Introduction
Asthma is a complex chronic respiratory disease
characterized by inflammation of the airways. The pathogenesis of
asthma involves recruiting various inflammatory cells, including
eosinophils, airway epithelial cells, macrophages and neutrophils
(1). Suppression of airway
inflammation by inhaled glucocorticoids is currently the mainstay
of asthma treatment. Despite significant advances in asthma
treatment, patients with increased airway neutrophilic infiltration
tend to experience more severe symptoms and are resistant to
corticosteroid therapy (2). Asthma
can be divided into Type 2-high endotype and Type 2-low endotype
(containing neutrophilic and paucigranulocytic types) (3). Previous studies have reported that
neutrophilic asthma is characterized by airway inflammation caused
predominantly by neutrophils infiltration, increased secretion of
Th1/Th17 cytokines and heightened susceptibility to pulmonary
infections (4,5). However, since neutrophilic asthma is
rare in asthma, the development of effective diagnosis and
treatment is hampered by its unclear pathogenesis. Consequently,
further research should be conducted to investigate the pathogenic
mechanisms of neutrophilic asthma and to identify potential
therapeutic targets that could be effective in its treatment.
Ferroptosis, originally proposed in 2012, has
recently been identified as a new form of cell death (6-9).
In contrast to other forms of programmed cell death, ferroptosis
maintains the structural integrity of the cell nucleus without
accumulation of chromosome edges, condensation, blebbing in the
plasma membrane, or formation of apoptotic bodies (10). Excessive iron accumulation within
cells, lipid peroxidation and oxidative stress are the key
indicators of ferroptosis (11).
In recent years, an increasing amount of research has shown that
ferroptosis is closely associated with the pathogenesis of asthma.
Tang et al (12)
demonstrated that inhalation of house dust mites can markedly
reduce SLC7A11 and glutathione peroxidase 4 (GPX4) activity in lung
tissue of mouse, increase the level of reactive oxygen species
(ROS) and lipid peroxidation and then induce ferroptosis. Yang and
Shang (13) reported that
ferroptosis inhibitors can alleviate airway inflammation by
inhibiting iron release and lipid peroxidation in the lung tissue
of asthmatic mice. Bao et al (14) discovered that Lip-1 can attenuate
airway inflammation in neutrophilic asthma by regulating
ferroptosis regulator levels (including GPX4, PTGS2 and SLC7A11),
resulting in the relief of asthma symptom. Although more and more
studies link ferroptosis to the pathogenic mechanisms of asthma,
the role of ferroptosis in the pathogenesis of neutrophilic asthma
remains to be elucidated.
Hypoxia-inducible factor 1 (HIF-1) is a
transcriptional activator that regulates physiological responses
and oxygen homeostasis when exposed to hypoxic conditions. HIF-1α,
one of the HIF subunits, is an essential component of the HIF-1
signaling pathway (15,16). Fu and Zhang (17) indicated that the HIF-1α signaling
pathway may be associated with the progression of chronic
obstructive pulmonary disease. When HIF-1α accumulates in
low-oxygen environments, it moves towards the nucleus, where it
binds to hypoxia-responsive elements and stimulates transcription
of associated genes in response to cell ischemia and hypoxia. The
HIF-1 signaling pathway and related genes play an important role in
ferroptosis of other diseases (18). The role of HIF in ferroptosis might
exist difference in different diseases and whether HIF-1α was
involved in ferroptosis of neutrophilic asthma remains unclear.
The aim of the present study was to elucidate the
role of ferroptosis-related key genes and the HIF-1 signaling
pathway in the pathophysiology of neutrophilic asthma through
bioinformatics analysis and in vivo experiments. The
association between ferroptosis and neutrophilic asthma was
confirmed and the differentially expressed key genes and signaling
pathways associated with ferroptosis were identified. The
small-molecule drugs that may protect against neutrophilic asthma
by preventing ferroptosis were predicted. By screening the key
genes and signaling pathways associated with ferroptosis, the
present study deepens the understanding of ferroptosis involved in
the pathogenesis of neutrophilic asthma and provides a theoretical
basis for the diagnosis and treatment of neutrophilic asthma.
Materials and methods
Experimental mice
A total of 12, 4-6-week-old, female BALB/c mice
(weight, 18-20 g) were provided by Changsha Tianqin Biotechnology
Co., Ltd. The mice were maintained in a special pathogen-free (SPF)
laboratory environment and were free to drink water and eat
food at an ambient temperature of ~23˚C and a humidity of
40-50%. The present study was approved by the Ethics
Committee of Guangxi Medical University (approval no. 202210101),
the experimental process strictly adhered to the Guidelines for the
Care and Use of Laboratory Animals issued by the Ministry of
Science and Technology of the People's Republic of China (19).
Mouse model with neutrophil-dominated
airway inflammation construction
The experimental mice were randomized into two
groups: control (Con), neutrophilic asthma (Neu), each consisting
of six mice. The Neu group was sensitized by subcutaneous injection
of ovalbumin (20 µg; OVA), complete Freund's adjuvant (75
µl; CFA) and phosphate-buffered saline (25 µl; PBS) on the
first day. Subsequently, OVA was diluted into a 1% OVA aerosol
solution with sterile PBS dilution and mice were placed in a
confined space ~20x30x20 cm in size and aerosolized with 1% OVA
using an ultrasonic nebulizer in the closed chamber for 30 min each
day (days 22-24). The Con group was sensitized and challenged with
PBS. The aforementioned mouse model of neutrophil-dominated airway
inflammation was constructed according to previous literature
(20,21).
Airway responsiveness testing
Airway responsiveness was assessed 12 h after the
last challenge using a previously established method according to
our research group (22). After
acclimatization for 5 min the laboratory, each mouse was exposed to
methacholine (MCh; MilliporeSigma) at different
concentrations (PBS, 6.25, 12.5, and 25 mg/ml) for 3 min each.
Specific airway resistance (sRaw) was used to indicate airway
responsiveness.
Cells count and cytokine detection of
bronchoalveolar lavage fluid
According to previous literature (23), after 24 h of the final challenge,
mice were anesthetized by intraperitoneal injection of 50 mg/kg
0.3% sodium pentobarbital and, after several minutes of immobility
and no response to gentle stimulation, the mice were sacrificed by
cervical dislocation. Specifically, the characteristics with
cessation of breathing, dilated pupils and cardiac arrest in mice
were confirmed as mortality. The behaviors (such as shortness of
breath, restlessness and scratching of the mouth, nose and feet) in
mice were observed and recorded during the artificial period of the
neutrophil-dominated airway inflammation mouse model. The
observation period for the model mice was 25 days. Then, the
sacrificed mice were used to collect alveolar lavage fluid under
endotracheal intubation. After ligation of one side of the trachea,
the bilateral lungs were flushed three times with 0.5 ml of cold
PBS through the unligated trachea. Bronchoalveolar lavage fluid
(BALF) was collected by centrifugation at 400 x g for 10 min at
4˚C. After resuspension of isolated cells in PBS, cells were
counted with a hemocytometer and sorted with Wright-Giemsa staining
solution (Beijing Solarbio Science & Technology Co., Ltd.) at
room temperature for 10 min to calculate different cell
proportions. Enzyme-linked immunosorbent assay (ELISA) kits were
used to measure IL-17A (cat. no. SEA063Mu; Cloud-Clone Corp.) and
interferon-γ (IFN-γ) levels (cat. no. SEA049Mu; Cloud-Clone Corp.)
Measure-specific protocols in BALF were as follows.
Pathological staining and
immunofluorescence of lung tissue
After fixation with 4% paraformaldehyde for 12 h at
room temperature, the left lung tissue was subjected to paraffin
embedding with different concentrations of alcohol and xylene, and
then cut into 3-µm sections. Subsequently, the obtained samples
were stained with hematoxylin-eosin (HE) for 3 min and periodic
acid-Schiff (PAS) for 15 min at room temperature to assess
inflammatory cell infiltration and goblet cell proliferation and
release, respectively. The percentage of goblet cells staining
positive for PAS was scored using a semiquantitative scoring system
(0-4 points; none, few, one circle, 2-4 circles and deep ring of
inflammatory cell infiltration, respectively) (24). Goblet cell hyperplasia and mucus
production were identified by PAS staining. The proportion of
goblet cells positive for PAS staining was assessed using a
semi-quantitative scoring system. 0 points for no goblet cells
detected, 25-50% goblet cells detected, 2 points with 3 points
representing 50-75% of goblet cells detected. More than 75%
detected goblet cells scored 4 points (25). Immunohistochemical staining was
used to detect the expression of neutrophil marker and then further
assess neutrophil infiltration in lung tissue (26). The steps of the immunostaining
procedure were as follows: First, the paraffin sections were
dewaxed into water, then 20X Tris-EDTA antigen retrieval solution
(pH 9.0) was used for antigen retrieval solution for 20 min. After
blocking endogenous peroxidase activity for 25 min at room
temperature with 3% hydrogen peroxide, sections were blocked with
3% BSA (cat. no. GC305010; Wuhan Servicebio Technology Co., Ltd.)
for 30 min at room temperature. After shaking off the blocking
solution, the sections were covered with Ly6G (cat. no. GB11229;
1:500) and myeloperoxidase (MPO; cat. no. GB12224; 1:1,000) (both
Wuhan Servicebio Technology Co., Ltd.) prepared in PBS. The
sections were placed flat in a moistened box and incubated at 4˚C
overnight. The tissue was then covered separately with HRP-labeled
goat anti-rabbit IgG (cat. no. GB23303; 1:200) and goat anti-mouse
IgG (cat. no. GB23301; 1:200) (both Wuhan Servicebio Technology
Co., Ltd.) and then incubated for 50 min at room temperature.
Freshly prepared chromogenic DAB solution (cat. no. G1212; Wuhan
Servicebio Technology Co., Ltd.) was used for the chromogenic DAB
reaction. After hematoxylin counterstaining for 3 min at room
temperature, each section was dehydrated, mounted and observed
under a white light microscope.
RNA-Seq data analysis
For mRNA sequencing, lung tissues were collected
from three mice in each group. The starting material for RNA sample
preparation was 1 µg of lung tissue RNA. Illumina HiSeq 2500/4000
(Gene Denovo Biotechnology Co., Ltd.) was used for sequencing of
the RNA library. Transcript reconstruction was performed using
Stringtie v1.3.1 (27,28). Gene concentrations in each sample
were calculated using RSEM (27).
The distance relationship of the samples was examined using
principal component analysis (PCA). Differentially expressed genes
(DEGs) were identified using the R package DESeq2 (version 1.30.1)
(29) of R software (version
4.0.3, http://rproject.org/) (29) at screening thresholds of P<0.05
and |logFC|>1. The cluster profile package was applied in the
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) analysis (30,31) with P<0.05, which corresponds to
the significance threshold. To avoid biases arising from the
exclusive use of crossover gene enrichment, Gene Set Enrichment
Analysis (GSEA) was applied (30)
according to the gene expression profile to examine the differences
in biological processes between the different groups. The Omicsmart
platform (http://www.omicsmart.com) was used
for all bioinformatic analyses. The enrichment results were
visualized in the form of bubble diagrams.
Screening and biological function
analysis of differentially expressed (DE)-ferroptosis-related genes
(FRGs)
FRGs were obtained from the FerrDB database
(http://zhounan.org/ferrdb) and two
datasets (GSE143303 and GSE108417) were obtained from the Gene
Expression Omnibus database (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The GSE108417
dataset included lung tissue from three healthy mice and three
house dust mite-sensitized neutrophilic asthma mouse models. The
GSE143303 dataset, on the other hand, consisted of bronchial
biopsies from 13 healthy individuals, nine patients with
neutrophilic asthma and 38 patients with non-neutrophilic asthma
(22 eosinophilic and 16 paucigranulocytic asthma). DEGs analysis
was performed using the GEO2R online approach, which was considered
significant at P<0.05 and |logFC|>0. DE-FRGs were identified
using the R4.2.3 package Venn (version 1.11) (32). KEGG pathway analysis was performed
on the KOBAS 3.0 platform (http://kobas.cbi.pku.edu.cn/). The Pathview website
(https://pathview.uncc.edu/analysis)
was used to create a graphical representation of the pathway.
Screening and verification of key
DE-FRGs
To identify the common DE-FRGs in different
neutrophilic asthma mouse models, the R package Venn was used to
intersect FRGs with DEGs from RNA-seq analysis and dataset
GSE108417. The shared DE-FRGs were then used to construct
protein-protein interaction networks (PPIs) using the STRING
database (33) (version 12.0;
https://string-db.org/) with a pooled score
threshold of 0.4. The most important DE-FRGs were then identified.
The limma (version 3.54.2) (34)
and ggpubr (version 0.6.0) (35)
packages were used to evaluate these key DE-FRGs based on the
external validation set GSE143303. Furthermore, the receiver
operating characteristic (ROC) curve of the model was drawn and the
area under the curve (AUC) values were determined using the
validation set and R package pROC (version 1.18.0) (36).
Immune infiltration analysis of key
DE-FRGs
The GSEA package was used to determine immune cell
infiltration levels in lung tissue from mice with neutrophilic
asthma and bronchial biopsies from patients with neutrophilic
asthma. The proportion of immune cell infiltration was visualized
using the ggplot2 (version 3.4.2) (37) R-package accumulation histogram.
Furthermore, the relationship between DE-FRGs and infiltrating
immune cells was examined using Spearman's rank correlation.
Transcription factor and ceRNA network
prediction
The online database TRRUST (version2) (https://www.grnpedia.org/trust/) was used to
predict transcription factors for key DE-FRGs. Regulatory networks
between key genes and transcription factors were established using
Cytoscape (version 3.8.2) (38).
TargetScan (version 8.0, http://www.targetscan.org), miRDB (version 6.0)
(39) and the miRWalk (version 3)
(40) database were used for miRNA
prediction of these key DE-FRGs. Subsequently, the Spongescan
database (http://spongescan.rc.ufl.edu/) (41) was used to predict the long
non-coding (lnc)RNAs that interacted with miRNA and Cytoscape was
used to construct and visualize the competing endogenous (ce)RNA
network.
Target drug prediction and molecular
docking
A comprehensive database developed by DSigDB
(http://tanlab.ucdenver.edu/DSigDB)
can be used to identify targeted drugs associated with DEGs
(42). The present study used the
DSigDB website to predict potential drug target associated with key
DE-FRGs. The predicted results were imported into Cytoscape
software to construct and visualize the protein-drug interaction
network. Molecular docking simulation was performed using AutoDock
Tool 1.5.7(43) to clarify the
interaction between therapeutic target drugs and their
corresponding protein structures of target genes. Targeted
therapeutics were downloaded from the DrugBank database (version 5;
https://go.drugbank.com/) (44). The PDB profiles of the target
proteins were retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/) (45). Analysis and visualization of the
docking results were rendered by PyMOL Molecular Graphics System
(version 4.6) (46).
Transmission electron microscopy
Lung tissues from mice was preserved and kept in 3%
glutaraldehyde at 4˚C. After fixation in 1% osmium tetroxide for 2
h, the tissues were subjected to gradient ethanol dehydration and
epoxy resin embedding. Ultrathin sections (60-90 nm) were prepared
and stained with uranyl acetate and lead citrate. Finally, a
JEM-1400FLASH transmission electron microscope (JEOL, Ltd.) was
used to observe the morphology of mouse bronchial epithelial
cells.
Determination characteristics of
ferroptosis in lung tissue
To further demonstrate the occurrence of ferroptosis
in neutrophilic asthma, the expression of malondialdehyde (MDA) and
reduced glutathione (GSH) in lung tissues of mice was examined.
Lung tissue was processed into lung homogenate using a tissue
homogenizer according to the reagent instructions. The total
protein concentration was determined using the Enhanced BCA Protein
Assay Kit (cat. no. P0010S; Beyotime Institute of Biotechnology).
The MDA and GSH contents were detected according to the
instructions of the MDA kit (cat. no. A003-1; Nanjing Jiancheng
Bioengineering Institute) and the GSH and GSSG Assay Kit (cat. no.
S0053; Beyotime Institute of Biotechnology), respectively.
Reverse transcription-quantitative
(RT-q) PCR
TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total tissue RNAs. A reverse
transcription kit (Takara Bio, Inc.) was used for reverse
transcription. Subsequently, TB Green® Premix Ex
Taq™ II (Takara Bio, Inc.) was utilized for the
implementation of qPCR. Real-time PCR was performed on a 7500
Real-Time PCR System (Applied Biosystems). The following PCR
cycling parameters were adopted for amplification: 95˚C for 30 sec,
then 40 cycles of denaturation at 95˚C for 3 sec, and annealing and
extension at 60˚C for 30 sec. Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) expression was used to normalize the
amplified products. All the procedures were performed according to
the manufacturers' protocols. Additionally, the 2-ΔΔCq
method and real-time quantitative PCR were used to analyze relative
gene expression data in order to determine the relative expression
levels (fold-change) (47). These
experiments were replicated three times. Primer sequences were
designed and synthesized by China Gensys Biotechnology Co., Ltd.
and the Beijing TsingKe Biological Technology Company (Table SI).
Western blotting
Total proteins from lung tissue were extracted with
RIPA buffer (Beijing Solarbio Science & Technology Co., Ltd.).
The protein content was measured using a BCA kit (Beyotime
Institute of Biotechnology). Soluble protein (40 µg) was added to
each lane, and 10 and 12.5% gels were used for SDS-PAGE. Next
protein imprinting to a PVDF membrane was achieved by using
Invitrolon polyvinylidene fluoride filter paper. The membranes were
blocked with 5% defatted milk in TBST (1% Tween-20) solution for
1-1.5 h at room temperature. Subsequently, Primary antibodies
against HIF-1α (cat. no. 340462; 1:1,000; Zenbio), heme oxygenase 1
(HO-1; Abmart, TA5393; 1:1,000) and GPX4 (cat. no. T56959; 1:1,000;
Abmart) were then added and incubated overnight at 4˚C. After
washing three times with 1X TBST, the membranes were incubated with
a goat anti-rabbit IgG H&L secondary antibody (cat. no.
bs-0295G; 1:10,000; BIOSS) for 60-90 min at room temperature. After
washing with TBST three times in the dark, an Odyssey system
(LI-COR Biosciences) was used for protein band detection. The
proteins were analyzed and visualized using ImageJ software
(National Institutes of Health).
Statistical analysis
GraphPad Prism 9.0.0 software (Dotmatics) was
adopted for statistical analysis. The Shapiro-Wilk test was
performed to examine whether the measured data were normally
distributed. Results between groups were compared by independent
t-test or K-W test and expressed as mean ± standard deviation (SD).
One-way ANOVA was performed after the normality test and post hoc
LSD was performed to distinguish differences between groups. Data
conforming to abnormal distribution were compared by Mann-Whitney U
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Mice with neutrophil-dominated airway
inflammation induced by OVA and CFA
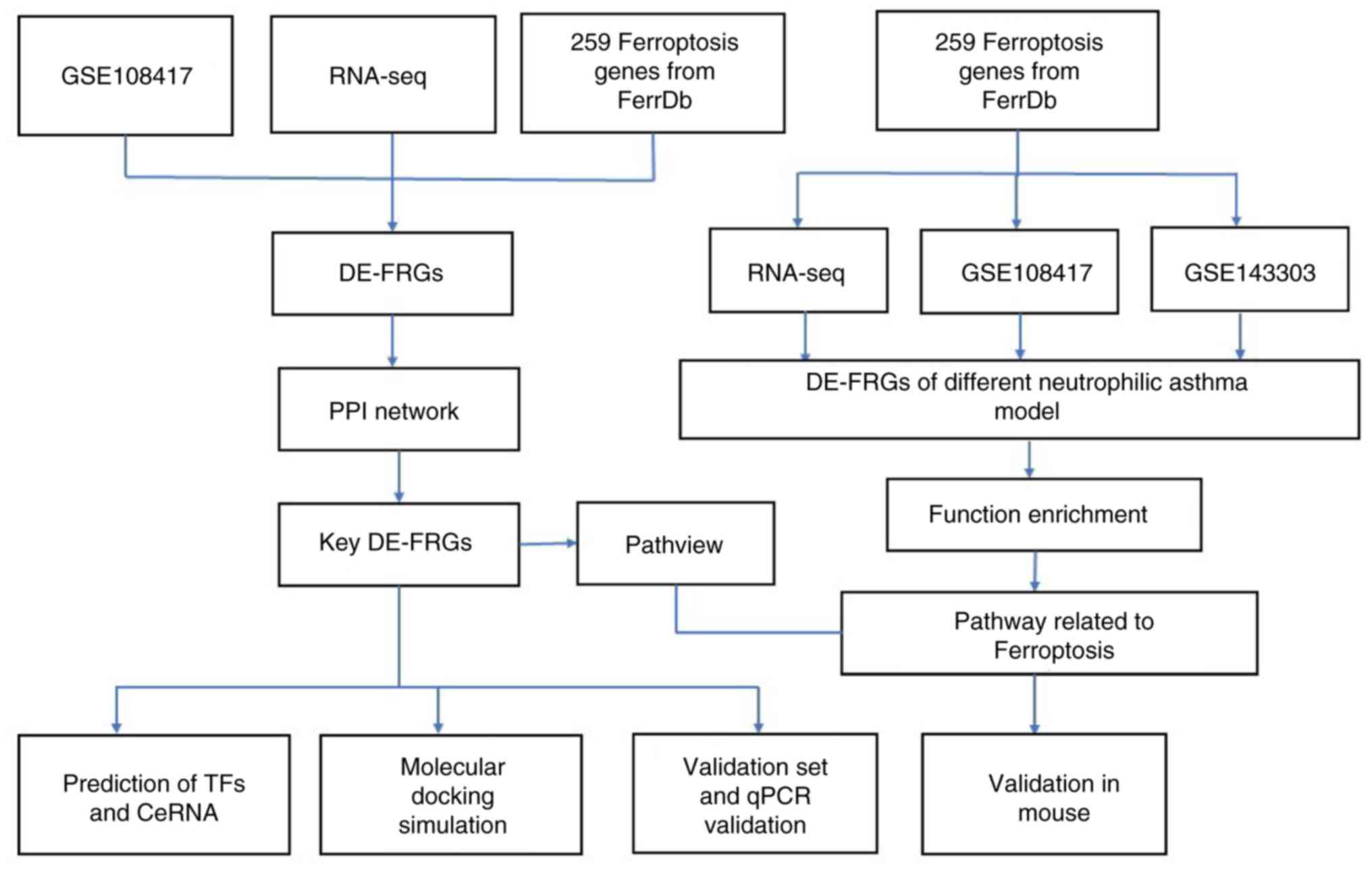
Fig. 1 displays the
schematic diagram of this study. Fig.
2A shows the established asthma mouse model with
neutrophil-dominated airway inflammation induced by OVA in
combination with CFA. It can be seen that the specific airway
resistance (sRaw) in mice increased with the increased dose of
nebulized methacholine in Pulmonary function tests. After
inhalation of Mch aerosol at different doses (6/12.5/25 mg/ml), the
Neu group exhibited significant airway hyperresponsiveness compared
with the Con group in Fig. 2B. In
addition, the total number of cells in BALF and the proportion of
neutrophils were markedly increased in the Neu group compared with
the Con group (Fig. 2C and
D). As shown in HE staining, there
was greater infiltration of inflammatory cells around the airways,
mucus secretion and goblet cell proliferation than the control
group (Fig. 2E and F). The PAS staining results (Fig. 2E and G) of the Neu group also showed greater
airway inflammation compared with the Con group. Furthermore,
significantly increased expression of neutrophil markers for Ly6G
and MPO can be found in the lung tissue of neutrophilic asthma mice
compared with the Con group (Fig.
2H-J). According to ELISA results, IL-17A and IFN-γ cytokines
in the BALF of neutrophilic asthmatic mice were significantly
higher compared with those in the control group (Fig. 2K and L). Previous studies (48,49)
reported that overexpression of IFN-γ and IL-17A, as a common
phenotype in severe asthma, is closely associated with airway
neutrophil infiltration and poor response to corticosteroids. The
results in Fig. 2 showed that an
asthma mouse model with the typical feature of neutrophilic
inflammation was successfully established.
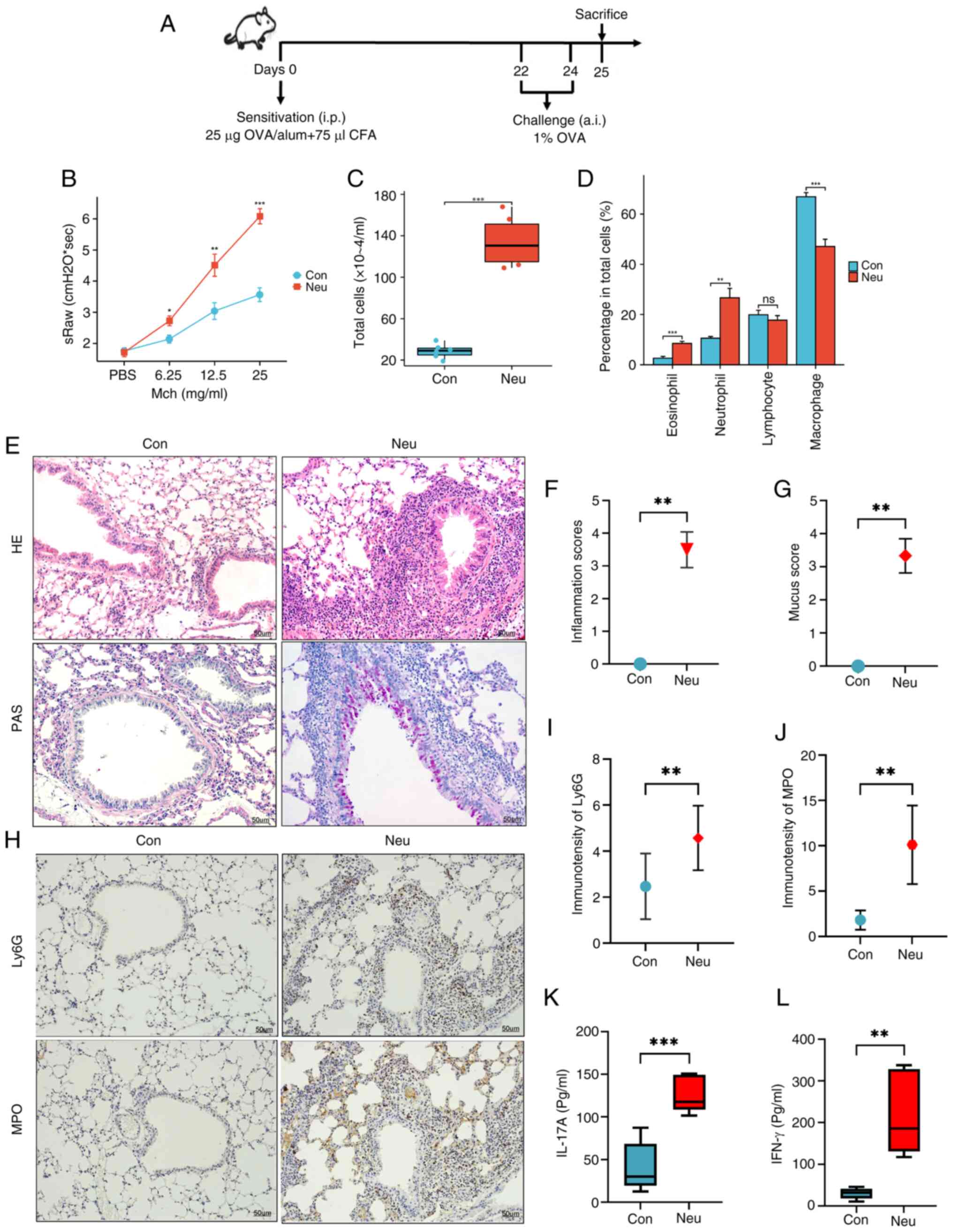 | Figure 2Construction of the neutrophilic
asthma mouse model. (A) Model construction protocol. (B) sRaw
stimulated by different doses of Mch (mg/ml); (C) Total number of
inflammatory cells in BALF. (D) Percentages of different types of
cells in BALF; (E) Lung tissues were stained with HE and PAS. (F)
HE inflammation score. (G) PAS-positive fraction. Magnification,
x200. (H) IHC was used to observe the expression of neutrophil
markers Ly6G and MPO in lung tissue of mice; Magnification, x200
(blue refers to the nucleus, brown is the marker protein). (I)
Expression of LY6G in lung tissue. (J) Expression of MPO in lung
tissue. (K and L) IFN-γ and IL-17A secretion within BALF (n=6).
Statistical significance vs. the control group is indicated
(*P<0.05, **P<0.01 and
***P<0.005). sRaw, specific airway resistance Mch,
methacholine; BALF, bronchoalveolar lavage fluid; HE,
hematoxylin-eosin; PAS, periodic acid-Schiff; IHC,
immunohistochemistry; Con, Control; Neu, neutrophilic asthma. |
Identification and functional
enrichment analysis of DEGs based on RNA-seq data
RNA-seq data were obtained and read length count
values (RPKM) were normalized. The difference between the control
and asthma groups can be effectively distinguished from Fig. 3A by PCA data with good quality. A
total of 1,639 DEGs were identified. The top 50 DEGs are depicted
in the heatmap (Fig. 3B), while
the volcano plot shows all DEGs (Fig.
3C). GO enrichment analysis in Fig. 3D showed that the most abundant DEGs
were clearly associated with immune-related biological processes,
such as immune system, immune response and inflammatory response.
KEGG pathway analysis revealed that the NF-kB, Th17, Th1 and Th2
cell differentiation and chemokine signaling pathways were the main
pathways involved (Fig. 3E). In
addition, GSEA showed that biological processes, such as IFN-γ
production, activation of an immune response and signaling pathways
(including NF-kB, NOD-like receptor and HIF-1) are also associated
with the pathogenesis of neutrophilic asthma (Fig. 3F and G).
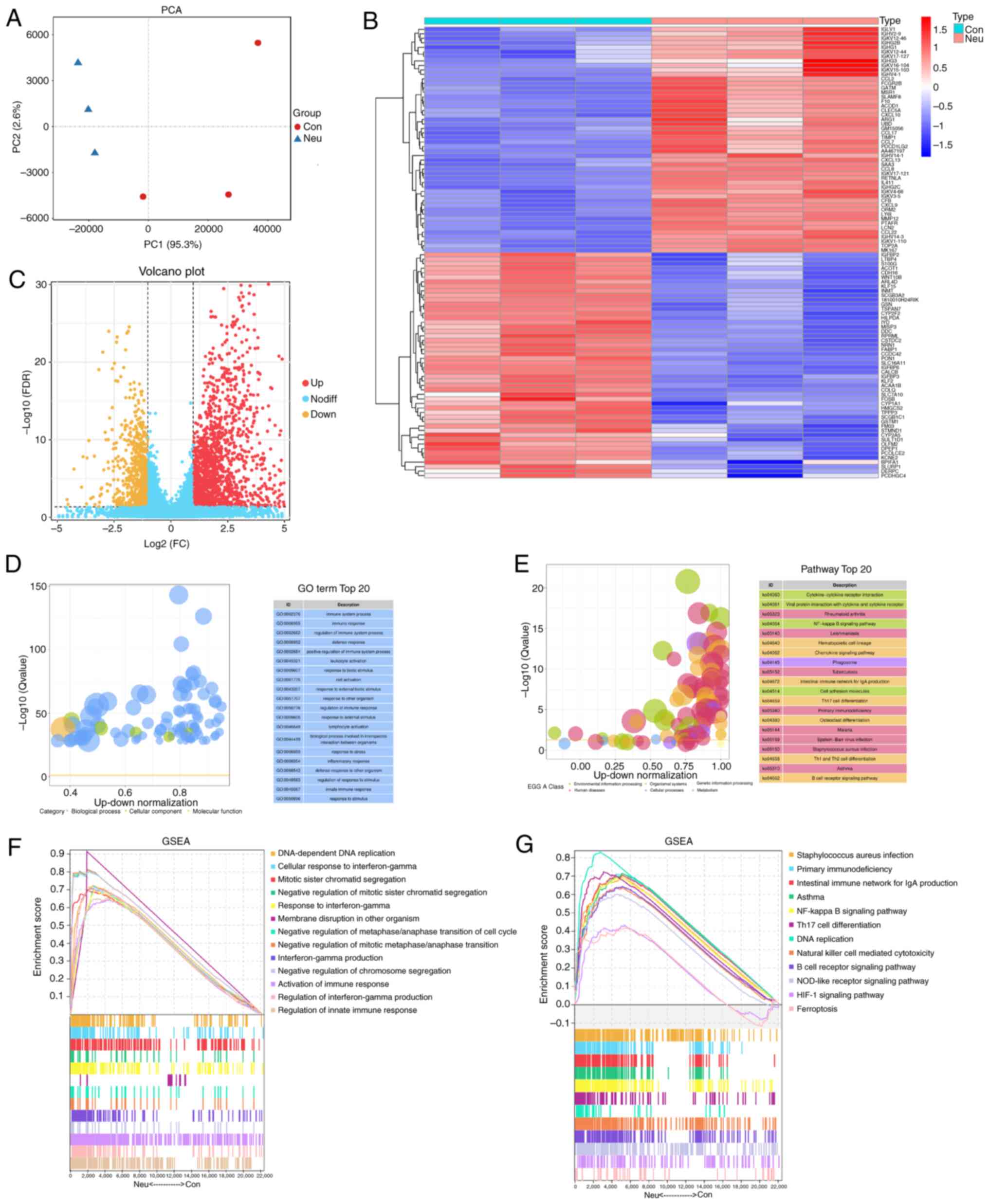 | Figure 3Identification and enrichment
analysis of DEGs. (A) PCA plot showing clustering of control
samples (yellow) and Neu samples (blue). (B) Heatmap of DEGs, where
red and blue stand for genes with up- and down-regulation,
separately. (C) Volcano plot of DEGs, where red and blue stand for
genes with up- and down-regulation, separately. (D) GO annotation.
(E) KEGG analysis. GSEA of (F) GO terms and (G) KEGG pathways.
DEGs, differentially expressed genes; PCA, principal component
analysis; Neu, neutrophilic asthma; GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment
Analysis; Con, Control. |
DE-FRGs and biological functions
analysis
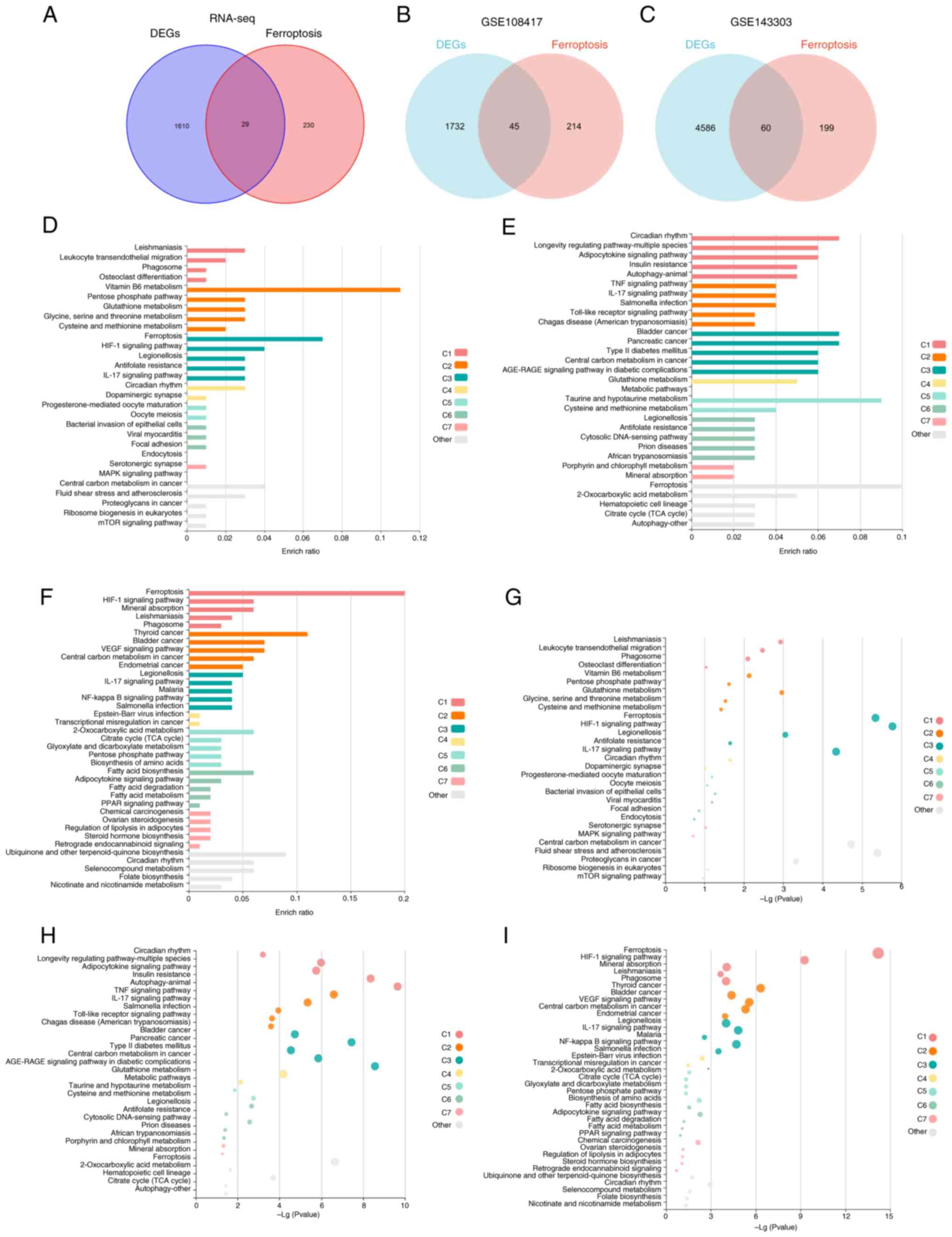
To obtain DE-FRGs in different neutrophilic asthma
models, 259 FRGs from the FerrDB database were used to intersect
with RNA-sequencing data, GSE108417 and GSE143303, separately
(Fig. 4A-C). Simultaneously, KEGG
enrichment of DE-FRGs was performed in three datasets using the
online KOBAS tool. The analysis revealed that ferroptosis, HIF-1,
NOD-like receptor and IL-17 signaling pathways were enriched in the
three neutrophilic asthma models (Fig.
4D-I; Table SII, Table SIII and Table SIV). It can be hypothesized that
these signaling pathways may be closely associated with the
progression of ferroptosis in neutrophilic asthma. According to the
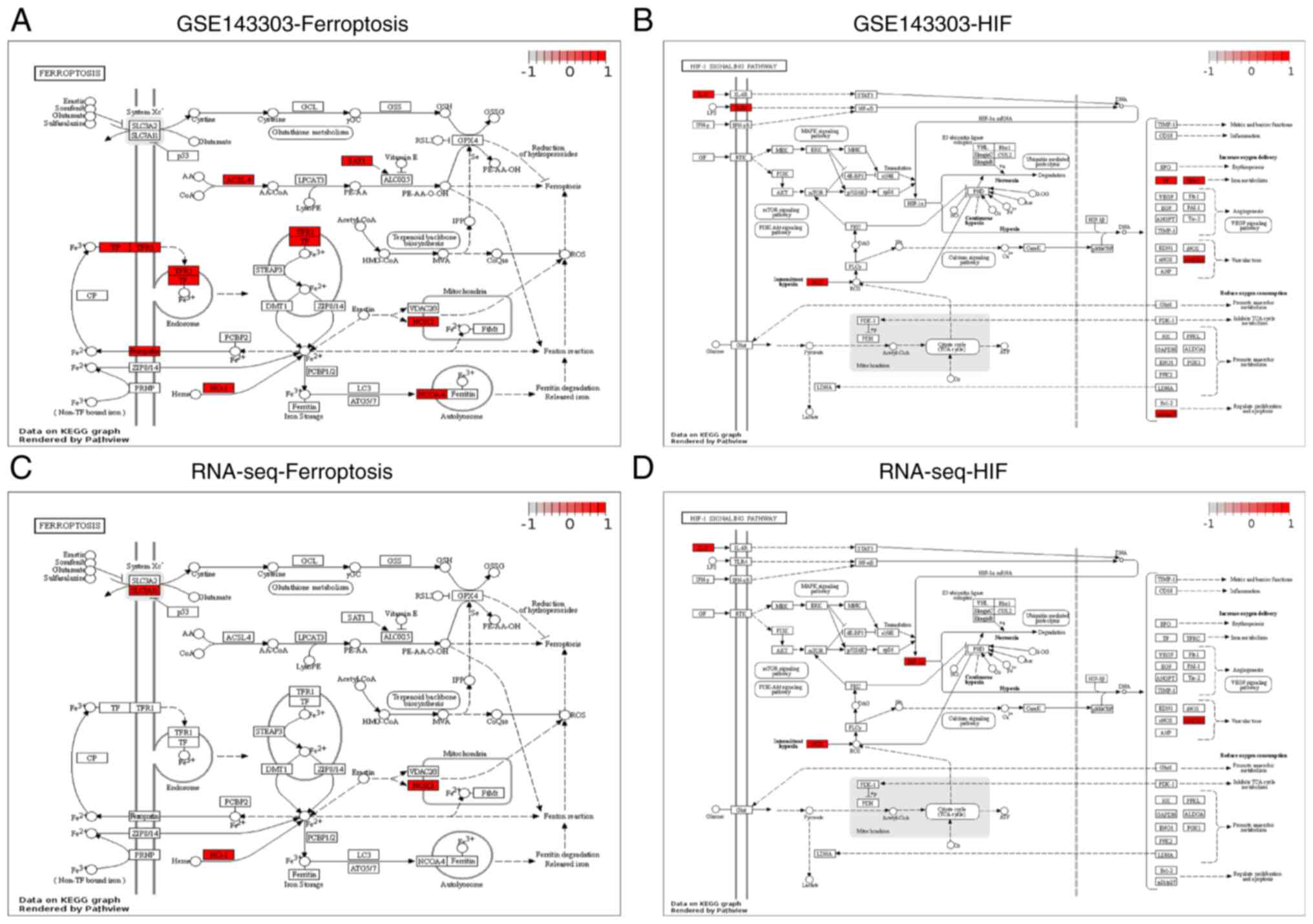
DE-FRGs identified in the GSE143303 dataset and the RNA-Seq data,
the ferroptosis pathway (Fig. 5A
and C) and the HIF-1 pathway
(Fig. 5B and D) were mediated Pathview mapped. Several
DE-FRGs have been found to be associated with ferroptosis.
Furthermore, it can be seen from both the GSE143303 dataset and the
RNA-seq dataset that HIF-1 exerts an important influence on
ferroptosis through the modulation of HMOX1.
Screening and verification of key
ferroptosis-related differential genes
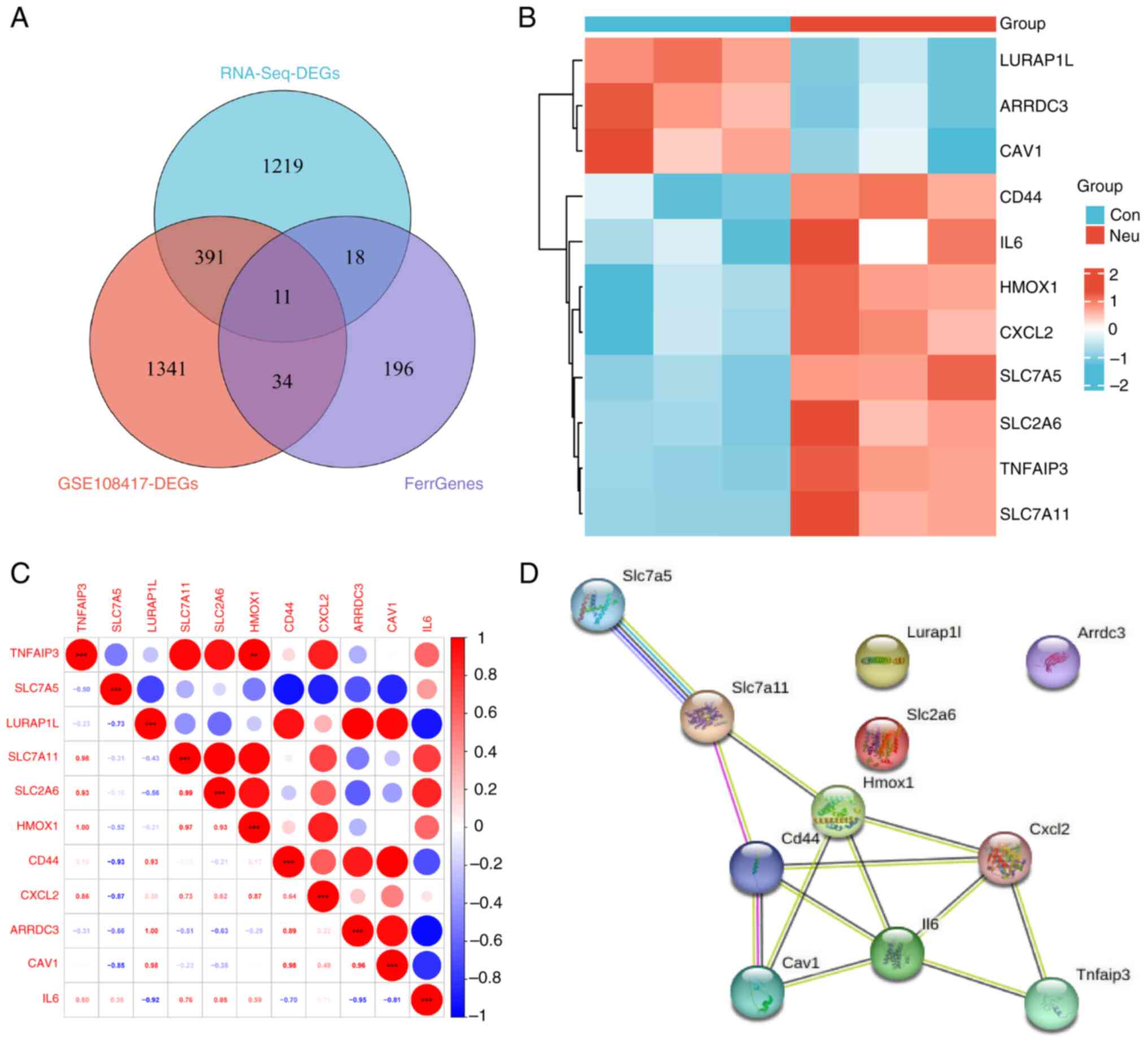
By intersecting DE-FRGs from the transcriptome
dataset with those from the GSE108417 dataset, 11 key DE-FRGs were
identified (Fig. 6A). Among them,
eight upregulated and three downregulated genes can be seen in
Fig. 6B. Subsequently, correlation
analysis revealed that the expression of HMOX1 was positively
associated with TNFAIP3 (Fig. 6C).
The PPI network in Fig. 6D showed
that eight genes involved in ferroptosis have tightly interactive
network relationships. As a result, it can be speculated that these
key DE-FRGs (IL-6, CXCL2, HMOX1, CD44, SLC7A11, SLC7A5, CAV1 and
TNFAIP3) may play an essential role in ferroptosis in mice with
neutrophilic asthma.
External validation of key
DE-FRGs
Based on the validation dataset GSE143303, the
present study sought to validate the expression of these key
DE-FRGs in bronchial biopsy tissues from patients with neutrophilic
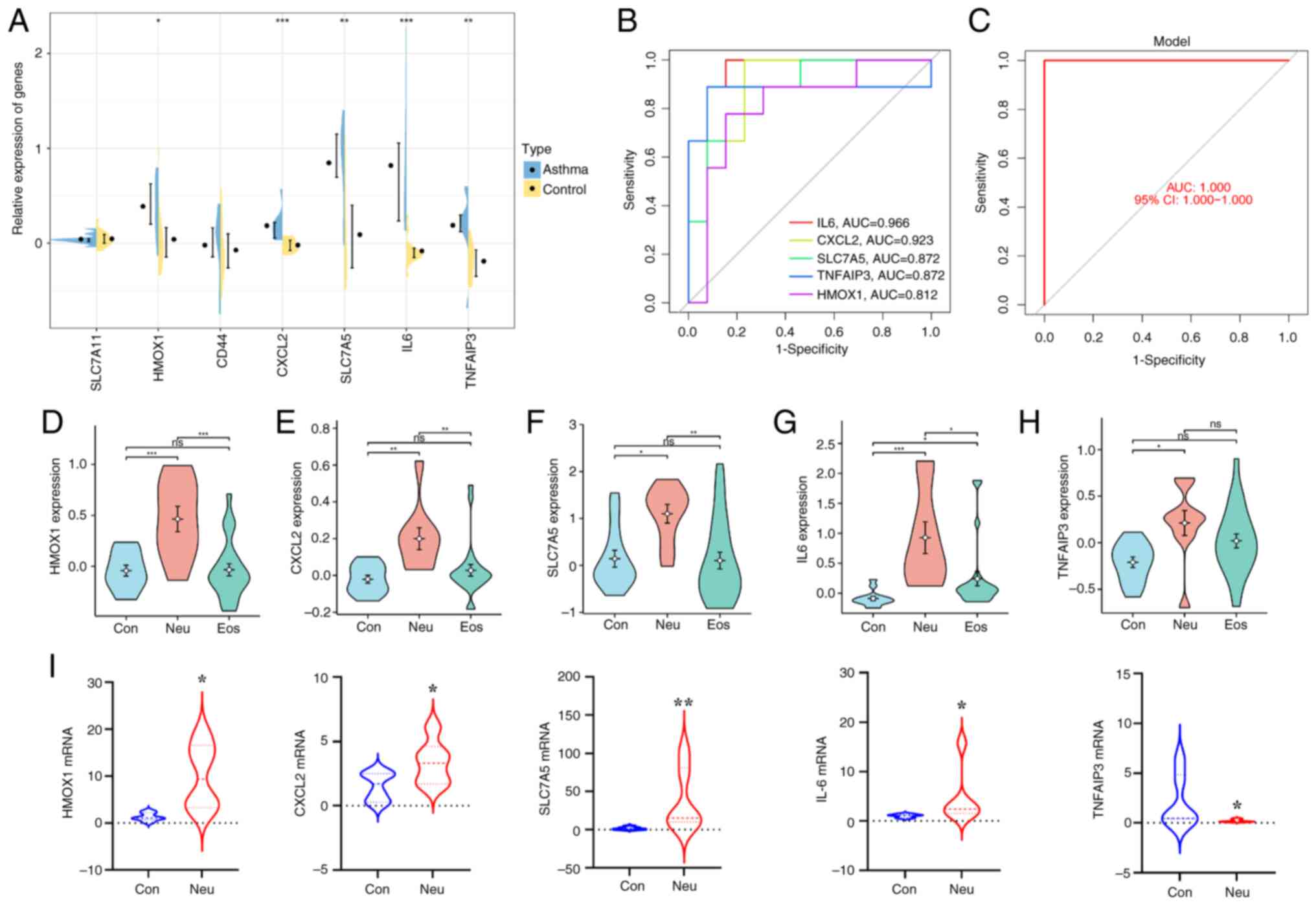
asthma. As consequence, the expression of IL6, CXCL2, HMOX1, SLC7A5
and TNFAIP3 was significantly increased in bronchial biopsies from
neutrophilic asthma compared with normal subjects (P<0.05).
There were not clearly significant differences in CD44 and SLC7A11,
whereas CAV1 was not detected in this dataset (Fig. 7A). To further investigate the
potential of these five DEGs as diagnostic biomarkers in
neutrophilic asthma, an ROC curve was constructed. As shown in
Fig. 7B, the AUC values of IL6,
CXCL2, TNFAIP3, SLC7A5 and HMOX1 were 0.966, 0.923, 0.872, 0.872
and 0.812, respectively, indicating high diagnostic efficacy. It
can also be seen from Fig. 7C that
the top five DE-FRGs were significantly different between patients
with neutrophilic asthma and healthy control participants based on
the established logistic regression model and the AUC value
(1.000). In addition, the expression results of these genes in
normal, neutrophilic and eosinophilic asthma in Fig. 7D-H showed that there were
significant differences between patients with neutrophilic (Neu)
and eosinophilic asthma (Eos) in the expression of HMOX1, CXCL2,
SLC7A5 and IL-6. Collectively, these results suggest that the four
DE-FRGs may play an important role in neutrophilic asthma.
Identification of key DE-FRGs in
neutrophilic asthma
To verify the bioinformatics analysis, the
expression of hub ferroptosis-related differential genes in
vivo was further examined. The qPCR results showed that the
mRNA expression levels of HMOX1, CXCL2, SLC7A5 and IL-6 in the lung
tissue of the asthmatic mice (Fig.
7I) were significantly higher than those of the control group.
However, the expression level of TNFAIP3 in the lung tissue of
asthmatic mice was lower than that of control mice. The results
demonstrated the reliability of the bioinformatics analysis.
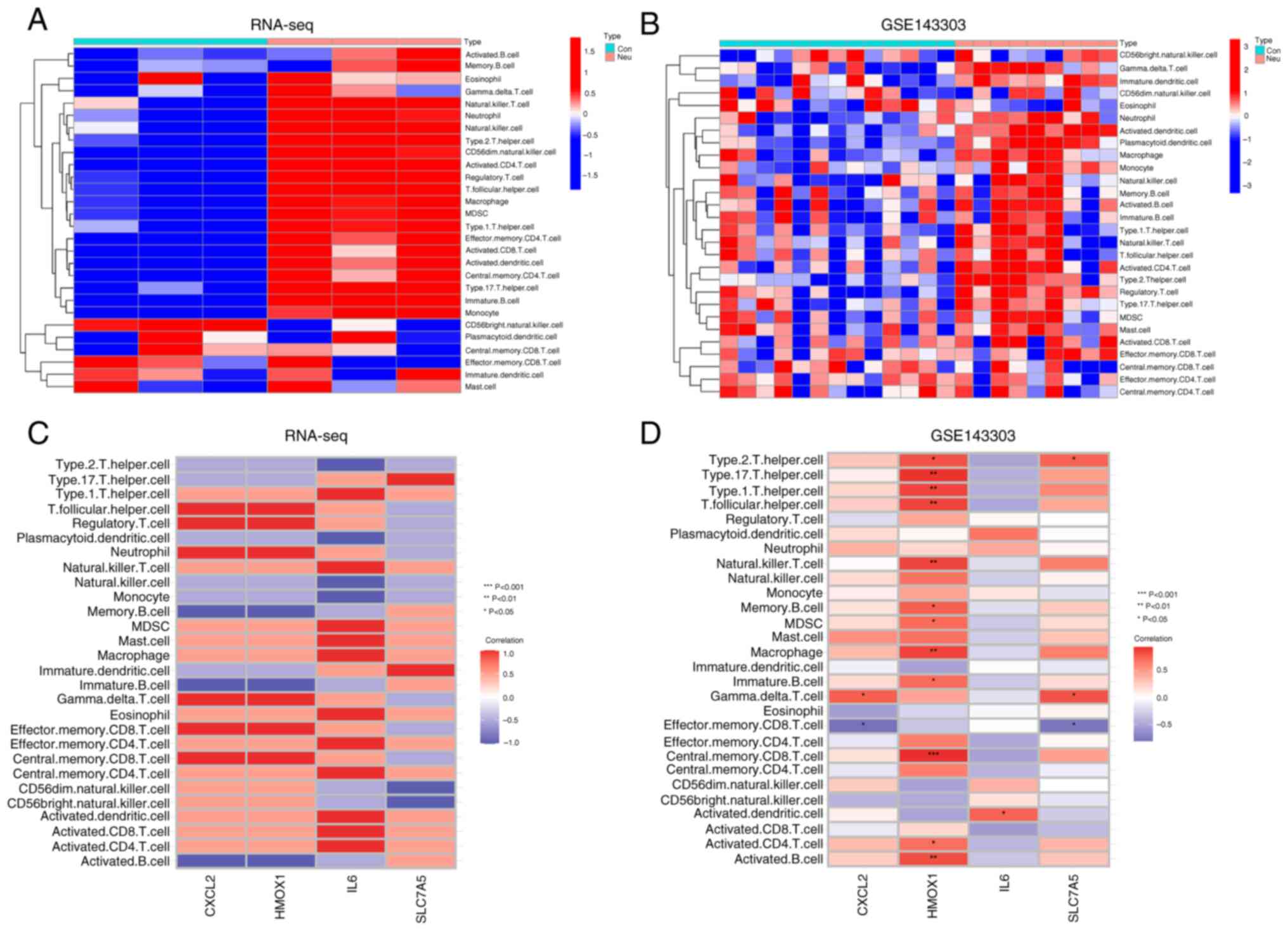
Analysis of the immune
microenvironment in neutrophilic asthma
The present study performed single-sample GSEA
(ssGSEA) to understand more clearly the immune microenvironment in
neutrophilic asthma and analyzed the correlations between key
DE-FRGs and immune cells. It can be found that both the lung tissue
of neutrophilic asthmatics mice (Fig.
8A) and the bronchial biopsy tissue of neutrophilic asthmatics
patients (Fig. 8B) were associated
with various immune cells (including macrophages, neutrophils,
Type2 T helper cells, or Type17 T helper cells). In mouse lung
tissue, DE-FRGs associated with infiltrating cells without a
significant difference (Fig. 8C),
possibly due to the small sample size. In human bronchial biopsy
tissues, HMOX1 was markedly positively associated with immune
cells, such as macrophages, Type1/Type2/Type17 T helper cells,
active CD4 T cells and CD8 T cells in central memory. CXCL2 showed
a positive relationship with gamma delta T cells. IL-6 showed a
significant positive relationship with activated dendritic cells.
SLC7A5 had a dramatic positive relationship with gamma delta T
cells and a negative relationship with effector CD8 T cells
(Fig. 8D). These results further
confirmed the involvement of various immune cells in the
progression of neutrophilic asthma and that DE-FRGs are associated
with various immune cells.
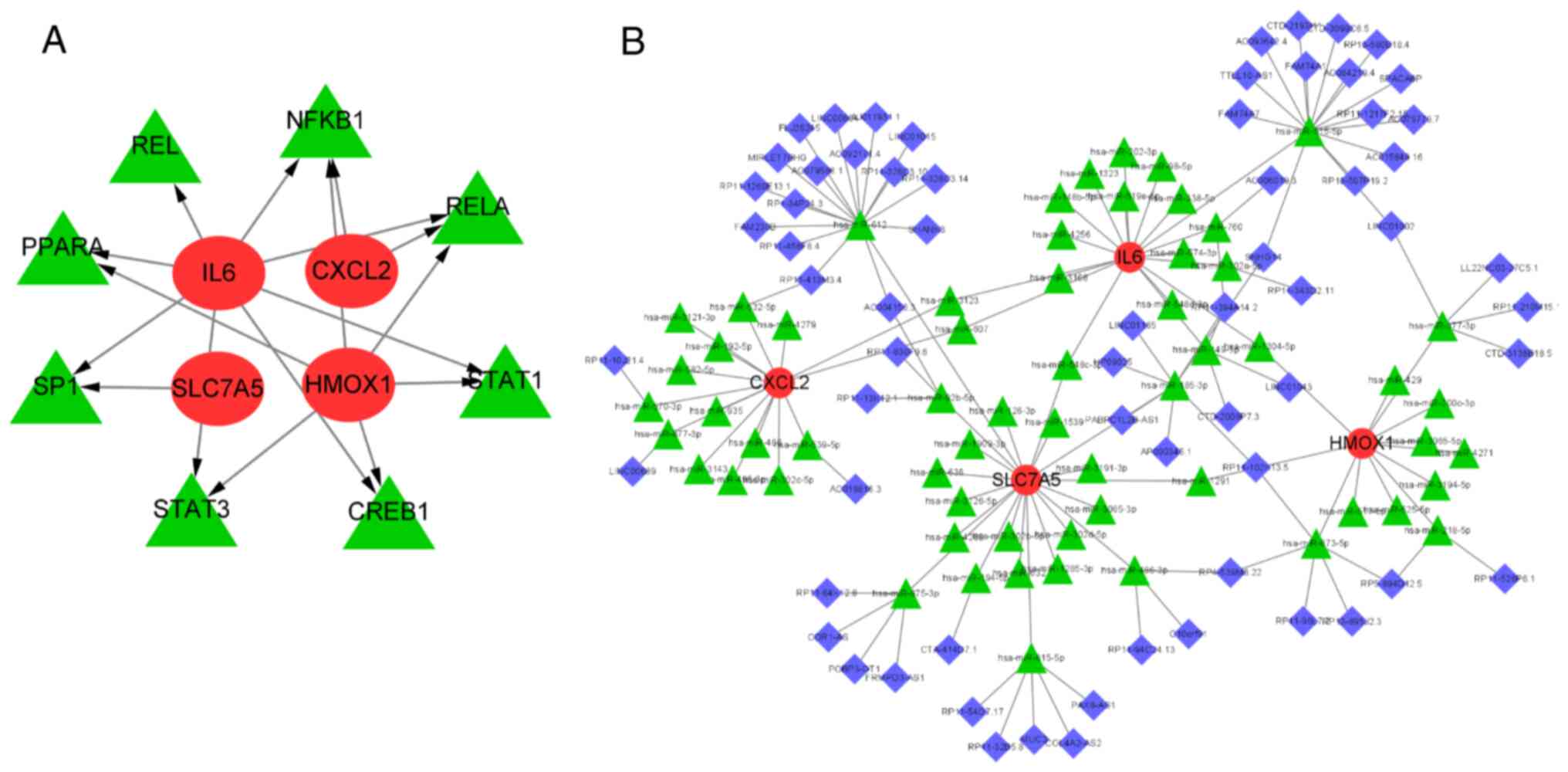
Prediction of key DE-FRGs
transcription factors and CeRNA network construction
To investigate the possible regulation mechanisms of
these DE-FRGs at the transcriptional and post-transcriptional
levels, transcription factor and ceRNA prediction and network
construction were performed. A total of eight transcription factors
(TFs) were predicted using the TRRUST online database (https://www.grnpedia.org/trust/). The mRNA-TF
network was constructed using Cytoscape, as shown in Fig. 9A. It can be seen that RELA and
NF-κB were simultaneously regulated by most major DE-FRGs,
suggesting that the two TFs may be involved in ferroptosis in
neutrophilic asthma at the transcriptional level. Then, StarBase,
miRDB and miRanda databases were used to predict miRNAs that might
have regulatory relationships between these genes and a total of 66
miRNAs were predicted. Finally, 75 lncRNAs interacting with these
66 common miRNAs were predicted by the Spongescan database and the
ceRNA network of DE-FRGs, miRNAs and lncRNAs was constructed
(Fig. 9B).
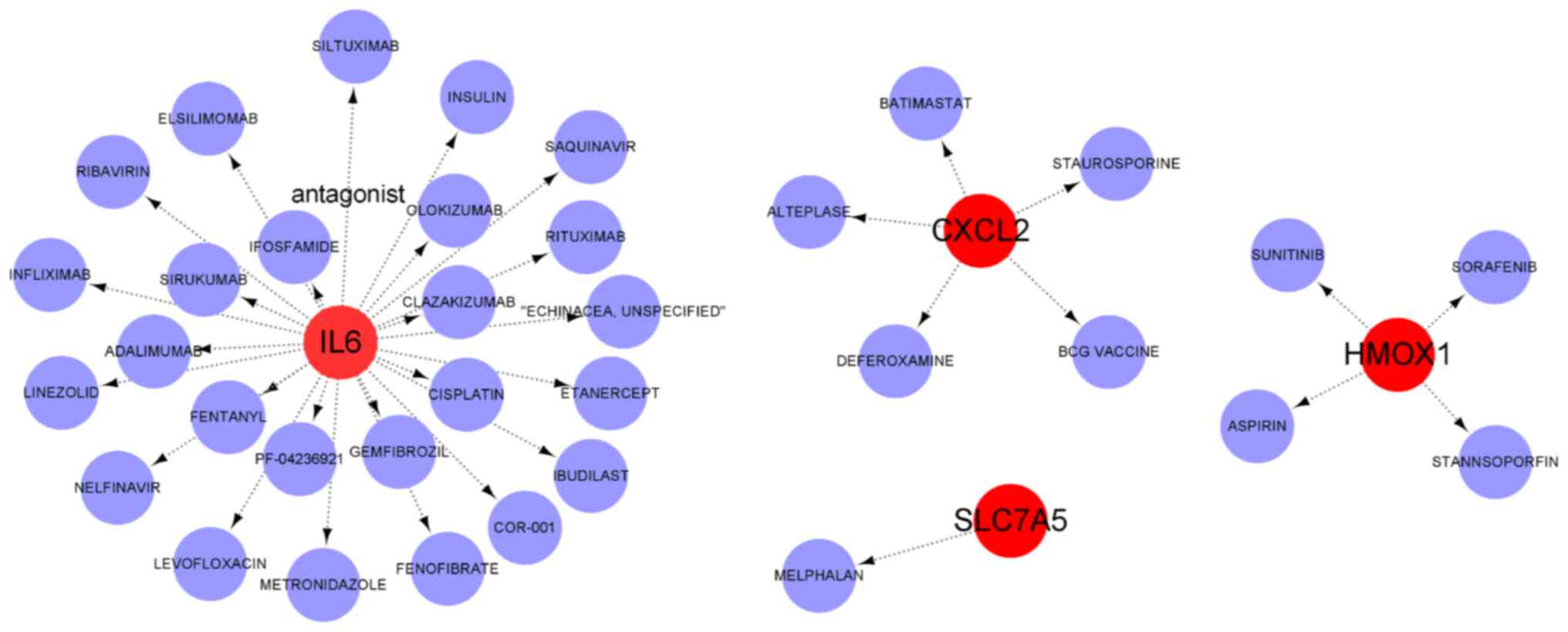
Drug prediction and molecular docking
simulation
The DSigDB database was used to predict the
potential therapeutic drugs associated with hub DE-FRGs. As a
result, 37 target drugs with potential therapeutic effects were
obtained. Among them, there were 25 potential therapeutic drugs
associated with key DE-FRGs in IL6. Of these, four small-molecule
drugs (SORAFENIB, SUNITINIB, STANNSOPORFIN and ASPIRIN) were found
to correlate with HMOX1 and five target drugs (BCG VACCINE,
BATIMASTAT, STAUROSPORINE, ALTEPLASE and DEFEROXAMINE) were
associated with CXCL2. In addition, SLC7A5 exists as a (MELPHALAN)
protein-drug interaction network. Subsequently, a chemical mRNA
network was constructed and visualized using Cytoscape (Fig. 10). Of these target drugs,
deferoxamine has been reported in previous studies to inhibit
HDM-induced airway inflammation in mice by regulating ferroptosis
(50). However, further research
was needed to determine clinical effectiveness in patients with
asthma. Subsequently, through molecular docking simulations, 35
small molecule drugs that can target ferroptosis-related proteins
to exert anti-inflammatory activities in the neutrophilic airways
were further studied to explore the molecular interactions between
these potential therapeutic agents and the main proteins of
neutrophil asthma. Simulated docking results of 13 drugs with a
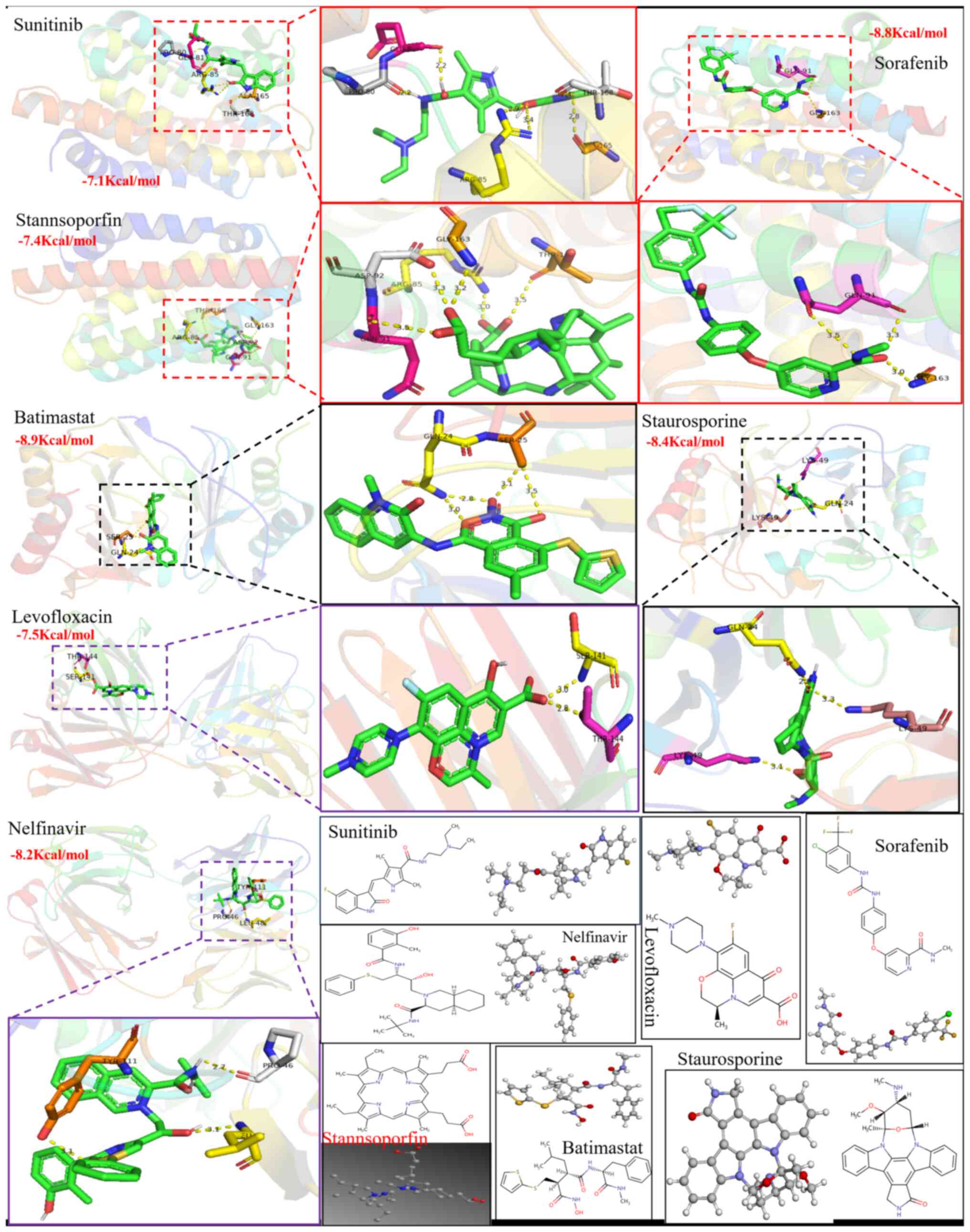
docking score of ≤-6 Kcal/mol are shown in Table I. Fig. 11 also shows that the 2D/3D
structural diagrams of seven candidate drug molecules (Sorafenib,
Stanannsopofin, Sunitinib, Batimastat, Staurosporine, Levofloxacin
and Nelfinavir) and their intermolecular interaction have a lower
affinity of ≤-7 Kcal/mol. Four potential therapeutics (Sunitinib,
Batimastat, Staurosporine and Nelfinavir) were found to interact
with the corresponding primary proteins and had a higher binding
affinity energy (≤-8 Kcal/mol). Interestingly, further analysis may
reveal that two binding residues (Arg85 and Gly163) were mutual
intermolecular interaction sites in the structure of HMOX1, while
the Gln24 residue is present in the binding residue on the CXCL2
sequence. Deng et al (51)
also emphasized that the binding affinity energy (>-6 kcal/mol)
triggered high binding affinity of the potential therapeutic drug
with the target molecules. Thus, these data showed that 13 drugs
had a higher stable binding to proteins.
 | Table IDocking score (≤-6 kcal/mol) of 13
potential therapeutic drugs in which they combined with the primary
proteins in neutrophilic asthma. |
Table I
Docking score (≤-6 kcal/mol) of 13
potential therapeutic drugs in which they combined with the primary
proteins in neutrophilic asthma.
| Primary
proteins | HMOX1
(Kcal/mol) | CXCL2
(Kcal/mol) | IL6 (Kcal/mol) |
|---|
| Potential
therapeutic agents | Sorafenib (-8.8),
Stanannsopofin (-7.4), Sunitinb (-7.1) | Batimastat (-8.9),
Deferoxamine (-6.3), Staurosporine (-8.4) | Fenofibrate (-6.8),
Fentanyl (-6.8), Gemfibrozil (-6.5), Ibudilast (-6.1), Levofloxacin
(-7.5), Linezolid (-6.8), Nelfinavir (-8.2) |
Validation of HIF-1 pathway associated
with ferroptosis in neutrophilic asthma
The significance of the HIF-1 signaling pathway in
the development of neutrophilic asthma is confirmed by the
aforementioned bioinformatics analyses. The present study then
further verified the occurrence of ferroptosis in neutrophilic
asthma. Fig. 12A shows the change
in mitochondrial morphology in mouse airway epithelial cells. It
can be seen that the ciliated epithelial cells of mitochondria
morphology in the Neu group exhibited a smaller size and a slightly
higher membrane and inner membrane density than those in the Con
group, while the cristae of mitochondria morphology were reduced in
the Neu group. These features are consistent with those observed in
ferroptosis (52). Lipid
peroxidation and impaired REDOX responses were further confirmed as
important features of ferroptosis. The results in Fig. 12B showed that the MDA
concentration in the lung tissue of the Neu group was significantly
higher than that of the Con group. The degree of lipid peroxidation
is positively associated with the MDA content. In Fig. 12C, a reduced GSH concentration can
be observed, leading to an impaired redox response in the lung
tissue of the Neu group. Additionally, it was found that the mRNA
expression of GPX4 decreased in the lung tissue of the Neu group
(Fig. 12D). The western blotting
detection (Fig. 12E and F) was consistent with the qPCR results.
Further investigations were carried out on the expression of HO-1
and HIF-1α proteins in the HIF-1 signaling pathway. It was found
that there was a significant increase in HIF-1α and HO-1 protein
levels in neutrophilic asthmatic mice (Fig. 12E and H) compared with the control group.
Therefore, it can be concluded that the HIF-1 signaling pathway may
be involved in ferroptosis during the pathogenesis of neutrophilic
asthma. The results were consistent with the biological information
analysis.
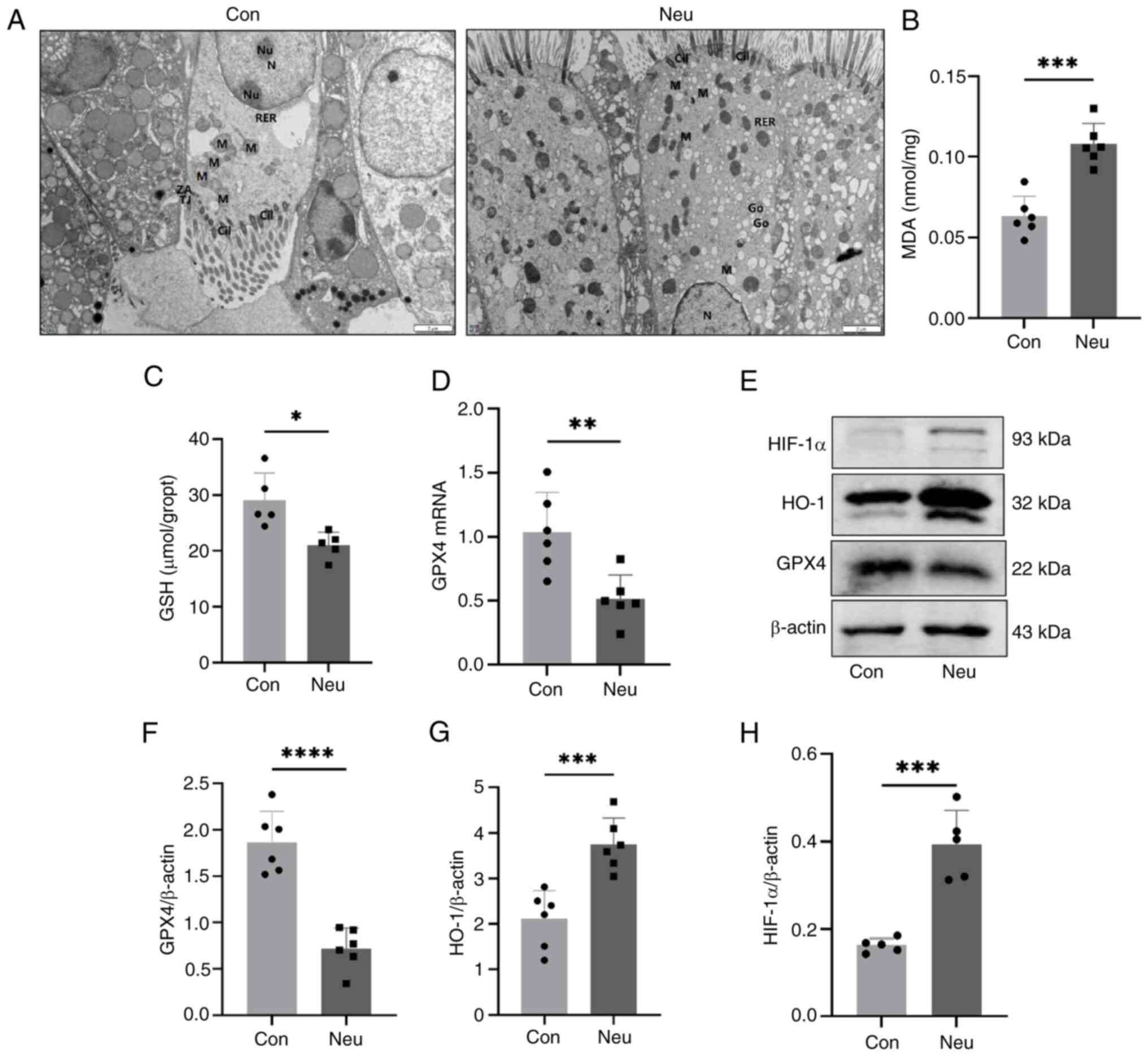 | Figure 12HIF-1 pathway is involved in the
ferroptosis of neutrophilic asthma. (A) Transmission electron
microscopy images (Scale bar, 2 µm). mRNA expression of (B) MDA,
(C) GSH and (D) GPX4; (E-H) GPX4 and proteins associated with the
HIF-1α/HO-1 pathway in mice lung tissue (*P<0.05,
**P<0.01 ***P<0.005 and
****P<0.001). MDA, malondialdehyde; GSH, glutathione;
GPX4, glutathione peroxidase; Mi, mitochondria; N, nucleus; Cil,
Cilia; HIF-1, hypoxia-inducible factor 1; HO-1, heme oxygenase
1. |
Discussion
Ferroptosis is considered a special form of cell
death that is closely associated with inflammatory diseases
(53,54). Unfortunately, understanding of the
role of ferroptosis in neutrophilic airway inflammation is still
limited. Therefore, exploring central mechanisms associated with
ferroptosis in neutrophilic asthma would be of great
importance.
The present study focused on the transcriptomic
analysis and validation of key genes and biological pathway
associated with ferroptosis in neutrophilic asthma. It first
created an asthmatic mouse model with neutrophilic airway
inflammation using OVA and CFA. The mouse model with the
characteristics of neutrophilic airway inflammation (including
airway hyperresponsiveness, infiltration of neutrophils and
inflammatory cells around the airway and hypersecretion of IL-17A
and IFN-γ), demonstrating the mouse model of neutrophilic asthma
was successfully constructed. DE-FRGs were then picked out in
different models of neutrophilic asthma. Furthermore, KEGG
enrichment in three datasets showed that these DE-FRGs were
enriched in ferroptosis, HIF-1, NOD-like receptor and IL-17
signaling pathways. In addition, the Pathview analysis of GSE143303
and RNA-seq dataset showed that HIF-1 may have an important
influence on ferroptosis by modulation of HMOX1. Simultaneously, 11
common DE-FRGs were screened in neutrophilic asthmatic mice by
intersecting the transcriptome dataset with those from the
GSE108417 dataset. The key DE-FRGs were then identified using the
PPI analysis. Of these, five key DE-FRGs (IL6, HMOX1, CXCL2, SLC7A5
and TNFAIP3) with good diagnostic value differed markedly in the
validation set. To further validate the bioinformatic analysis, the
expression levels of these five key genes were further analyzed by
qPCR in mouse lung tissues. Among them, IL6, HMOX1, CXCL2 and
SLC7A5 were upregulated in the neutrophilic model, which was
consistent with the bioinformatics analysis.
Previous studies have shown that these key
ferroptosis genes play an important role in the process of
ferroptosis. IL-6 was found to be involved in angiotensin
II-induced ferroptosis (55).
Zhang et al (56)
emphasized that ferroptosis development in cardiac tissues of
hypertensive mice is associated with the IL-6/STAT3 signaling
pathway. Other research showed that IL-6 can exacerbate airway
inflammation by promoting ROS expression and lipid peroxidation and
disrupting cellular iron homeostasis, leading to ferroptosis in
bronchial epithelial cells (57).
For CXCL2, Yi et al (58)
have demonstrated that overexpression of CXCL2 is strongly
associated with raised expression levels of ROS, Fe2+
and MDA in hepatocytes. Inhibition of CXCL2 expression has been
shown to reverse the reduced expression of GPX4, diminish ROS
production and decrease lipid peroxidation (58). Jin et al (59) found that the ferroptosis inducer
RLS3 can boost CXCL2 expression induced by HHP. A similar change
can be observed in the lung tissue of our engineered asthmatic mice
where CXCL2 mRNA levels were markedly increased. Several studies
have focused on the association between SLC7A5 and airway
inflammation. Inhibition of SLC7A5 expression can inhibit IL-17
production and neutrophil infiltration by interfering with the
incorporation of large neutral amino acids (60). SLC7A5 was expected to be a new
target for the treatment of Th17-mediated and steroid-resistant
severe asthma (60). Furthermore,
there is a positive relationship between SLC7A5 level and HIF-1α
(61). Consistent with previous
studies, the results of the present study also identified SLC7A5 as
an important DE-FRG with increased expression in the Neu group.
Additionally, HO-1, as a rate-limiting enzyme for heme metabolism
(62), is not only an
anti-inflammatory protective factor but also a fundamental
regulator of ferroptosis. HO-1-derived iron can increase
intracellular lipid peroxidation and lead to ferroptosis. Previous
research has shown that high levels of HO-1 expression in
doxorubicin-stimulated cardiac tissue are linked to iron deposition
in cardiac mitochondria (63).
Tang et al (64) also
reported that excessive activation of HO-1 induces the accumulation
of ferrous ions in retinal pigment epithelium (RPE) cells, thereby
triggering ferroptosis. By contrast, inhibition of HO-1 expression
could effectively prevent RPE ferroptosis. In the present study,
the expression level of HO-1 increased in the lung tissue of
neutrophilic asthmatic mice. Immune infiltration analysis also
indicated that HO-1 was linked to various immune cells, which was
consistent with previous studies (65).
Based on the aforementioned analysis, Cytoscape was
used to construct mRNA-TF and ceRNA networks and eight TFs, 66
miRNA and 75 lncRNA were predicted that could regulate DE-FRGs
providing a direction for the research of transcription and ceRNA
post-transcriptional regulatory mechanism of these genes.
Simultaneously, the DSigDB database was applied to further predict
potential therapeutic agents of key DE-FRGs. As consequence, 35
potential therapeutic drugs were identified. Through molecular
docking simulation, small molecule drugs targeting HMOX1, CXCL2,
IL6 and SLC7A5 with anti-inflammatory effects were further studied
to elucidate interaction mechanism between ferroptosis-related
signaling pathway proteins and target drug prediction. A total of
13 small-molecule drugs exhibited higher binding stability with a
binding affinity energy of ≤-6 Kcal/mol. Further investigation
revealed that seven of the 13 small-molecule drugs had
conformations that bind to the corresponding target protein [heme
oxygenase 1 (HMOX1), chemokine ligand 2 (CXCL2) and IL6] with a
binding affinity energy of ≤-7 Kcal/mol. It was found that the most
common interaction residues of HMOX1 and CXCL2 with potential
therapeutic drugs were Gln91, Gly163 and Gln24, respectively. This
provides a new direction for drug selection in neutrophilic asthma
targeting ferroptosis.
The potential mechanism of ferroptosis in
neutrophilic asthma was further explored. First, the expression of
the ferroptosis signature gene GPX4 and the concentrations of MDA
and GSH were detected in mice. GPX4 has an important effect in
inhibiting lipid peroxidation by reducing phospholipid
hydroperoxides. Reduced expression of GPX4 is a hallmark of
ferroptosis (66). It was found
that the expression of GPX4 in the lung tissue of mice with
neutrophilic asthma was lower compared with that of the Con group.
Simultaneously, lipid peroxidation and oxidative stress occurred in
the Neu group. It was demonstrated that ferroptosis occurred in the
lung tissue of mice with neutrophilic asthma. Next, the enriched
signaling pathways of ferroptosis-related genes was examined to
understand the potential regulatory mechanisms of ferroptosis in
neutrophilic asthma. According to the enrichment analysis, DE-FRGs
were mainly enriched in ferroptosis, HIF-1, IL-17 and the NOD-like
receptor pathway. Among them, previous studies have shown that
HIF-1, as an important factor regulating the hypoxic response
(67), was shown to
transcriptionally regulates the expression of the ferroptosis gene
HMOX1(68). Moreover, other
studies have demonstrated that the HIF-1 signaling pathway is
involved in the regulation of ferroptosis. Wu et al
(69) indicated that DEHP exposure
induces ferroptosis in mouse testicular tissues via the HIF-1α/HO-1
pathway. Additionally, Feng et al (70) also reported that ferroptosis can
exacerbate DN-induced tubular injury via the HIF-1α/HO-1 pathway.
Based on the increased HIF-1α expression in the bronchial fluid and
lung tissue of asthmatics, as well as in the nasal fluid of
rhinitis patients, it can be proven that HIF-1 is directly involved
in the development of asthma (71). However, HIF-1α expression as well
as its regulatory mechanism in neutrophilic asthma remains unclear
and whether it is involved in the occurrence of ferroptosis in
neutrophilic asthma remains to be elucidated. Therefore, through
bioinformatics analysis combined with experimental validation, it
was found that the protein levels of HO-1 and HIF-1α were increased
in asthmatic lung tissue, which is closely associated with
ferroptosis. It was hypothesized that HIF-1α may play an important
role in the progression of ferroptosis in neutrophilic asthma by
regulating the transcription of HMOX1. These results shed new light
on the role of ferroptosis in the treatment of neutrophilic
asthma.
The present study also had some limitations. First,
it only use animal models to verify these ferroptosis-related
genes. More clinical samples will be needed in the future to
further test the hypothesis. Second, although the present study
clarified the expression of signature genes and the HIF pathway of
ferroptosis in neutrophilic asthma, their role in neutrophilic
asthma and the underlying regulatory mechanism are not yet clear.
In the future, in-depth studies will be conducted to explore the
regulatory role of HIF-1α/HO-1 pathway proteins in ferroptosis of
neutrophilic asthma and the mechanism of interaction between
pathway proteins to further elucidate the pathogenesis of
neutrophilic asthma. It is worth considering that clinical samples
for future work could be used to further verify these
ferroptosis-related genes as markers or targets in the diagnosis or
treatment of neutrophilic asthma.
Collectively, the present study provided evidence
for the occurrence of ferroptosis in neutrophilic asthma and used
bioinformatic methods to discover FRGs associated with neutrophilic
asthma. These genes exhibited good diagnostic value and their
expression was markedly different from that of eosinophilic asthma.
The results of the present study suggested that the four hub
DE-FRGs and the HIF-1 signaling pathway was closely associated with
ferroptosis pathogenesis in neutrophilic asthma. The present study
provided a novel insight into understanding the pathogenesis of
neutrophilic asthma. The identified four hub DE-FRGs and HIF-1
signaling pathway may be proposed as a potential target for the
diagnosis or treatment of neutrophilic asthma.
Supplementary Material
Primer Sequences in reverse
transcription-quantitative PCR.
KEGG enrichment of differentially
expressed ferroptosis-related genes in RNA-seq data.
KEGG enrichment of differentially
expressed ferroptosis-related genes in GSE108417 dataset.
KEGG enrichment of differentially
expressed ferroptosis-related genes in GSE143303 dataset.
Acknowledgements
Not applicable.
Funding
Funding: The present study was approved by Guangxi Natural
Science Foundation (approval no. 2020GXNSFDA238003).
Availability of data and materials
Datasets provided in the present work are available
from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The raw
sequence data reported in this paper have been deposited in the
Genome Sequence Archive (72) in
National Genomics Data Center (73), China National Center for
Bioinformation/Beijing Institute of Genomics, Chinese Academy of
Sciences (GSA: CRA015119) that are publicly accessible at
https://ngdc.cncb.ac.cn/gsa.
Authors' contributions
CL contributed to the conception and design of the
present study. LL and ZL performed bioinformatics analyses and
interpreted the results, and wrote the manuscript. SP and SW
supervised the experiments and performed the construction of the
asthma mouse model. LL, YL and QXS analyzed the database, prepared
the diagrams and revised the article. YL and QXS confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Animal experiments were approved by the Ethics
Committee of Guangxi Medical University (approval no.
202210101).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hammad H and Lambrecht BN: The basic
immunology of asthma. Cell. 184:1469–1485. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ray A and Kolls JK: Neutrophilic
inflammation in asthma and association with disease severity.
Trends Immunol. 38:942–954. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhao L, Gao J, Chen G, Huang C, Kong W,
Feng Y and Zhen G: Mitochondria dysfunction in airway epithelial
cells is associated with type 2-low asthma. Front Genet.
14(1186317)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Doe C, Bafadhel M, Siddiqui S, Desai D,
Mistry V, Rugman P, McCormick M, Woods J, May R, Sleeman MA, et al:
Expression of the T helper 17-associated cytokines IL-17A and
IL-17F in asthma and COPD. Chest. 138:1140–1147. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Green RH, Brightling CE, Woltmann G,
Parker D, Wardlaw AJ and Pavord ID: Analysis of induced sputum in
adults with asthma: Identification of subgroup with isolated sputum
neutrophilia and poor response to inhaled corticosteroids. Thorax.
57:875–789. 2002.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jiang YH, Wu SY, Wang Z, Zhang L, Zhang J,
Li Y, Liu C, Wu WZ and Xue YT: Bioinformatics analysis identifies
ferroptosis-related genes in the regulatory mechanism of myocardial
infarction. Exp Ther Med. 24(748)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shao L, Fang Q, Ba C, Zhang Y, Shi C,
Zhang Y and Wang J: Identification of ferroptosis-associated genes
in chronic kidney disease. Exp Ther Med. 25(60)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hu T, Yu WP, Zou HX, Chai ZH, Le SY, Hu
FJ, Wang YC, Huang H, Lai SQ and Liu JC: Role of dysregulated
ferroptosis-related genes in cardiomyocyte ischemia-reperfusion
injury: Experimental verification and bioinformatics analysis. Exp
Ther Med. 26(534)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhou Q, Li T, Qin Q, Huang X and Wang Y:
Ferroptosis in lymphoma: Emerging mechanisms and a novel
therapeutic approach. Front Genet. 13(1039951)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang Q, Xiong Z, Wang B, Wang W and Zheng
H: Ferroptosis and preeclampsia: Genetic analysis of potential
biomarkers and therapeutic targets. Biochem Genet. 62:853–875.
2024.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tang W, Dong M, Teng FZ, Cui J, Zhu X,
Wang W, Wuniqiemu T, Qin J, Yi L, Wang S, et al: Environmental
allergens house dust mite-induced asthma is associated with
ferroptosis in the lungs. Exp Ther Med. 22(1483)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yang N and Shang Y: Ferrostatin-1 and
3-methyladenine ameliorate ferroptosis in OVA-induced asthma model
and in IL-13-challenged BEAS-2B cells. Oxid Med Cell Longev.
2022(9657933)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bao C, Liu C, Liu Q, Hua L, Hu J, Li Z and
Xu S: Liproxstatin-1 alleviates LPS/IL-13-induced bronchial
epithelial cell injury and neutrophilic asthma in mice by
inhibiting ferroptosis. Int Immunopharmacol.
109(108770)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhao X, Gao S, Ren H, Sun W, Zhang H, Sun
J, Yang S and Hao J: Hypoxia-inducible factor-1 promotes pancreatic
ductal adenocarcinoma invasion and metastasis by activating
transcription of the actin-bundling protein fascin. Cancer Res.
74:2455–2464. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gui D, Li Y, Chen X, Gao D, Yang Y and Li
X: HIF-1 signaling pathway involving iNOS, COX-2 and caspase-9
mediates the neuroprotection provided by erythropoietin in the
retina of chronic ocular hypertension rats. Mol Med Rep.
11:1490–1496. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fu X and Zhang F: Role of the HIF-1
signaling pathway in chronic obstructive pulmonary disease. Exp
Ther Med. 16:4553–4561. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Dong H, Zhang C, Shi D, Xiao X, Chen X,
Zeng Y, Li X and Xie R: Ferroptosis related genes participate in
the pathogenesis of spinal cord injury via HIF-1 signaling pathway.
Brain Res Bull. 192:192–202. 2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Qian JW, Wang C, Wang B, Yang J, Wang Y,
Luo F, Xu J, Zhao C, Liu R and Chu Y: The IFN-γ/PD-L1 axis between
T cells and tumor microenvironment: Hints for glioma
anti-PD-1/PD-L1 therapy. J Neuroinflamm. 15(290)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen L, Hou W, Liu F, Zhu R, Lv A, Quan W
and Mao S: Blockade of NLRP3/caspase-1/IL-1β regulated Th17/treg
immune imbalance and attenuated the neutrophilic airway
inflammation in an ovalbumin-induced murine model of asthma. J
Immunol Res. 2022(9444227)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bogaert P, Naessens T, De Koker S, Hennuy
B, Hacha J, Smet M, Cataldo D, Di Valentin E, Piette J, Tournoy KG
and Grooten J: Inflammatory signatures for eosinophilic vs
neutrophilic allergic pulmonary inflammation reveal critical
regulatory checkpoints. Am J Physiol Lung Cell Mol Physiol.
300:L679–L690. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen X, Jiang X, Lu Y, Yao Y, Lu J, Zhi Q,
Lai L, Liang J and Li C: Aerosol inhalation of Mycobacterium bovis
can reduce the Th2 dominant immune response induced by ovalbumin
sensitization. Am J Transl Res. 14:3430–3438. 2022.PubMed/NCBI
|
|
23
|
Li L, Sun Q, Xiao H, Zhang Q, Xu S, Lai L,
Li Z and Li C: Aerosol inhalation of heat-killed clostridium
butyricum CGMCC0313-1 alleviates allergic airway inflammation in
mice. J Immunol Res. 2022(8447603)2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xiao H, Zhang QN, Sun QX, Li LD, Xu SY and
Li CQ: Transcriptomic analysis reveals a link between hippo
signaling pathway and macrophages in lungs of mice with OVA-induced
allergic asthma. J Inflamm Res. 15:423–437. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dong L, Wang Y, Zheng T, Pu Y, Ma Y, Qi X,
Zhang W, Xue F, Shan Z, Liu J, et al: Hypoxic hUCMSC-derived
extracellular vesicles attenuate allergic airway inflammation and
airway remodeling in chronic asthma mice. Stem Cell Res Ther.
12(4)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jia M, Fu H, Jiang X, Wang L, Xu J, Barnes
PJ, Adcock IM, Liu Y, He S, Zhang F, et al: DEL-1, as an
anti-neutrophil transepithelial migration molecule, inhibits airway
neutrophilic inflammation in asthma. Allergy. 79:1180–1194.
2024.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and ballgown. Nat Protoc.
11:1650–1667. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Dusa A: venn: Draw Venn Diagrams R
package. R Core Team, Vienna, 2020.
|
|
33
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261.
2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kassambara A: ggpubr: ‘ggplot2’ based
publication ready plots. R package version 0.4. 0, 2020. https://CRAN.R-project.org/package=ggpubr.
|
|
36
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12(77)2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ginestet C: ggplot2: Elegant graphics for
data analysis. J R Stat Soc Ser A. 174:245. 2011.
|
|
38
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43 (Database Issue):D146–D152. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Furió-Tarí P, Tarazona S, Gabaldón T,
Enright AJ and Conesa A: spongeScan: A web for detecting microRNA
binding elements in lncRNA sequences. Nucleic Acids Res. 44
(W1):W176–W180. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee
S, Jeon M, Kang J and Tan AC: DSigDB: Drug signatures database for
gene set analysis. Bioinformatics. 31:3069–3071. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Morris GM, Goodsell DS, Halliday RS, Huey
R, Hart WE, Belew RK and Olson AJ: Automated docking using a
Lamarckian genetic algorithm and an empirical binding free energy
function. J Comput Chem. 19:1639–1662. 1998.
|
|
44
|
Wishart DS, Feunang YD, Guo AC, Lo EJ,
Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al:
DrugBank 5.0: A major update to the DrugBank database for 2018.
Nucleic Acids Res. 46 (D1):D1074–D1082. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Tian YN, Zhou YH, Li L, Huang C, Lin L, Li
C and Ye Y: Effect of substrate composition on physicochemical
properties of the medium-long-medium structured triacylglycerol. J
Sci Food Agric. 104:942–955. 2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Raundhal M, Morse C, Khare A, Oriss TB,
Milosevic J, Trudeau J, Huff R, Pilewski J, Holguin F, Kolls J, et
al: High IFN-γ and low SLPI mark severe asthma in mice and humans.
J Clin Invest. 125:3037–3050. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
McKinley L, Alcorn JF, Peterson A, Dupont
RB, Kapadia S, Logar A, Henry A, Irvin CG, Piganelli JD, Ray A and
Kolls JK: TH17 cells mediate steroid-resistant airway inflammation
and airway hyperresponsiveness in mice. J Immunol. 181:4089–4097.
2008.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zeng Z, Huang H, Zhang J, Liu Y, Zhong W,
Chen W, Lu Y, Qiao Y, Zhao H, Meng X, et al: HDM induce airway
epithelial cell ferroptosis and promote inflammation by activating
ferritinophagy in asthma. FASEB J. 36(e22359)2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Deng B, Liao F, Liu Y, He P, Wei S, Liu C
and Dong W: Comprehensive analysis of endoplasmic reticulum
stress-associated genes signature of ulcerative colitis. Front
Immunol. 14(1158648)2023.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Choi W, Wu Y, Li Y and Dong J: Network
pharmacology prediction and molecular docking analysis reveal the
mechanism of modified Bushen Yiqi formulas on chronic obstructive
pulmonary disease. J Gene Med. 26(e3607)2024.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Cao F, Luo A and Yang C: G6PD inhibits
ferroptosis in hepatocellular carcinoma by targeting cytochrome
P450 oxidoreductase. Cell Signal. 87(110098)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang S, Song Y, Xu F, Liu H, Shen Y, Hu L,
Fu Y and Zhu L: Identification and validation of
ferroptosis-related genes in lipopolysaccharide-induced acute lung
injury. Cell Signal. 108(110698)2023.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen D, Li Z, Bao P, Chen M, Zhang M, Yan
F, Xu Y, Ji C, Hu X, Sanchis D, et al: Nrf2 deficiency aggravates
angiotensin II-induced cardiac injury by increasing hypertrophy and
enhancing IL-6/STAT3-dependent inflammation. Biochim Biophys Acta
Mol Basis Dis. 1865:1253–1264. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhang Z, Tang J, Song J, Xie M, Liu Y,
Dong Z, Liu X, Li X, Zhang M, Chen Y, et al: Elabela alleviates
ferroptosis, myocardial remodeling, fibrosis and heart dysfunction
in hypertensive mice by modulating the IL-6/STAT3/GPX4 signaling.
Free Radical Bio Med. 181:130–142. 2022.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Han F, Li S, Yang Y and Bai Z:
Interleukin-6 promotes ferroptosis in bronchial epithelial cells by
inducing reactive oxygen species-dependent lipid peroxidation and
disrupting iron homeostasis. Bioengineered. 12:5279–5288.
2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Yi Q, Liang Q, Liu Y, Gong Z and Yan Y:
Application of genomic selection and experimental techniques to
predict cell death and immunotherapeutic efficacy of
ferroptosis-related CXCL2 in hepatocellular carcinoma. Front Oncol.
12(998736)2022.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Jin R, Yang R, Cui C, Zhang H, Cai J, Geng
B and Chen Z: Ferroptosis due to cystathionine γ Lyase/hydrogen
sulfide downregulation under high hydrostatic pressure exacerbates
VSMC dysfunction. Front Cell Dev Biol. 10(829316)2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Hayashi K, Saeki M, Miura K, Yamasaki N,
Matsuda M, Shimora H, Nabe T, Shimizu Y, Fujita T, Endou H and
Kaminuma O: JPH203, a LAT1 inhibitor, alleviates steroid-resistant
murine airway inflammation mediated by Th17 cells. Allergy.
78:2780–2783. 2023.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Törnroos R, Tina E and Eremo AG: SLC7A5 is
linked to increased expression of genes related to proliferation
and hypoxia in estrogen-receptor-positive breast cancer. Oncol Rep.
47(17)2022.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Consoli V, Sorrenti V, Grosso S and
Vanella L: Heme oxygenase-1 signaling and redox homeostasis in
physiopathological conditions. Biomolecules. 11(589)2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kwon MY, Park E, Lee SJ and Chung SW: Heme
oxygenase-1 accelerates erastin-induced ferroptotic cell death.
Oncotarget. 6:24393–24403. 2015.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Tang Z, Ju Y, Dai X, Ni N, Liu Y, Zhang D,
Gao H, Sun H, Zhang J and Gu P: HO-1-mediated ferroptosis as a
target for protection against retinal pigment epithelium
degeneration. Redox Bio. 43(101971)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wong TH, Chen HA, Gau RJ, Yen JH and Suen
JL: Heme oxygenase-1-expressing dendritic cells promote Foxp3+
regulatory T cell differentiation and induce less severe airway
inflammation in murine models. PLoS One.
11(e0168919)2016.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Feng FH, He SS, Li XL, He JK and Luo LX:
Mitochondria-mediated ferroptosis in diseases therapy: From
molecular mechanisms to implications. Aging Dis. 15:714–738.
2024.PubMed/NCBI View Article : Google Scholar
|
|
67
|
McGettrick AF and O'Neill LAJ: The role of
HIF in immunity and inflammation. Cell Metab. 32:524–536.
2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zhongyin Z, Wei W, Juan X and Guohua F:
Isoliquiritin apioside relieves intestinal
ischemia/reperfusion-induced acute lung injury by blocking
Hif-1α-mediated ferroptosis. Int Immunopharmacol.
108(108852)2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Wu Y, Wang J, Zhao T, Chen J, Kang L, Wei
Y, Han L, Shen L, Long C, Wu S and Wei G: Di-(2-ethylhexyl)
phthalate exposure leads to ferroptosis via the HIF-1α/HO-1
signaling pathway in mouse testes. J Hazard Mater.
426(127807)2022.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Feng X, Wang S, Sun Z, Dong H, Yu H, Huang
M and Gao X: Ferroptosis enhanced diabetic renal tubular injury via
HIF-1α/HO-1 Pathway in db/db mice. Front Endocrinol (Lausanne).
12(526390)2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Huerta-Yepez S, Baay-Guzman GJ, Bebenek
IG, Hernandez-Pando R, Vega MI, Chi L, Riedl M, Diaz-Sanchez D,
Kleerup E, Tashkin DP, et al: Hypoxia inducible factor promotes
murine allergic airway inflammation and is increased in asthma and
rhinitis. Allergy. 66:909–918. 2011.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Chen T, Chen X, Zhang S, Zhu J, Tang B,
Wang A, Dong L, Zhang Z, Yu C, Sun Y, et al: The genome sequence
archive family: Toward explosive data growth and diverse data
types. Genomics Proteomics Bioinformatics. 19:578–583.
2021.PubMed/NCBI View Article : Google Scholar
|
|
73
|
National Genomics Data Center Members and
Partners. Database resources of the national genomics data center
in 2020. Nucleic Acids Res. 48 (D1):D24–D33. 2020.PubMed/NCBI View Article : Google Scholar
|