer from
any symptoms |
| 8 | Female | 10 | Healthy | Did not suffer from
any symptoms |
Extraction of genomic DNA
Following the manufacturer's instructions, DNA was
extracted from fecal samples using a QIAMP mini kit designed for
stool purification (QIAamp DNA Mini Kit; Qiagen GmbH). A DeNovix
DS-11 FX Spectrophotometer/Fluorometer nanodrop (Thermo Fisher
Scientific, Inc.) was used to assess the integrity and purity of
the sample.
Bioinformatics analysis
The Beijing Genomics Institute sent eight samples of
genomic DNA (four from the patients with ASD and four from the
healthy individuals) for bioinformatics analysis. The DNBSEQ
platform was used to test the samples initially. The total number
of detected gene catalogs was 673,091, which were functionally
annotated by seven databases, including BacMet (Antibacterial
Biocide and Metal Resistance Genes Database; version 20180311;
http://bacmet.biomedicine.gu.se/), CARD
(The Comprehensive Antibiotic Resistance Database; version 3.0.9;
https://card.mcmaster.ca/), KEGG (Kyoto
Encyclopedia of Genes and Genomes; version 101; http://www.genome.ad.jp/kegg/), eggNOG (evolutionary
genealogy of genes: Non-supervised Orthologous Groups; version 5.0;
http://eggnog6.embl.de/), COG (Clusters of
Orthologous Groups; version 20201125; https://www.ncbi.nlm.nih.gov/research/cog-project/),
(Swiss-Prot; release-2021_04; https://www.uniprot.org/), and CAZy
(Carbohydrate-Active enZYmes Database; version 20211013; http://www.cazy.org/). The average output of each
sample was 6.04 gigabyte of data and the average assembly length
was 134.31 megabytes. Following taxonomy analysis, the number of
species found was as follows: Kingdom level, 2; phylum level, 25;
class level, 34; order level, 84; family level, 257; genus level,
1,314; and species level, 4,339.
The library was sequenced on a DNBSEQ-400RS using a
DNA Library Prep Kit (cat. no. 1000017571, MGI Tech Co., Ltd.). The
main steps of sequencing and library preparation were as follows:
The concentration of the sample was detected using a Qubit
Fluorometer (Invitrogen; Thermo Fisher Scientific, Inc.), and the
integrity and purity of samples were assessed by agarose gel
electrophoresis (concentration of agarose gel, 1%; 150 V;
electrophoresis time, 40 min). A total of 1 µg genomic DNA was
randomly fragmented using an ultrasonicator (M220; Covaris, Inc.).
75.0 W (Peak incident power), 5.0% duty factor, 200 cycles per
burst, 50 sec treatment time), 20.0˚C temperature, 50 µl sample
volume, and 200-400 bp fragments of genomic DNA were selected by
2.8 µm of Dynabeads M-280 Streptavidin magnetic beads (cat. no.
112-05D; Invitrogen; Thermo Fisher Scientific, Inc.). Next, the
fragments were end-repaired and then 3' adenylated, then adaptors
were ligated to the ends of these 3' adenylated fragments. PCR was
used [98˚C/1 min, 11 cycles of (98˚C/10 sec, 60˚C/30 sec, 72˚C/30
sec), 72˚C/5 min and 4˚C/hold] to amplify fragments with adaptors
from the previous step, and then PCR products were purified using
the magnetic beads. The double-stranded PCR products were
heat-denatured and circularized by the splint oligo sequence. The
single-strand circle DNA was formatted as the final library. The
library was amplified using phi29 to make a DNA nanoball (DNB)
which contained >300 copies of one molecule. The DNBs were
loaded into the patterned nanoarray, and pair-end 100/150 base
reads were generated by combinatorial probe-anchor synthesis.
Since a certain percentage of low-quality linker
sequences may have been present in the original sequencing data,
SOAPnuke software (version 1.5.0; https://github.com/BGI-flexlab/SOAPnuke) was used to
filter out the low-quality data to obtain high-quality clean data.
MEGAHIT (version 1.1.3; https://github.com/voutcn/megahit) was used to put
together clean, filtered data, and to remove sequences <300 bp,
as well as for statistical analysis and gene prediction.
MetaGeneMark (version 3.38; http://exon.gatech.edu/index.html) was used to perform
metagenomic gene prediction for the assembled scaffold, CD-HIT
(version 4.6.4; http://weizhong-lab.ucsd.edu/cd-hit/) was used to
cluster predicted genes, and redundant sequences were removed to
create the gene catalog. Then, Venn diagrams were created between
various samples or groups by aligning reads with non-redundant gene
catalogs using Salmon (version 1.3.0; https://github.com/COMBINE-lab/salmon). Kraken
(version 2.1.2; https://github.com/DerrickWood/kraken2) was used to
perform the taxonomy annotation for the metagenomic data, to
determine the species abundance and to compare or contrast various
samples or groups. Bar charts, nonmetric multidimensional scaling
dimension reduction analysis, principal component analysis (PCA),
principal coordinates analysis (PCoA), anosim analysis of
similarity of different species and multivariate statistical
analysis of linear discriminant analysis were all employed in the
metagenomic taxonomy analysis. The analysis of species diversity in
a single sample is called α diversity. The functional database was
screened for the absolute abundance profile, the top 10 abundant
genes were assigned functional Circos (https://circos.ca), and the distribution of the
corresponding functional genes in each group was visually
displayed.
Results
To identify the taxonomic classification of the GM
and demonstrate the differences in gut microbe abundance between
children with ASD and their healthy control siblings, mWGS was
conducted on the GMs of both children with ASD and their healthy
control siblings. Tables III and
IV show the statistics of the raw
metagenomics data (filtering and assembly). Using a Venn diagram,
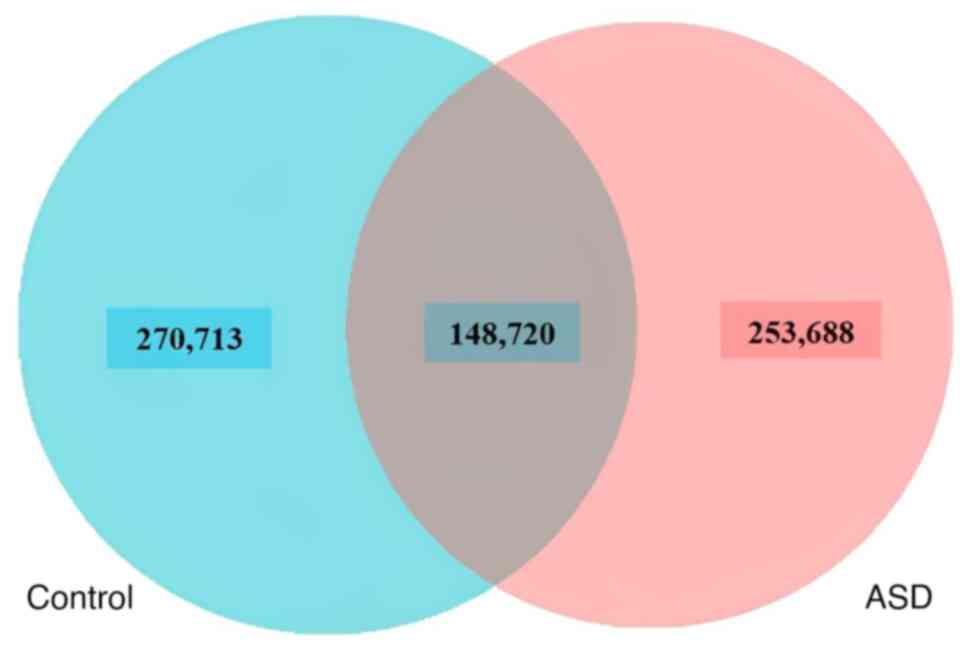
it can be seen that there were 253,688 unique genes found in
children with autism, compared with 270,713 unique genes in healthy
control subjects. The overlap area, or core genes, between the
samples of ASD and healthy control subjects was 148,720, indicating
that there were also shared genes between the two groups of
children (Fig. 1). To determine
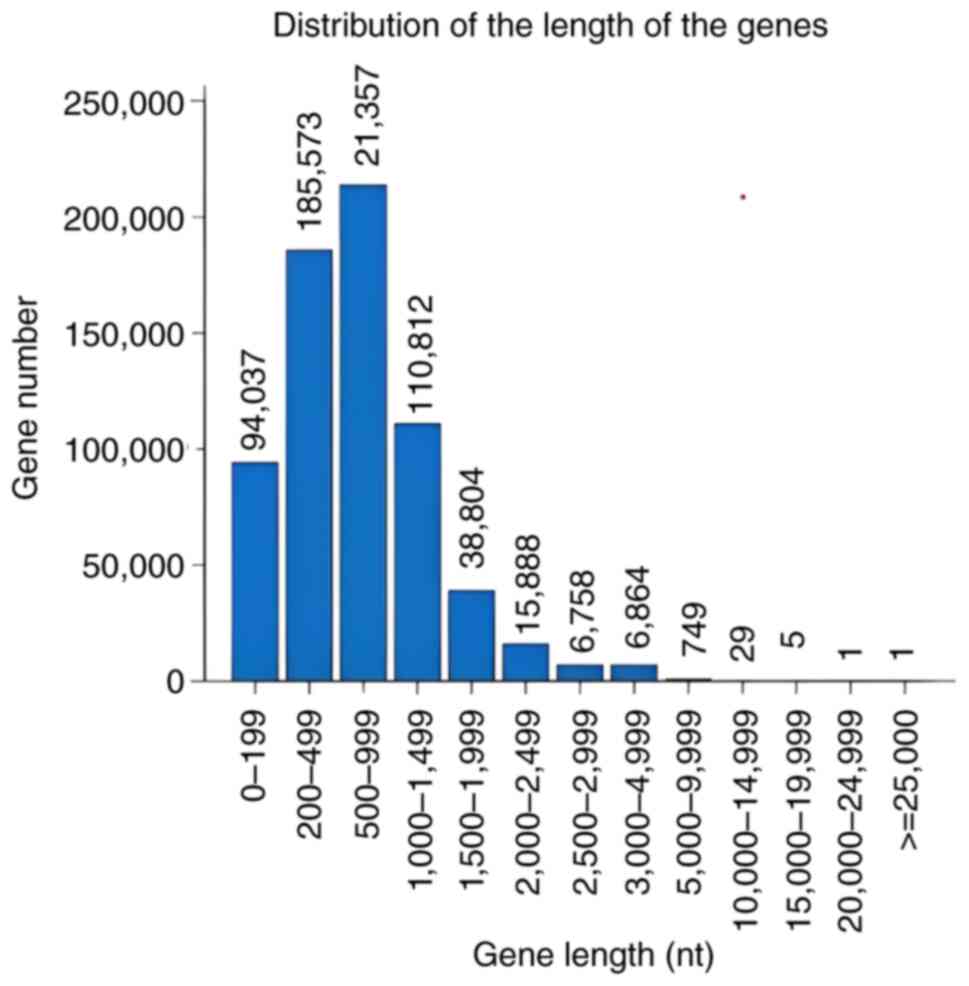
the functional regions of genes and the distribution of gene length
for gene prediction, the assembled genomic sequences based on gene
structure information was used. The longer gene query was 213,570
base pairs, while the shorter gene query was 1 (Fig. 2).
 | Table IIIFiltering statistics for the DNA
samples collected from fecal samples of four children with autism
(A) and four healthy control siblings (C). |
Table III
Filtering statistics for the DNA
samples collected from fecal samples of four children with autism
(A) and four healthy control siblings (C).
| Sample ID | Raw data size,
bp | Clean data, bp
(Filtering) | Raw data, % | Clean read (remove
host) | Raw data (%) |
|---|
| A1 | 6,310,231,200 | 6,182,568,000 | 97.98 | 6,182,139,600 | 97.97 |
| A2 | 6,047,304,900 | 5,965,336,200 | 98.64 | 5,964,390,300 | 98.63 |
| A3 | 6,047,304,900 | 5,954,599,500 | 98.47 | 5,954,453,400 | 98.46 |
| A4 | 6,310,231,200 | 6,144,631,500 | 97.38 | 6,144,401,400 | 97.37 |
| C1 | 6,310,231,200 | 6,169,231,200 | 97.77 | 6,169,022,700 | 97.76 |
| C2 | 6,047,304,900 | 5,961,072,900 | 98.57 | 5,960,247,900 | 98.56 |
| C3 | 6,047,304,900 | 5,955,091,200 | 98.48 | 5,954,439,900 | 98.46 |
| C4 | 6,047,304,900 | 5,963,012,400 | 98.61 | 5,931,360,300 | 98.08 |
 | Table IVAssembly statistics for the DNA
samples collected from fecal samples of four children with autism
(A) and four healthy control siblings (C) by using megahit
software. |
Table IV
Assembly statistics for the DNA
samples collected from fecal samples of four children with autism
(A) and four healthy control siblings (C) by using megahit
software.
| Sample ID | Contig Number | Assembly length,
bp | N50, bp | N90, bp | Max, bp | Min, bp | Average size,
bp | Mapping rate % |
|---|
| A1 | 19,193 | 84,328,730 | 29,340 | 1,367 | 757,805 | 300 | 4,393 | 89.2 |
| A2 | 68,535 | 147,395,260 | 5,125 | 797 | 365,378 | 300 | 2,150 | 66.34 |
| A3 | 41,520 | 123,293,288 | 8,915 | 1,120 | 690,186 | 300 | 2,969 | 75.1 |
| A4 | 48,415 | 148,672,006 | 12,445 | 1,041 | 445,303 | 300 | 3,070 | 79.87 |
| C1 | 67,622 | 180,343,949 | 6,774 | 989 | 438,212 | 300 | 2,666 | 73.41 |
| C2 | 14,596 | 71,410,148 | 34,067 | 1,566 | 618,837 | 300 | 4,892 | 83.87 |
| C3 | 49,572 | 158,802,107 | 12,427 | 1,144 | 398,112 | 300 | 3,203 | 77.5 |
| C4 | 57,057 | 160,194,825 | 6,948 | 1,066 | 420,902 | 300 | 2,807 | 73.81 |
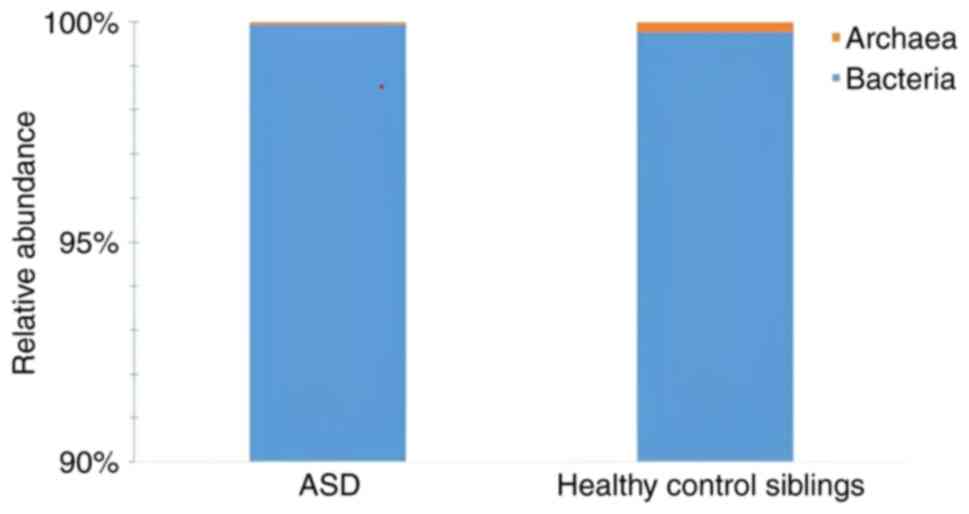
The findings showed that all samples had the highest
evenness (high community biodiversity) and richness at the kingdom
level. Autistic samples with high evenness displayed higher levels
of richness (Fig. 3). Compared
with their siblings, the microbiomes of children with autism had
undergone more changes. In addition, when compared with the ASD
samples, the healthy control sibling samples had a higher level of
Archaea and a lower level of Bacteria at the kingdom level
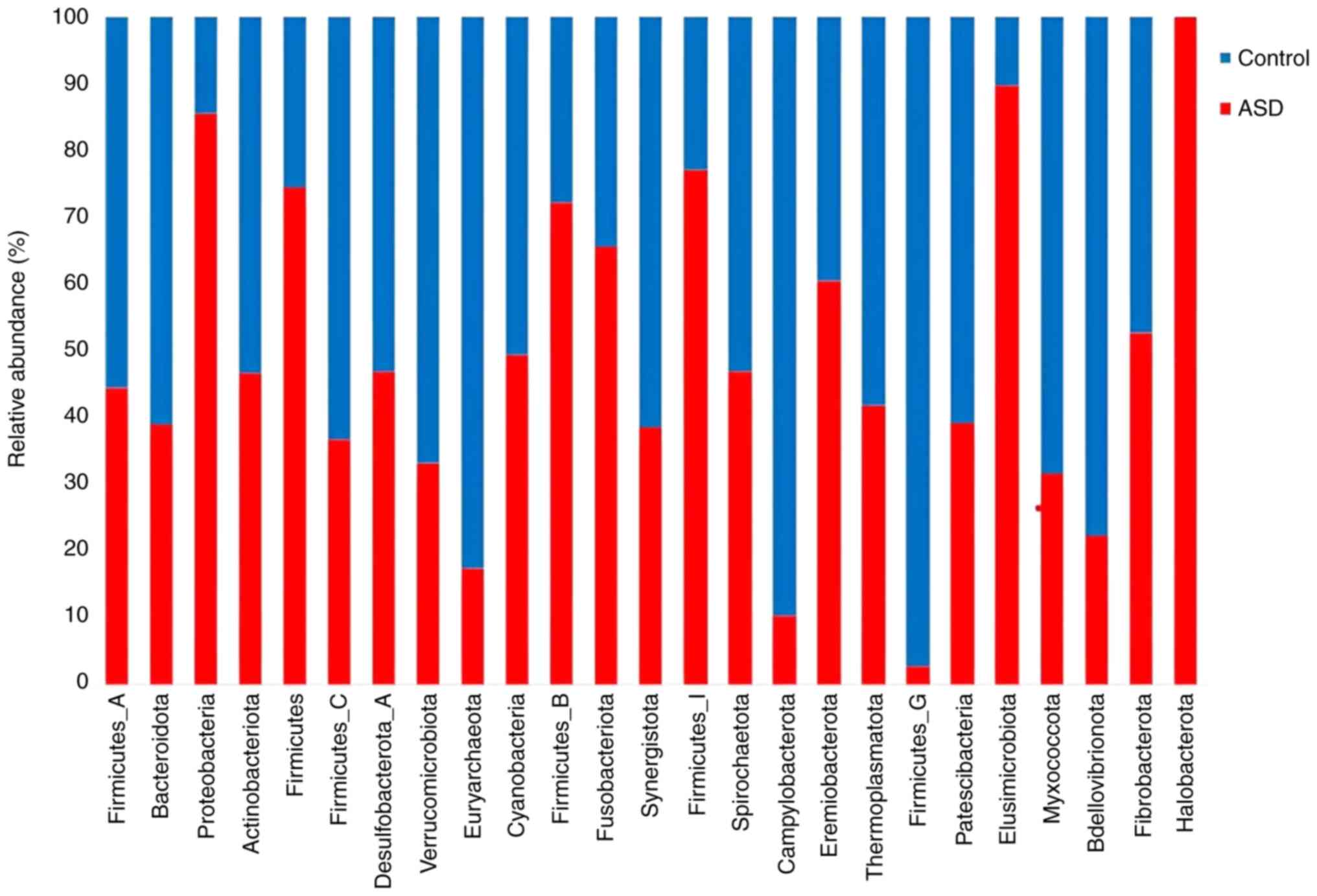
(Fig. 3). At the phylum level, all
samples had a propensity for high richness and low evenness between
the two groups. The species richness was higher in all samples from
healthy control siblings. The phyla proteobacteria and
Firmicutes were the most abundant in the ASD samples.
Several phyla, including Bdellovibrionota,
Verrucomicrobiota, and Bacteroidota, showed a higher
abundance in the healthy control sibling samples (Fig. 4).
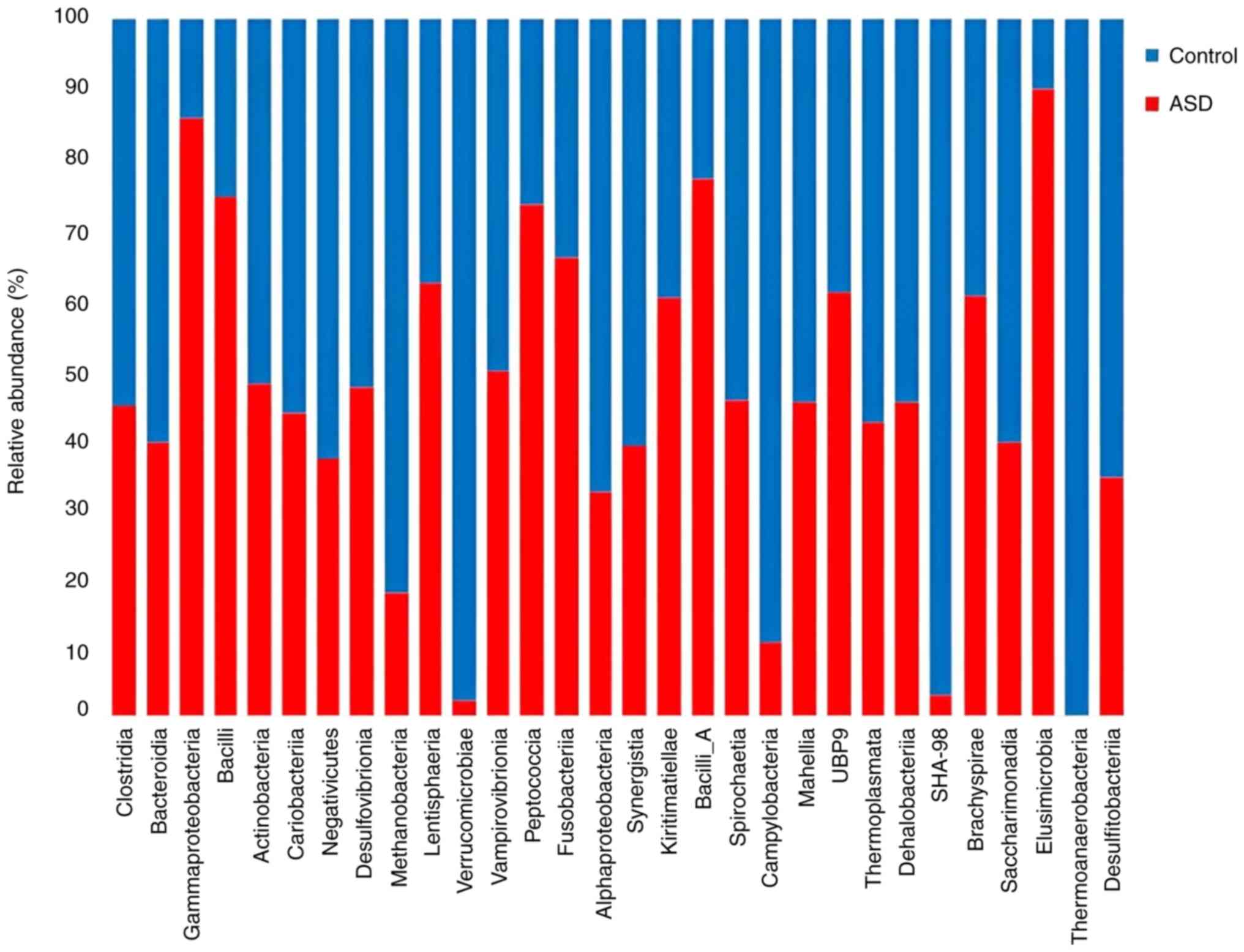
Additionally, there were differences in the
diversity of the healthy control and ASD samples at the class
level. When compared with healthy controls, ASD samples had higher
richness and lower evenness, which shows that children with autism
had more pronounced changes in their microbiome diversity.
Furthermore, only children in good health contained members of the
Thermoanaerobacteria class. Methanobacteria and
Verrucomicrobiae were more abundant in the healthy control
samples than they were in the ASD samples. ASD samples had a
noticeably higher concentration of Gammaproteobacteria,
Bacilli, and Elusimicrobia (Fig. 5). In comparison with the normal
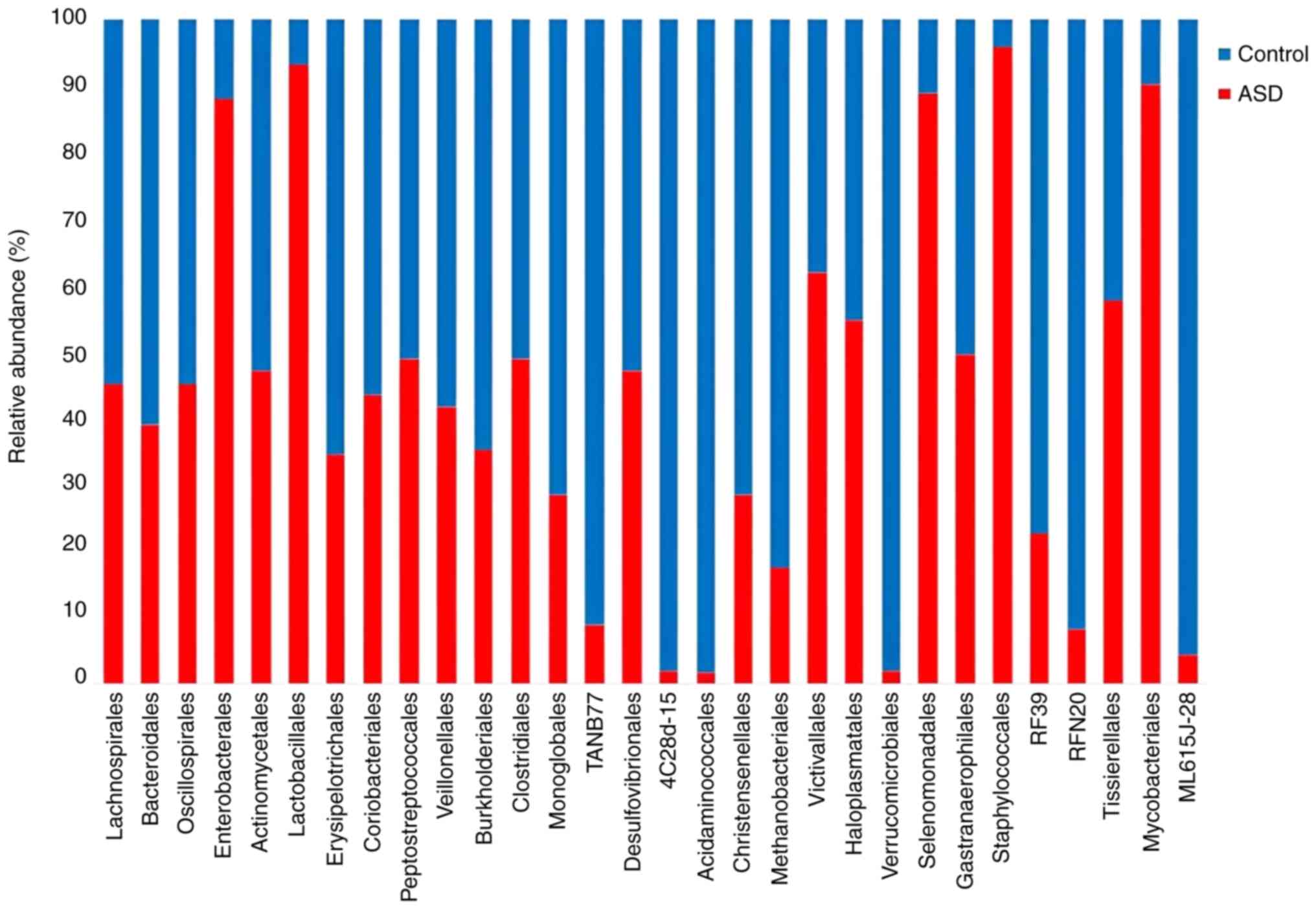
controls, all ASD samples displayed a higher abundance and evenness
at the order level. The Enterobacterales,
Staphylococcales, Halomicrobiota, and Mycobacteriales
were the orders with the highest abundance in the ASD samples.
Verrucomicrobiales, Acidaminococcales, and
4C28d-15 exhibited the highest abundance orders in the
healthy controls (Fig. 6).
Moreover, ASD children had a higher abundance of
Enterobacterales and Lactobacillales at order level.
However, Lachnospirales was found in fluctuated abundance in
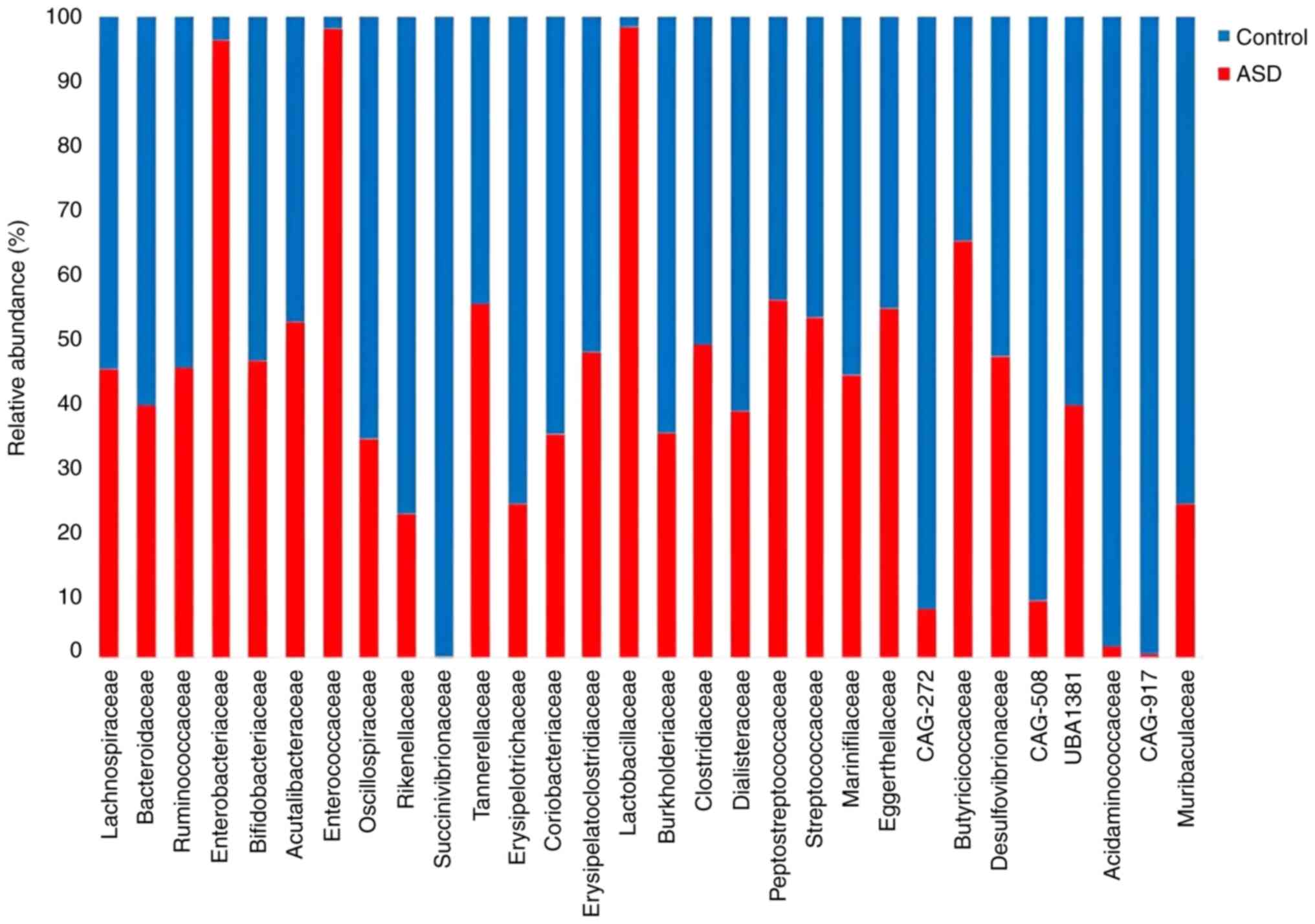
both ASD and controls. Results at the family level revealed that
children with ASD had a discernible change in diversity.
Furthermore, as shown in Fig. 7,
the Lactobacillaceae, Enterobacteriaceae, and
Peptostreptococcaceae families were more abundant in the
children with ASD than they were in the healthy controls, whereas
the Succinivibrionaceae and Acidaminococcaceae
families were more abundant in the controls. These findings showed
that ASD samples and their healthy control siblings had higher
richness and lower evenness across all samples at both the genus
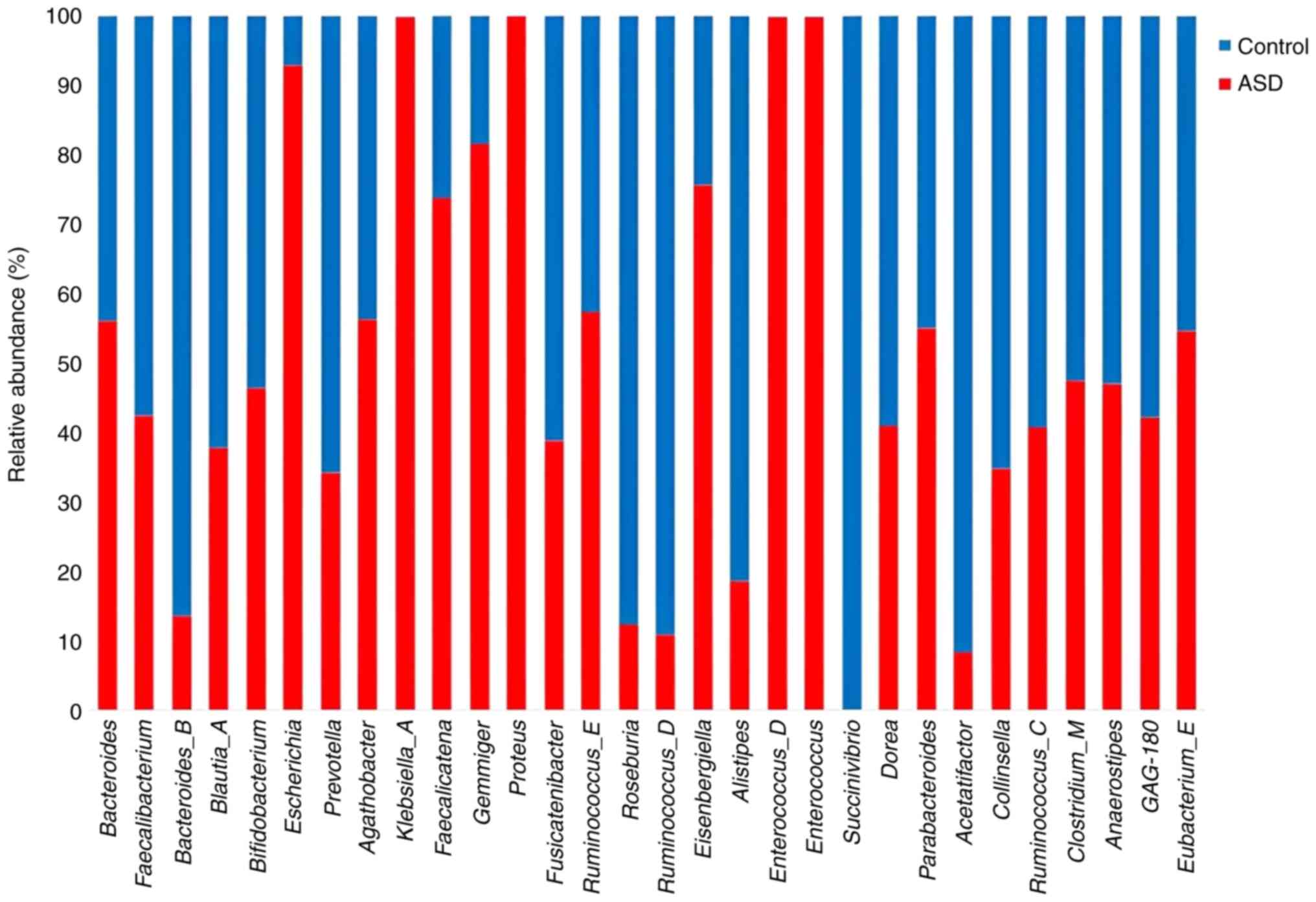
and species levels. Bacteroidetes_B, Bifidobacterium,
Prevotella, and Blautia were more abundant in the
healthy control samples compared with the ASD samples while
Parabacteroides and Proteus were more abundant in the
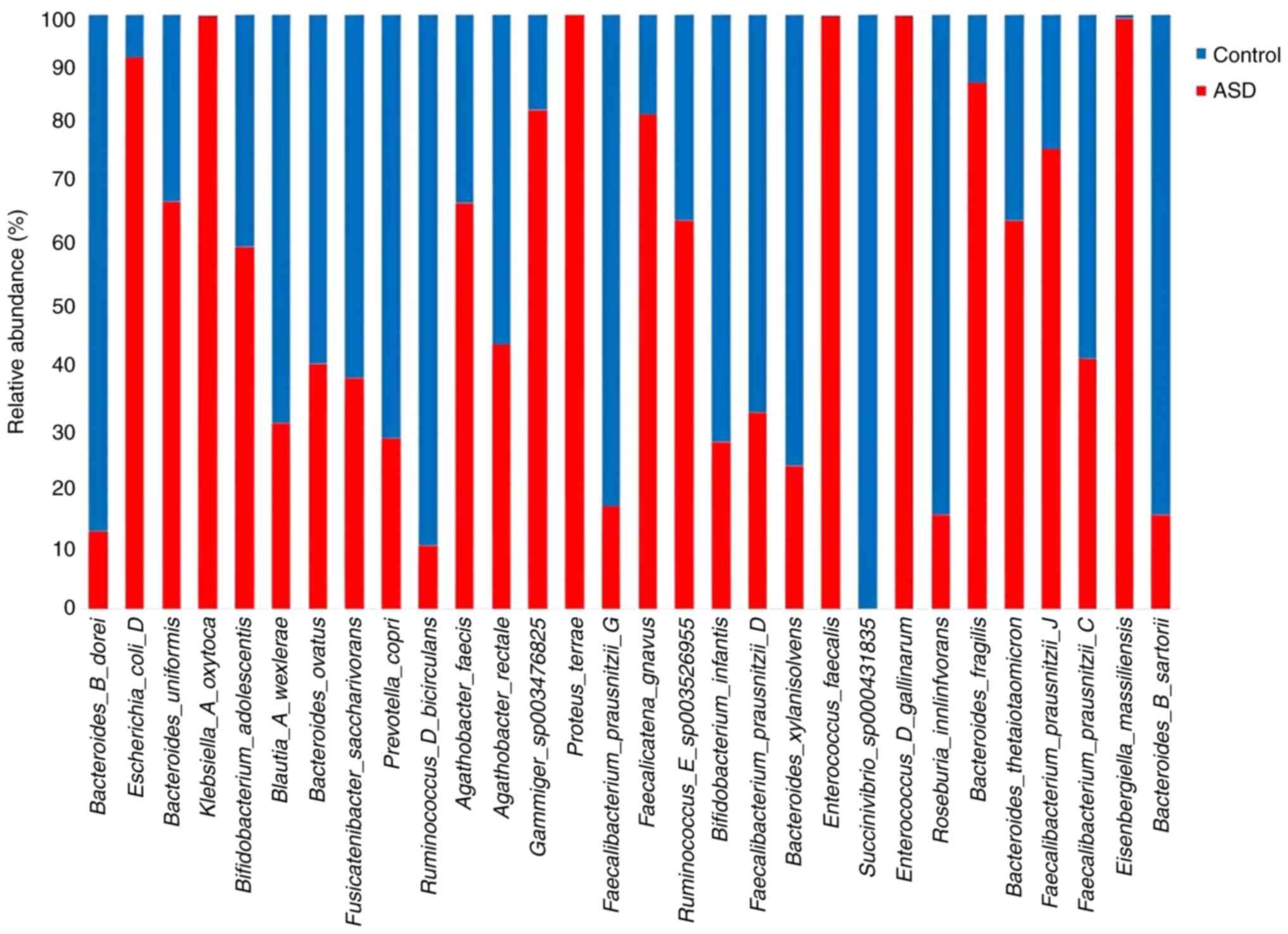
ASD samples (Fig. 8).
Bacteroides uniformis, Bacteroides fragilis,
Klebsiella A oxytoca, and Faecalibacterium prausnitziia
J were more abundant in the children with ASD at species level,
whereas Roseburia inulinivorans and Bifidobacterium
infantis were higher in the healthy control samples (Fig. 9).
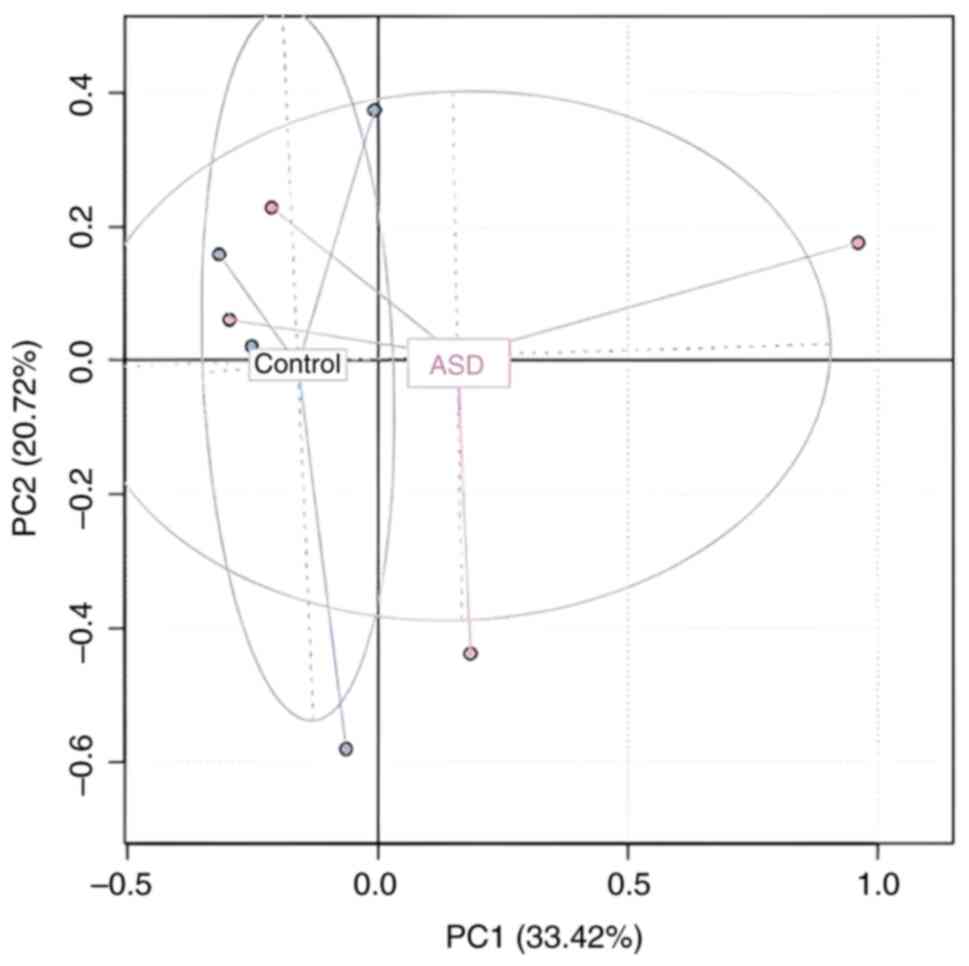
Principal coordinate analysis (PCoA) plot with
Bray-Curtis dissimilarity both depicted β diversity (36). The difference in species diversity
between two or more communities is referred to as β diversity. To
describe β-diversity patterns, PCA based on Bray Curtis distances
was used. This divides samples based on a single condition, whether
it is an ASD or a healthy condition. The results show that among
ASD, the biggest data changes were seen at the species level,
whereas relatively small changes were seen among samples of healthy
controls. It is notable that the PCA1 axis was able to completely
distinguish between healthy controls and ASD Although some
pathological samples were present with the control, the general
separation between samples from the children with ASD and healthy
controls across one condition is shown in Fig. 10, despite the overlap between the
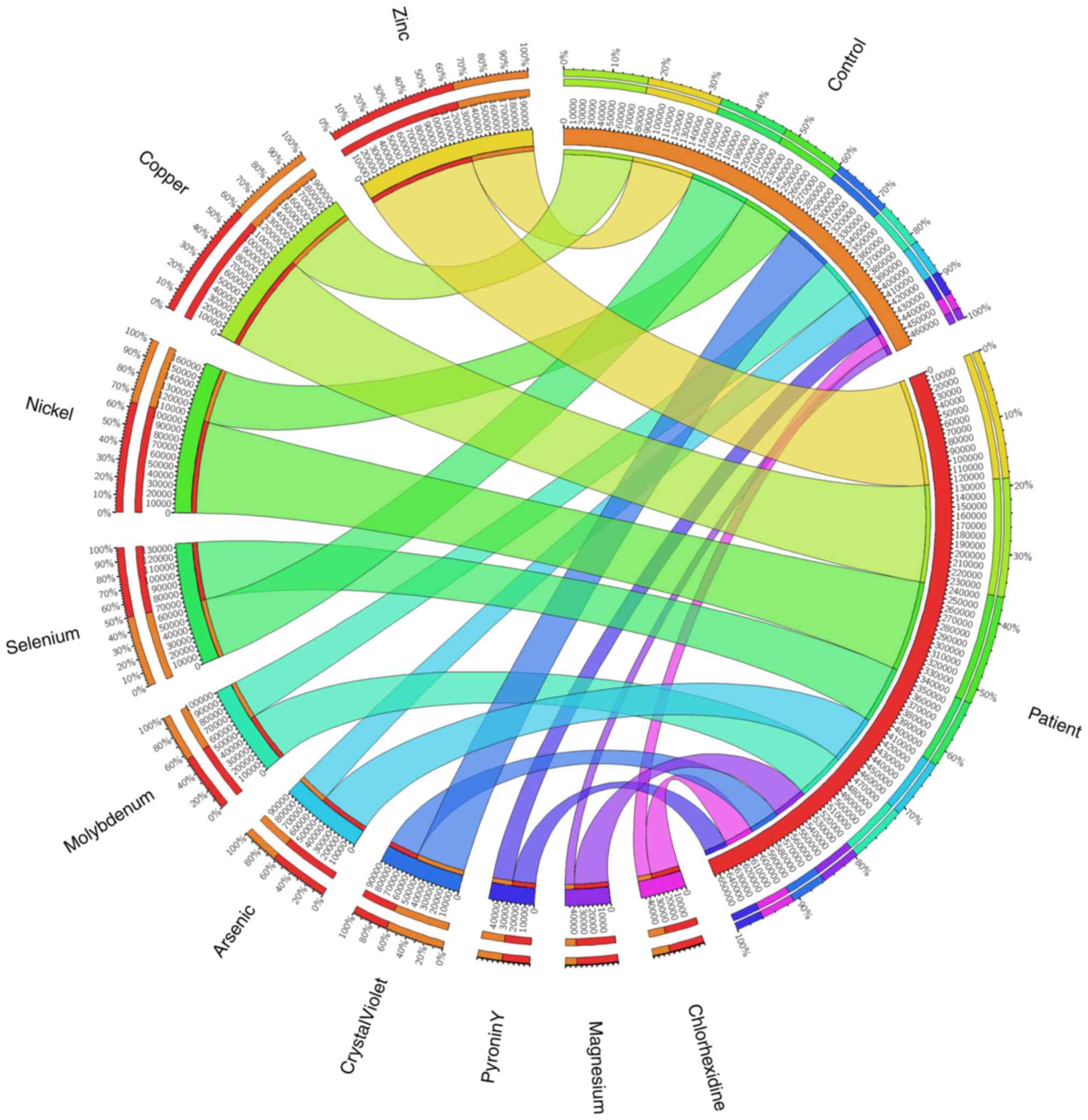
ASD and healthy control samples being separated. The findings also
revealed the presence of a few minerals and dyes, which are listed
in Fig. 11 in order of decreasing
abundance. The findings indicated that compared with their healthy
control siblings, children with ASD exhibited higher levels of zinc
(Zn), copper (Cu), and nickel (Ni), suggesting that these elements
may have some association with autism.
The results of the association between mineral
concentrations and gene abundance that affects the way these
minerals work is shown in Fig.
11. Of note, 124,053.237 genes or 60% of the Zn genes were
found in the ASD samples, compared with 72,194.72 genes or 40% of
the Zn genes in the control samples. For Cu, the numbers were
~113,018.838 genes (55%) and 81,013.569 genes (50%), respectively,
in the control and ASD samples. For Ni, the numbers were
~105,305.895 genes in ASD samples (60%) compared with 61,909.547
genes in controls (45%). For selenium, the numbers were 66,569.415
genes and 67,635.147 genes, and for pyronin Y they were 26,602.361
and 23,183.965, in the ASD and control samples respectively,
suggesting approximately equal amounts (~50%). According to the
findings, the amount of molybdenum increased in the ASD group by
55%, arsenic by 60%, magnesium by 80% and chlorhexidine by 90%.
When compared with the control sample, the abundance of crystal
violet was 60% while in the ASD samples it was 30%.
Discussion
The present study aimed to classify and identify
molecular biomarkers for ASD by examining differences in the
composition of the gut microbiota between children with autism and
their healthy siblings. Children with ASD have a different GM
compared with healthy controls, according to the results of the
present study, which showed higher levels of bacteria in children
with autism compared with the healthy controls. The GMs of children
with ASD were more diverse, rich and biomass-rich than those of
typically developing children, which is consistent with earlier
research by Finegold et al (37) and De Angelis et al (38).
The findings at the phylum level showed that
children with ASD had a higher abundance of the phylum
Proteobacteria, which is associated with host inflammation
(39). Animal studies found that
Proteobacteria produce lipopolysaccharides (LPS), which
result in a decreased level of glutathione in the brain and this is
a primary cause of immune dysregulation in individuals with ASD
(40,41). Additionally, there was a higher
abundance of Firmicutes in the samples from the patients
with autism, which is consistent with a prior study by Tomova et
al (42).
Bdellovibrionota and Verrucomicrobia phyla were more
abundant in healthy control samples. Bdellovibrionota is an
obligate predator that can engulf and kill other gram-negative
bacteria (43). It may thus serve
as a protective agent against pathogens, resulting in a decrease in
the number of harmful bacteria in the gut of healthy control
siblings (44). Additionally, the
most abundant phyla in healthy control samples were
Verrucomicrobiota, which is consistent with a previous study
by Zou et al (45). Members
of the Verrucomicrobiota phylum are mucin-degrading bacteria
that contribute to glucose homeostasis and intestinal health
(46). Furthermore, the results of
the present study showed that the Bacteroidota phylum was
more abundant in healthy individuals than in patients with ASD,
which is consistent with the findings of Settanni et al
(47). Bacteroidota is
responsible for the digestion of polysaccharides, thus a reduction
in this phylum results in mucosal dysbiosis in the gut and abnormal
digestion of carbohydrates in children with autism (48,49).
Results at the class level showed that
Thermoanaerobacteria were only present in the healthy
control groups. Thermoanaerobacteria play a role in the
fermentation of both carbohydrates and polysaccharides by producing
L-lactic acid, H2, CO2, acetic acid and
ethanol (50-52).
Methanobacteria were more abundant in healthy control
siblings and they produce methane as a metabolic by-product
(53). A reduction in the number
of microorganisms producing methane is a major mechanism of
hydrogen disposal in the human colon, which can be associated with
excess abdominal gas in irritable bowel syndrome (54). Additionally, there were more
Verrucomicrobiae in healthy controls than in the ASD
samples. As aforementioned, the Verrucomicrobiae class is a
mucin-degrading bacteria residing in the intestinal mucosa, which
plays a role in glucose homeostasis and intestinal health and
serves as an interface between host tissues and the human GM
(55). Children with ASD had
higher levels of Gammaproteobacteria and Bacilli,
which is consistent with the study by Plaza-Díaz et al
(56). Gammaproteobacteria
contains most of the human pathogens; for example,
Salmonella and Escherichia coli, some of these genera
exist in symbiosis with hydrothermal vent-dwelling animals while
others are methane oxidizers (57). Bacilli are known to cause
diarrhea, nausea, vomiting and abdominal pain (58), which may be associated with GI
symptoms in children with ASD.
At the order level, Enterobacterales were
more abundant in ASD samples. This order includes several harmful
gram-negative bacteria that are responsible for numerous enteric
infections including several of the more familiar pathogens, such
as Salmonella and Escherichia coli (59). Furthermore, the preponderance of
Staphylococcales was higher in ASD samples than healthy
control samples. Most members of Staphylococcales can cause
several types of infection; for example, skin lesions, food
poisoning, endocarditis and urinary tract infections (60). Additionally, a higher abundance of
the Mycobacteriales order was recorded in children with ASD.
Mycobacteriales contain one of the most important human
pathogens, Mycobacterium tuberculosis which is the causative
agent of tuberculosis (TB) (61).
A previous study found that maternal infection with TB during
pregnancy was sufficient to affect the development of the brain in
the offspring and this contributed to impaired social interactions
and enhanced systemic inflammation (62).
Enterobacteriaceae were more abundant in ASD
samples at the family level which is consistent with previous
studies by Plaza-Díaz et al (56) and De Angelis et al (38). In addition, a study found that the
higher abundance of Enterobacteriaceae in inflammatory bowel
diseases (IBD) caused inflammation that could be triggered by GM
imbalances such as those observed in patients with ASD (63). Lactobacillaceae were found
in greater abundance in ASD samples than in the healthy controls,
which is consistent with a study by Pulikkan et al (64). A higher abundance of
Lactobacillaceae is associated with a decrease in gut
microbial-derived bacterial metabolites such as SCFAs;
consequently, lower levels of SCFAs in individuals with ASD lead to
imbalances in their behavior, immune system function and brain
function (65,66). Peptostreptococcaceae were
more abundant in ASD samples. A study found that
Peptostreptococcaceae levels were associated with social
deficit symptoms (67).
Peptostreptococcaceae may thus have an impact on both
behavior and brain function, which could be related to cognitive
symptoms and social deficits in children with autism (68). However, healthy control samples
demonstrated a higher abundance of Succinivibrionaceae.
Members of this family are important for carbohydrate metabolism,
as they ferment glucose to produce large quantities of succinic and
acetic acid (69). Acetic acid
plays a role in relieving constipation (70), thus constipation is more common in
children with autism than in healthy control siblings.
Acidaminococcaceae were more abundant in healthy control
samples, in agreement with a previous that illustrated a higher
abundance of Acidaminococcaceae in the gut of healthy
individuals (71).
The presence of the Parabacteroides genus was
markedly higher in the ASD group. Parabacteroides are
closely associated with obesity, metabolic syndrome, and IBD
(72,73). Bacteroidetes were highly
abundant in the healthy control samples. The majority of the
Bacteroidetes genus produces propionic acid and SCFAs as
metabolic byproducts (37). When
SCFAs and propionic acid were injected into the cerebral ventricles
of rats, MacFabe et al (74) noticed that the rats exhibited
chemical, pathological and biological changes that were typical of
ASD, such as hyperactivity, abnormal motor movements and repetitive
behaviors, as well as exhibiting seizures (74). The Proteus genus was found
to be more abundant in ASD samples. Several gastrointestinal
conditions have been linked to an abundance of bacteria from the
Proteus genus, including Crohn's disease, gastroenteritis,
and appendicitis (75), which
suggests that an abundance of bacteria from the Proteus
genus could be related to GI symptoms in children with ASD.
Bifidobacterium was found to be a less abundant genus in
children with ASD, consistent with previous studies by
Iglesias-Vázquez et al (76) and Finegold et al (37). Certain Bifidobacterium
species showed decreased levels in children with autism compared
with healthy individuals; bacteria from this species produce
γ-aminobutyric acid (77), which
is closely associated with glutamate metabolism and is a major
excitatory neurotransmitter in the brain (78). According to previous studies, there
is a correlation between lower levels of glutamate concentrations
and the behavioral, anxiety and social disorder characteristics of
ASD (79,80). Prevotella was found in a
lower abundance in ASD samples than in the healthy control samples,
which is consistent with a previous study by Kang et al
(81). Additionally,
Blautia was found to be higher in healthy control siblings.
Blautia is a butyric acid-producing bacterium that helps to
remove gas from the intestine (82). Roseburia inulinivorans and
Bifidobacterium infantis were found to be less abundant at
the species level in ASD samples. A previous study discovered a
higher abundance of Roseburia inulinivorans in the
intestines of healthy individuals, where it plays a significant
role in butyrate formation from a variety of dietary polysaccharide
substrates in the large intestines (83). Furthermore, B. infantis is a
gut bacterium that plays a role in reducing intestinal inflammation
in infants with severe acute malnutrition (84) and may thus be used as a supplement
to improve the gut health of children with autism. The abundance of
Klebsiella A oxytoca was higher in children with ASD. K.
oxytoca is an intestinal pathobiont and the causative agent of
antibiotic-associated hemorrhagic colitis (85). Under conditions of gut dysbiosis,
K. oxytoca exerts pathogenic potential, such as in
conditions observed in patients with ASD (86). Additionally, K. oxytoca
exhibits natural resistance to penicillin and contributes to the
transmission of antibiotic-resistance genes to other bacteria
(87,88); this linking may explain the
relationship between children with children with ASD and the
increased probability of exhibiting antibiotic resistance compared
with healthy control siblings. The presence of Bacteroides
fragilis was found to be higher in children with autism, B.
fragilis produces LPS, a major virulence factor that can serve
as a potent poison under specific circumstances (37).
Regarding mineral concentration in the two groups of
this study, Zn was more abundant in the ASD samples compared with
the healthy controls, consistent with a previous study by Hawari
et al (89) but in
disagreement with a study by Faber et al (90), the latter of which found that Zn
deficiency was the primary cause of mood and behavioral disorders
in humans. Children with ASD in the study by Faber et al
(90) were found to have Zn
deficiency; the amount of Zn in their nails, plasma, and hair was
measured, and it was found that the levels of Zn were lower in ASD
compared with healthy controls (90). In addition, a study found that
sleep duration was favorably correlated with Zn levels and
adversely correlated with Cu levels (91). Sleep impairment has an effect on
the cognitive performance of children (92). However, several studies have
highlighted the numerous physiological functions of Zn, including
cell growth, differentiation and development (93-96).
Zn affects cognitive development and supports healthy brain
function by regulating differentiation, neurogenesis, and neuronal
migration (94,95). Zn, which may have an impact on the
brain-gut axis, is essential for gut and gastrointestinal system
function during neural development (96). Additionally, an increase in Cu
concentration has been linked to an increase in ASD severity
(97). The findings of the present
study corroborated those of the aforementioned studies; there was a
high ratio of Cu in the ASD samples compared with the controls. The
results also showed that Ni was more abundant in the ASD samples.
Organ dysfunction that results in various behavioral and
physiological disorders is associated with Ni homeostasis,
imbalanced due to an overload or a deficiency (98). The brain, lungs, kidneys, and liver
are just a few of the organs that are negatively affected by high
Ni levels (99-102).
The present study found that certain compounds,
including molybdenum, arsenic, and magnesium, were present at
slightly higher levels in the ASD samples. Cognitive function is
inversely associated with molybdenum (101). Arsenic is also a neurotoxic metal
that impairs cognitive function and has negative effects on brain
development as well as behavioral performance (103). Magnesium regulation of
glutamate-activated channels in neuronal membranes during
neurodevelopment is closely associated with the pathogenesis of ASD
(104). In both autistic and
healthy children, Se and pyronin Y (a cationic dye) were found in
comparable amounts. Se controls redox homeostasis, neuroimmune
processes and signal transduction pathways in brain tissues.
Pyronin Y and Se are both necessary for maintaining healthy
physiological processes and the growth of the brain (105).
The present study has shown that most of the
symptoms that children with autism suffer from are linked in one
way or another to the function of the microbiome. Certainly, more
research is required for the development of appropriate treatments
to assist individuals with ASD and improve the quality of their
lives.
An important area of research in the last decade
has been the function of the human GM in health and disease. To
classify and identify the significant pathogenic genera and species
in children with autism, as potential biomarkers for the detection
of autism, the present study compared the composition of the GM
between children with autism and their healthy siblings. The
results showed that individuals with ASD had a higher GM biomass,
diversity, and richness when compared with controls. These
differences included the presence of more pathogenic genera and
species, which may affect social interactions and behavioral
phenotypes associated with ASD. The relative percentage of the gene
abundance of each functional category in the samples revealed
significant differences between the two groups. The case and
control groups in the current study were only examined in four
samples; thus, the small sample size was a limitation to the
present study. Therefore, additional research using a larger cohort
of patients and controls is required to examine the interference of
the identified microbes with a variety of biological mechanisms to
confirm the findings and hypotheses presented in this study.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by Institutional Fund
Projects (grant no. IFPHI-083-248-2020). The authors gratefully
acknowledge technical and financial support from the Ministry of
Education and King Abdulaziz University, Jeddah, Saudi Arabia.
Availability of data and materials
The data generated in the present study may be
found in the European Nucleotide Archive under accession number
ERA29260632 or at the following URL: https://www.ebi.ac.uk/ena/browser/view/ERA29260632.
Authors' contributions
DA and AB conceived the present study. KA, SA, FB
and RA collected the patients' samples. DA and AAlo performed
experiments and wrote the original draft and FB and SA the second
draft. FB, KA, SA, AAlm, RA, AA, AAlh and AB reviewed and edited
the manuscript. DA and AAlo confirm the authenticity of all the raw
data. All authors have read and agreed to the final version of the
manuscript.
Ethics approval and consent to
participate
Research protocols conducted in the present study
were approved by the Biomedical Ethics Research Committee at King
Abdulaziz University (Jeddah, Saudi Arabia; approval no.
10-CEGMR-Bioeth-2021) and adhered to the guidelines of King
Abdulaziz University, Jeddah, Saudi Arabia, which were in
accordance with the declaration of Helsinki. Informed consent forms
were signed by the parents of all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bölte S, Girdler S and Marschik PB: The
contribution of environmental exposure to the etiology of autism
spectrum disorder. Cell Mol Life Sci. 76:1275–1297. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kral TVE, Eriksen WT, Souders MC and
Pinto-Martin JA: Eating behaviors, diet quality, and
gastrointestinal symptoms in children with autism spectrum
disorders: a brief review. J Pediatr Nurs. 28:548–556.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mukherjee SB: Autism spectrum
disorders-diagnosis and management. Indian J Pediatr. 84:307–314.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Heintz-Buschart A and Wilmes P: Human gut
microbiome: Function matters. Trends Microbiol. 26:563–574.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Schären OP and Hapfelmeier S: Robust
microbe immune recognition in the intestinal mucosa. Genes Immun.
22:268–275. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ellis JL, Karl JP, Oliverio AM, Fu X,
Soares JW, Wolfe BE, Hernandez CJ, Mason JB and Booth SL: Dietary
vitamin K is remodeled by gut microbiota and influences community
composition. Gut Microbes. 13:1–16. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bosco N and Noti M: The aging gut
microbiome and its impact on host immunity. Genes Immun.
22:289–303. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wu J, Wang K, Wang X, Pang Y and Jiang C:
The role of the gut microbiome and its metabolites in metabolic
diseases. Protein Cell. 12:360–373. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Davoli-Ferreira M, Thomson CA and McCoy
KD: Microbiota and microglia interactions in ASD. Front Immunol.
12(676255)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Han VX, Patel S, Jones HF and Dale RC:
Maternal immune activation and neuroinflammation in human
neurodevelopmental disorders. Nat Rev Neurol. 17:564–579.
2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kushak RI, Sengupta A and Winter HS:
Interactions between the intestinal microbiota and epigenome in
individuals with autism spectrum disorder. Dev Med Child Neurol.
64:296–304. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Morais LH, Schreiber HL and Mazmanian SK:
The gut microbiota-brain axis in behaviour and brain disorders. Nat
Rev Microbiol. 19:241–255. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zengeler KE and Lukens JR: Innate immunity
at the crossroads of healthy brain maturation and
neurodevelopmental disorders. Nat Rev Immunol. 21:454–468.
2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shreiner AB, Kao JY and Young VB: The gut
microbiome in health and in disease. Curr Opin Gastroenterol.
31:69–75. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mirzaei MK and Maurice CF: Ménage à trois
in the human gut: Interactions between host, bacteria and phages.
Nat Rev Microbiol. 15:397–408. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Saurman V, Margolis KG and Luna RA: Autism
spectrum disorder as a brain-gut-microbiome axis disorder. Dig Dis
Sci. 65:818–828. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lefter R, Ciobica A, Timofte D, Stanciu C
and Trifan A: A descriptive review on the prevalence of
gastrointestinal disturbances and their multiple associations in
autism spectrum disorder. Medicina (Kaunas). 56(11)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Penzol MJ, Salazar de Pablo G, Llorente C,
Moreno C, Hernández P, Dorado ML and Parellada M: Functional
gastrointestinal disease in autism spectrum disorder: A
retrospective descriptive study in a clinical sample. Front
Psychiatry. 10(179)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kang V, Wagner GC and Ming X:
Gastrointestinal dysfunction in children with autism spectrum
disorders. Autism Res. 7:501–506. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bresciani G, Da Lozzo P, Lega S, Bramuzzo
M, Di Leo G, Dissegna A, Colonna V, Barbi E, Carrozzi M and
Devescovi R: Gastrointestinal disorders and food selectivity:
Relationship with sleep and challenging behavior in children with
autism spectrum disorder. Children (Basel). 10(253)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lasheras I, Real-López M and Santabárbara
J: Prevalence of gastrointestinal symptoms in autism spectrum
disorder: A meta-analysis. An Pediatr (Engl Ed). 99:102–110.
2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li Q, Han Y, Dy ABC and Hagerman RJ: The
gut microbiota and autism spectrum disorders. Front Cell Neurosci.
11(120)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Breitwieser FP, Lu J and Salzberg SL: A
review of methods and databases for metagenomic classification and
assembly. Brief Bioinform. 20:1125–1136. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Mehra A, Arora G, Kaur M, Singh H, Singh B
and Kaur S: Gut microbiota and autism spectrum disorder: From
pathogenesis to potential therapeutic perspectives. J Tradit
Complement Med. 13:135–149. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mead J and Ashwood P: Evidence supporting
an altered immune response in ASD. Immunol Lett. 163:49–55.
2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen YC, Lin HY, Chien Y, Tung YH, Ni YH
and Gau SSF: Altered gut microbiota correlates with behavioral
problems but not gastrointestinal symptoms in individuals with
autism. Brain Behav Immun. 106:161–178. 2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao Y, Wang Y, Meng F, Chen X, Chang T,
Huang H, He F and Zheng Y: Altered gut microbiota as potential
biomarker biomarkers for autism spectrum disorder in early
childhood. Neuroscience. 523:118–131. 2023.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Korteniemi J, Karlsson L and Aatsinki A:
Systematic review: Autism spectrum disorder and the gut microbiota.
Acta Psychiatr Scand. 148:242–254. 2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhong JG, Lan WT, Feng YQ, Li YH, Shen YY,
Gong JH, Zou Z and Hou X: Associations between dysbiosis gut
microbiota and changes of neurotransmitters and short-chain fatty
acids in valproic acid model rats. Front Physiol.
14(1077821)2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kennedy MS and Chang EB: The microbiome:
Composition and locations. Prog Mol Biol Transl Sci. 176:1–42.
2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Feng P, Zhao S, Zhang Y and Li E: A review
of probiotics in the treatment of autism spectrum disorders:
Perspectives from the gut-brain axis. Front Microbiol.
14(1123462)2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Davies C, Mishra D, Eshraghi RS, Mittal J,
Sinha R, Bulut E, Mittal R and Eshraghi AA: Altering the gut
microbiome to potentially modulate behavioral manifestations in
autism spectrum disorders: A systematic review. Neurosci Biobehav
Rev. 128:549–557. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Naveed M, Zhou QG, Xu C, Taleb A, Meng F,
Ahmed B, Zhang Y, Fukunaga K and Han F: Gut-brain axis: A matter of
concern in neuropsychiatric disorders…! Prog Neuropsychopharmacol
Biol. Psychiatry. 104(110051)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Maiuolo J, Gliozzi M, Musolino V, Carresi
C, Scarano F, Nucera S, Scicchitano M, Oppedisano F, Bosco F, Ruga
S, et al: The contribution of gut microbiota-brain axis in the
development of brain disorders. Front Neurosci.
15(616883)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Muller PA, Matheis F, Schneeberger M,
Kerner Z, Jové V and Mucida D: Microbiota-modulated
CART+ enteric neurons autonomously regulate blood
glucose. Science. 370:314–321. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shi Y, Zhang L, Do KA, Peterson CB and
Jenq RR: aPCoA: Covariate adjusted principal coordinates analysis.
Bioinformatics. 36:4099–4101. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Finegold SM, Dowd SE, Gontcharova V, Liu
C, Henley KE, Wolcott RD, Youn E, Summanen PH, Granpeesheh D, Dixon
D, et al: Pyrosequencing study of fecal microflora of autistic and
control children. Anaerobe. 16:444–453. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
De Angelis M, Piccolo M, Vannini L,
Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni
ME, Gobbetti M and Francavilla R: Fecal microbiota and metabolome
of children with autism and pervasive developmental disorder not
otherwise specified. PLoS One. 8(e76993)2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shin NR, Whon TW and Bae JW:
Proteobacteria: Microbial signature of dysbiosis in gut
microbiota. Trends Biotechnol. 33:496–503. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhu Y, Carvey PM and Ling Z: Altered
glutathione homeostasis in animals prenatally exposed to
lipopolysaccharide. Neurochem Int. 50:671–680. 2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chauhan A and Chauhan V: Oxidative stress
in autism. Pathophysiology. 13:171–181. 2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Tomova A, Husarova V, Lakatosova S, Bakos
J, Vlkova B, Babinska K and Ostatnikova D: Gastrointestinal
microbiota in children with autism in Slovakia. Physiol Behav.
138:179–187. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li QM, Zhou YL, Wei ZF and Wang Y:
Phylogenomic insights into distribution and adaptation of
Bdellovibrionota in marine waters. Microorganisms.
9(757)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Varon M: Selection of predation-resistant
bacteria in continuous culture. Nature. 277:386–388. 1979.
|
|
45
|
Zou R, Xu F, Wang Y, Duan M, Guo M, Zhang
Q, Zhao H and Zheng H: Changes in the gut microbiota of children
with autism spectrum disorder. Autism Res. 13:1614–1625.
2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Espín JC, González-Sarrías A and
Tomás-Barberán FA: The gut microbiota: A key factor in the
therapeutic effects of (poly)phenols. Biochem Pharmacol. 139:82–93.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Settanni CR, Bibbò S, Ianiro G, Rinninella
E, Cintoni M, Mele MC, Cammarota G and Gasbarrini A:
Gastrointestinal involvement of autism spectrum disorder: Focus on
gut microbiota. Expert Rev Gastroenterol Hepatol. 15:599–622.
2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gyawali S and Patra BN: Trends in concept
and nosology of autism spectrum disorder: A review. Asian J
Psychiatr. 40:92–99. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Pulikkan J, Mazumder A and Grace T: Role
of the gut microbiome in autism spectrum disorders. Adv Exp Med
Biol. 1118:253–269. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lowe SE, Jain MK and Zeikus JG: Biology,
ecology, and biotechnological applications of anaerobic bacteria
adapted to environmental stresses in temperature, pH, salinity, or
substrates. Microbiol Rev. 57:451–509. 1993.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lynd LR, Weimer PJ, Van Zyl WH and
Pretorius IS: Microbial cellulose utilization: Fundamentals and
biotechnology. Microbiol Mol Biol Rev. 66:506–577. 2002.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wiegel J, Mothershed CP and Puls J:
Differences in xylan degradation by various noncellulolytic
thermophilic anaerobes and Clostridium thermocellum. Appl Environ
Microbiol. 49:656–659. 1985.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Shukla SK, Khan A and Rao TS: Microbial
fouling in water treatment plants. In: Microbial and Natural
Macromolecules. Elsevier, pp589-622, 2021.
|
|
54
|
Pozuelo M, Panda S, Santiago A, Mendez S,
Accarino A, Santos J, Guarner F, Azpiroz F and Manichanh C:
Reduction of butyrate- and methane-producing microorganisms in
patients with irritable bowel syndrome. Sci Rep.
5(12693)2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Anderson JR, Carroll I, Azcarate-Peril MA,
Rochette AD, Heinberg LJ, Peat C, Steffen K, Manderino LM, Mitchell
J and Gunstad J: A preliminary examination of gut microbiota,
sleep, and cognitive flexibility in healthy older adults. Sleep
Med. 38:104–107. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Plaza-Díaz J, Gómez-Fernández A, Chueca N,
Torre-Aguilar MJ, Gil Á, Perez-Navero JL, Flores-Rojas K,
Martín-Borreguero P, Solis-Urra P, Ruiz-Ojeda FJ, et al: Autism
spectrum disorder (ASD) with and without mental regression is
associated with changes in the fecal microbiota. Nutrients.
11(337)2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chandarana KA, Gohil K, Dwivedi MK and
Amaresan N: Culture-independent and culture-dependent approaches in
symbiont analysis. In: Microbial Symbionts. Elsevier, pp723-742,
2023.
|
|
58
|
Turnbull PC, Kramer J and Melling J:
Bacillus: Chapter 15. Medical microbiology, pp1-7, 1996.
|
|
59
|
Bujňáková D, Puvača N and Ćirković I:
Virulence factors and antibiotic resistance of
Enterobacterales. Microorganisms. 10(1588)2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Bhakdi S and Tranum-Jensen J: Alpha-toxin
of Staphylococcus aureus. Microbiol Rev. 55:733–751.
1991.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Gupta RS, Lo B and Son J: Phylogenomics
and comparative genomic studies robustly support division of the
genus Mycobacterium into an emended genus Mycobacterium and four
novel genera. Front Microbiol. 9(67)2018.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Manjeese W, Mvubu NE, Steyn AJ and Mpofana
T: Mycobacterium tuberculosis-induced maternal immune
activation promotes autism-like phenotype in infected mice
offspring. Int J Environ Res Public Health. 18(4513)2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Baldelli V, Scaldaferri F, Putignani L and
Del Chierico F: The role of Enterobacteriaceae in gut
microbiota dysbiosis in inflammatory bowel diseases.
Microorganisms. 9(697)2021.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Pulikkan J, Maji A, Dhakan DB, Saxena R,
Mohan B, Anto MM, Agarwal N, Grace T and Sharma VK: Gut microbial
dysbiosis in Indian children with autism spectrum disorders. Microb
Ecol. 76:1102–1114. 2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Derrien M and van Hylckama Vlieg JE: Fate,
activity, and impact of ingested bacteria within the human gut
microbiota. Trends Microbiol. 23:354–366. 2015.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Martín R, Miquel S, Benevides L,
Bridonneau C, Robert V, Hudault S, Chain F, Berteau O, Azevedo V,
Chatel JM, et al: Functional characterization of novel
Faecalibacterium prausnitzii strains isolated from healthy
volunteers: A step forward in the use of F. prausnitzii as a
next-generation probiotic. Front Microbiol. 8(1226)2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Milani C, Ticinesi A, Gerritsen J,
Nouvenne A, Lugli GA, Mancabelli L, Turroni F, Duranti S,
Mangifesta M, Viappiani A, et al: Gut microbiota composition and
Clostridium difficile infection in hospitalized elderly
individuals: A metagenomic study. Sci Rep. 6(25945)2016.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Labus JS, Hollister EB, Jacobs J, Kirbach
K, Oezguen N, Gupta A, Acosta J, Luna RA, Aagaard K, Versalovic J,
et al: Differences in gut microbial composition correlate with
regional brain volumes in irritable bowel syndrome. Microbiome.
5(49)2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Bryant MP and Small N: Characteristics of
two new genera of anaerobic curved rods isolated from the rumen of
cattle. J Bacteriol. 72:22–26. 1956.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Wang L, Cen S, Wang G, Lee YK, Zhao J,
Zhang H and Chen W: Acetic acid and butyric acid released in large
intestine play different roles in the alleviation of constipation.
J Funct Foods. 69(103953)2020.
|
|
71
|
Ma B, Liang J, Dai M, Wang J, Luo J, Zhang
Z and Jing J: Altered gut microbiota in Chinese children with
autism spectrum disorders. Front Cell Infect Microbiol.
9(40)2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Cui Y, Zhang L, Wang X, Yi Y, Shan Y, Liu
B, Zhou Y and Lü X: Roles of intestinal Parabacteroides in
human health and diseases. FEMS Microbiol Lett.
369(fnac072)2022.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Ho LKH, Tong VJW, Syn N, Nagarajan N, Tham
EH, Tay SK, Shorey S, Tambyah PA and Law ECN: Gut microbiota
changes in children with autism spectrum disorder: A systematic
review. Gut Pathog. 12(6)2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
MacFabe DF, Cain DP, Rodriguez-Capote K,
Franklin AE, Hoffman JE, Boon F, Taylor AR, Kavaliers M and
Ossenkopp KP: Neurobiological effects of intraventricular propionic
acid in rats: Possible role of short chain fatty acids on the
pathogenesis and characteristics of autism spectrum disorders.
Behav Brain Res. 176:149–169. 2007.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Hamilton AL, Kamm MA, Ng SC and Morrison
M: Proteus spp. as putative gastrointestinal pathogens. Clin
Microbiol Rev. 31:e00085–17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Iglesias-Vázquez L, Van Ginkel Riba G,
Arija V and Canals J: Composition of gut microbiota in children
with autism spectrum disorder: A systematic review and
meta-analysis. Nutrients. 12(792)2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Srikantha P and Mohajeri MH: The possible
role of the microbiota-gut-brain-axis in autism spectrum disorder.
Int J Mol Sci. 20(2115)2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Shen J: Modeling the glutamate-glutamine
neurotransmitter cycle. Front Neuroenergetics. 5(1)2013.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Horder J, Petrinovic MM, Mendez MA, Bruns
A, Takumi T, Spooren W, Barker GJ, Künnecke B and Murphy DG:
Glutamate and GABA in autism spectrum disorder-a translational
magnetic resonance spectroscopy study in man and rodent models.
Transl Psychiatry. 8(106)2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Wierońska JM, Stachowicz K, Nowak G and
Pilc A: The loss of glutamate-GABA harmony in anxiety disorders.
Anxiety disord. 24:135–156. 2011.
|
|
81
|
Kang DW, Park JG, Ilhan ZE, Wallstrom G,
Labaer J, Adams JB and Krajmalnik-Brown R: Reduced incidence of
Prevotella and other fermenters in intestinal microflora of
autistic children. PLoS One. 8(e68322)2013.PubMed/NCBI View Article : Google Scholar
|
|
82
|
McNabney SM and Henagan TM: Short chain
fatty acids in the colon and peripheral tissues: A focus on
butyrate, colon cancer, obesity and insulin resistance. Nutrients.
9(1348)2017.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Scott KP, Martin JC, Campbell G, Mayer CD
and Flint HJ: Whole-genome transcription profiling reveals genes
up-regulated by growth on fucose in the human gut bacterium
‘Roseburia inulinivorans’. J Bacteriol. 188:4340–4349.
2006.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Barratt MJ, Nuzhat S, Ahsan K, Frese SA,
Arzamasov AA, Sarker SA, Islam MM, Palit P, Islam MR, Hibberd MC,
et al: Bifidobacterium infantis treatment promotes weight
gain in Bangladeshi infants with severe acute malnutrition. Sci
Transl Med. 14(eabk1107)2022.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Högenauer C, Langner C, Beubler E, Lippe
IT, Schicho R, Gorkiewicz G, Krause R, Gerstgrasser N, Krejs GJ and
Hinterleitner TA: Klebsiella oxytoca as a causative organism
of antibiotic-associated hemorrhagic colitis. N Engl J Med.
355:2418–2426. 2006.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Herzog KA, Schneditz G, Leitner E, Feierl
G, Hoffmann KM, Zollner-Schwetz I, Krause R, Gorkiewicz G, Zechner
EL and Högenauer C: Genotypes of Klebsiella oxytoca isolates
from patients with nosocomial pneumonia are distinct from those of
isolates from patients with antibiotic-associated hemorrhagic
colitis. J Clin Microbiol. 52:1607–1616. 2014.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Sievert DM, Ricks P, Edwards JR, Schneider
A, Patel J, Srinivasan A, Kallen A, Limbago B and Fridkin S:
National Healthcare Safety Network (NHSN) Team and Participating
NHSN Facilities. Antimicrobial-resistant pathogens associated with
healthcare-associated infections: Summary of data reported to the
National healthcare safety network at the centers for disease
control and prevention, 2009-2010. Infect Control Hosp Epidemiol.
34:1–14. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
88
|
Molton JS, Tambyah PA, Ang BSP, Ling ML
and Fisher DA: The global spread of healthcare-associated
multidrug-resistant bacteria: A perspective from Asia. Clin Infect
Dis. 56:1310–1318. 2013.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Hawari I, Eskandar MB and Alzeer S: The
role of lead, manganese, and zinc in autism spectrum disorders
(ASDS) and attention-deficient hyperactivity disorder (ADHD): A
case-control study on Syrian children affected by the Syrian
crisis. Biol Trace Elem Res. 197:107–114. 2020.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Faber S, Zinn GM, Kern JC II and Kingston
HM: The plasma zinc/serum copper ratio as a biomarker in children
with autism spectrum disorders. Biomarkers. 14:171–180.
2009.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Ji X, Grandner MA and Liu J: The
relationship between micronutrient status and sleep patterns: A
systematic review. Public Health Nutr. 20:687–701. 2017.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Liu J, Zhou G, Wang Y, Ai Y, Pinto-Martin
J and Liu X: Sleep problems, fatigue, and cognitive performance in
Chinese kindergarten children. J Pediatr. 161:520–525.e2.
2012.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Kawamura T, Ogawa Y, Nakamura Y, Nakamizo
S, Ohta Y, Nakano H, Kabashima K, Katayama I, Koizumi S, Kodama T,
et al: Severe dermatitis with loss of epidermal Langerhans cells in
human and mouse zinc deficiency. J Clin Invest. 122:722–732.
2012.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Prasad KM, Watson AMM, Dickerson FB,
Yolken RH and Nimgaonkar VL: Exposure to herpes simplex virus type
1 and cognitive impairments in individuals with schizophrenia.
Schizophr Bull. 38:1137–1148. 2012.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Hagmeyer S, Haderspeck JC and Grabrucker
AM: Behavioral impairments in animal models for zinc deficiency.
Front Behav Neurosci. 8(443)2015.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Cezar LC, Kirsten TB, da Fonseca CCN, de
Lima APN, Bernardi MM and Felicio LF: Zinc as a therapy in a rat
model of autism prenatally induced by valproic acid. Prog
Neuropsychopharmacol Biol Psychiatry. 84:173–180. 2018.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Lakshmi Priya MD and Geetha A: Level of
trace elements (copper, zinc, magnesium and selenium) and toxic
elements (lead and mercury) in the hair and nail of children with
autism. Biol Trace Elem Res. 142:148–158. 2011.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Samal L and Mishra C: Significance of
nickel in livestock health and production. Int J Agro Vet Med Sci.
5:349–361. 2011.
|
|
99
|
Denkhaus E and Salnikow K: Nickel
essentiality, toxicity, and carcinogenicity. Crit Rev Oncol
Hematol. 42:35–56. 2002.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Costa-Pinto FA and Basso AS: Neural and
behavioral correlates of food allergy. Chem Immunol Allergy.
98:222–239. 2012.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Fiore M, Barone R, Copat C, Grasso A,
Cristaldi A, Rizzo R and Ferrante M: Metal and essential element
levels in hair and association with autism severity. J Trace Elem
Med Biol. 57(126409)2020.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Das KK, Das SN and Dhundasi SA: Nickel,
its adverse health effects & oxidative stress. Indian J Med
Res. 128:412–425. 2008.PubMed/NCBI
|
|
103
|
Wang M, Hossain F, Sulaiman R and Ren X:
Exposure to inorganic arsenic and lead and autism spectrum disorder
in children: A systematic review and meta-analysis. Chem Res
Toxicol. 32:1904–1919. 2019.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Saghazadeh A, Ahangari N, Hendi K, Saleh F
and Rezaei N: Status of essential elements in autism spectrum
disorder: Systematic review and meta-analysis. Rev Neurosci.
28:783–809. 2017.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Zhang C, Ge J, Lv M, Zhang Q, Talukder M
and Li JL: Selenium prevent cadmium-induced hepatotoxicity through
modulation of endoplasmic reticulum-resident selenoproteins and
attenuation of endoplasmic reticulum stress. Environ Pollut.
260(113873)2020.PubMed/NCBI View Article : Google Scholar
|