Introduction
As a prevalent gastrointestinal malignancy, gastric
cancer (GC) is distinguished by its high incidence, aggressive
progression profile and high mortality rates. There were
>968,000 novel cases of stomach cancer in 2022 and close to
660,000 deaths, ranking the disease as fifth in terms of both
incidence and mortality worldwide (1). Despite a decline in the global
prevalence and death rate of GC, rates remain high in Eastern Asian
countries (2). The majority of
patients with GC are typically diagnosed already at advanced stages
during consultations, resulting in a poor prognosis post-surgery.
This scenario poses considerable public health challenges (3). Consequently, the quest for
efficacious molecular markers for GC assumes paramount importance
in facilitating precise treatment design, extending patient
survival and enhancing their overall quality of life.
The primary approach for treating GC continues to
involve conventional surgical procedures, followed by postoperative
radiotherapy and chemotherapy (4).
However, the 5-year survival rate following surgery remains low
(<10%) (5). With the rapid
advancement of molecular biology research, GC treatment methods
have undergone continuous improvements, including the introduction
of immunotherapy, such as programmed cell death protein
(PD)-1/PD-L1 inhibitors and cytotoxic T lymphocyte antigen 4
inhibitors (6). Additionally, the
emergence of chemotherapeutic and molecular-targeted drugs, such as
oxaliplatin and herceptin, has instilled optimism among patients
with intermediate and advanced stages of GC. Although these
interventions have led to improved postoperative survival rates,
the prognosis for GC, particularly in cases in, remains poor due to
the severe toxic side effects, limited sensitivity and specificity
of drugs (7). This is especially
the case in patients at intermediate and advanced stages (7). The metastatic propensity of GC
represents a formidable obstacle in the treatment paradigm
(8).
Therefore, the exploration of effective strategies
to inhibit GC's metastatic spread and identification of molecular
markers for prognostic assessment is required.
E74-like factor 1 (ELF1), which is highly homologous
to the Drosophila E74 factor (9) is a transcription factor inducible by
ecdysone in Drosophila and a member of the Ets transcription
factor family (9). ELF1 is located
on chromosome 13q13 and consists of 619 amino acid residues,
possessing the ability of both transcriptional activation and
repression of target genes depending on the physiological context.
Posttranslational processing determines its subcellular
localization, biological activity and metabolic degradation
(10). The functional regulation
of ELF-1 is complex. ELF1 protein exists in a 80-kDa form in the
cytoplasm and enters the nucleus in a 98 kDa form after
phosphorylation and glycosylation (11). It is predominantly expressed in
lymphocytes, where its posttranslational modifications bestow ELF1
with regulatory functions, enabling it to bind to gene promoters or
enhancers critical for the selection, survival and maturation of
diverse immune cells (12). The
importance of ELF1 extends to its association with the development
and metastasis of various malignancies (13), such as glioma, oral squamous cell
carcinoma, endometrial carcinoma, nasopharyngeal carcinoma and
prostate cancer, and colon cancer. Long non-coding RNAs (lncRNAs)
are associated with cancer progression in GC (14). E2F transcription factor 1 has been
shown to activate the transcription of terminal
differentiation-induced non-coding RNA by binding to its promoter
region, thereby promoting the proliferation of GC cells and
inhibiting apoptosis (15).
lncRNAs NONHSAT057282 and NONHSAG023333 can regulate genes
associated with chemoresistance, such as GSTP1, BTG3, SOCS3, and
BRAC2, by interacting with the transcription factors ELF1 and E2F1.
ELF1 may become a new player in chemoresistance through its
interaction with different lncRNA interactions involved in tumor
therapy (16). So, it was
hypothesized that ELF1 may be associated with GC.
Tumor cells, together with extracellular matrix
(ECM), cancer-associated fibroblasts (CAF), vascular-associated
smooth muscle cells, pericytes, endothelial cells, mesenchymal stem
and immune cells collectively constitute the complex tumor
microenvironment (TME) (17). The
development, progression and ultimately the prognosis of
malignancies, are profoundly influenced by the characteristics of
tumor cell invasion and metastasis (18). A pivotal process in this cascade
involves ECM degradation, which is tightly regulated by MMPs and
tissue inhibitors of metalloproteinases (TIMPs) (19). Among MMPs, MMP-9 is of particular
importance as a key protease responsible for ECM degradation, where
it serves a crucial role in various types of cancers, such as
breast cancer (20,21). During cancer progression, ECM
homeostasis is dynamically disrupted by MMP9, enabling cancer
invasion and metastasis through the ECM barrier. MMP2 and MMP9 have
been previously found to be upregulated in GC tissues (22). MMP9 rs3918242 polymorphism has been
associated with the risk of various cancers, including lung,
prostate, breast, and colorectal cancers (23). The chromosomal location
(20q12-1q13) where MMP9 is located, has been identified as one of
the most common regions of genomic gain in GC (24). Therefore, the progression of GC is
highly likely to be influenced by altered MMP9 expression.
However, the precise expression pattern, interaction
with MMP9 and predictive value of ELF1 in GC remain elusive.
Therefore, the present study aimed to explore the expression
profile of ELF1 in GC, its relationship with MMP9 and its (combined
with MMP9) associations with various clinicopathological
parameters, survival and prognosis of patients with GC.
Bioinformatics and clinical sample analyses would be performed. In
addition, relevant mechanisms, such as the association of ELF1 with
epithelial-mesenchymal transition (EMT), angiogenesis and immune
infiltration, were explored.
Materials and methods
Clinical patient samples
Fresh GC and adjacent normal tissues (located >2
cm from the tumor margin) were randomly collected from 40 patients
post-GC surgery at the Affiliated Hospital of Nantong University
(Nantong, China) from November to December 2021. This cohort
included 28 males and 12 females, with a mean age of 66.25 years
(range, 36-92 years; Table SI).
The inclusion criteria were as follows: i) Pathological diagnosis
of GC; ii) clinical data and overall survival time were complete;
iii) no history of antitumor treatment before surgery and iv) no
combination of other organic disease and malignant tumors. The
exclusion criteria were as follows: i) incomplete clinical data and
overall survival time; ii) combined with other organic diseases and
malignant tumors; iii) a history of preoperative antitumor
treatment; and iv) use of pathological case data without the
patient's informed consent. Necrotic tissues were excised and blood
contaminants were rinsed with saline before the samples were frozen
at -80˚C for further analysis.
Additionally, 355 paraffin-embedded GC specimens and
corresponding adjacent normal tissues from the patients undergoing
surgery from January 2013 to December 31, 2015, preserved in the
Department of Pathology of Affiliated Hospital of Nantong
University (Nantong, China, were collected from January 2019 to
March 2019 for tissue microarrays for this study. The patients
included 241 males and 114 females, with a mean age of 63.63
(range, 26-90) years. The inclusion and exclusion criteria were as
aforementioned. The clinical characteristics collected included
sex, age, histological type, differentiation, invasive depth (T),
lymph node metastasis (N), distant metastasis (M), TNM stage,
microvascular invasion (MVI), lymphatic invasion, perineural
invasion, carcinoembryonic antigen (CEA), carbohydrate antigen 19-9
and Laurén classification (25).
The cases that were processed and stained with H&E were
pathologically confirmed on the basis of the latest WHO
classification and 8th edition of the TNM classification
recommended by the Union for International Cancer Control and
American Joint Committee on Cancer (26). None of the patients had undergone
any anticancer treatments, such as radiotherapy, chemotherapy or
immunotherapy, before surgery. Complete clinical data and
postoperative follow-up records (100% completion rate,) were
obtained prior to the study. Overall survival (OS) was defined as
the time from surgical resection to death or end of follow-up
(December 31, 2020).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis for ELF1 mRNA expression in GC
The expression of ELF1 mRNA in the 40 fresh human GC
tissue samples was validated through RT-qPCR. Total RNA was
extracted from tissues by using a TRIzol kit (Invitrogen; Thermo
Fisher Scientific, Inc.), which was then treated with DNase I (Cat
No. D7073; Beyotime Institute of Biotechnology). The purity and
concentration were analyzed using a NanoDrop ND-1000
spectrophotometer (Thermo Fisher Scientific, Inc.), followed by
cDNA synthesis with a PowerScript™ Reverse Transcriptase
kit (Bioland Scientific, LLC) according to the manufacturer's
protocol. qPCR amplification was then performed on a 7500 real-time
PCR system (Thermo Fisher Scientific, Inc.) using a
SYBR® Premix ExTaq™ kit (Takara Bio, Inc.).
The following thermocycling conditions were used: 95˚C for 5 min
for initial denaturation, followed by 40 cycles of 95˚C for 10 sec
(denaturation), 61˚C for 20 sec (annealing) and 70˚C for 40 sec
(extension). The specific primers for ELF1 and the internal
reference GAPDH were designed based on the NCBI gene sequence and
synthesized by Shanghai Yingjun Biotechnology Co., Ltd. The
specific forward primer for ELF1 was 5'-TGTCCAACAGAACGACCTAGT-3',
whilst the reverse primer was 5'-GGCAGGAAAAATAGCTGGATCAC-3'. The
length of the amplified target fragment was 88 bp. The forward
primer of the GAPDH gene was 5'-GGAGCGAGATCCCTCCAAAAT-3', whereas
the reverse primer was 5'-GGCTGTTGTCATACTTCTCATGG-3'. The length of
the amplified target fragment was 197 bp. The results were analyzed
by using the 2-ΔΔCq method (27) with each sample assayed in
triplicate.
Tissue microarray
(TMA)-immunohistochemistry (IHC) for protein expression levels in
GC
Postoperative GC and adjacent normal tissues from
the 355 patients were fixed in 10% neutral formalin at room
temperature for 24 h. Core tissue biopsy samples (0.2 cm in
diameter) obtained from the paraffin-embedded blocks were arranged
in fresh paraffin blocks using the Quick-Ray Manual Tissue
Microarrayer Full Set (cat. no. UT06; Unitma, Co., Ltd.). In total,
12 tissue microarrays comprising 710 samples were ultimately
prepared. Sections of 5-µm thickness were analyzed for ELF1, MMP9,
CD19, CD3, CD4, CD8 and CD56 protein expression levels by using
immunohistochemistry. Sections were deparaffinized by immersion in
xylene and rehydrated in gradient ethanol in separate batches,
washed in PBS (0.01 M, pH=7.0), boiled (98˚C, 20 min) under
pressure in citrate buffer (0.01 M, pH=6.0; antigen recovered) and
incubated with 5% goat-blocking serum (cat. no. SL039; Beijing
Solarbio Science & Technology Co., Ltd.) in PBS for 30 min at
37˚C to block non-specific binding. Subsequently, mouse antihuman
ELF1 polyclonal antibody (dilution 1:400; cat. no. 22565-1-AP;
ProteinTech Group, Inc.), MMP9 polyclonal antibody (dilution 1:300;
cat. no. 10375-2-AP; ProteinTech Group, Inc.), mouse anti-human
CD19 monoclonal (ready to use; cat. no. ZM-0038; ZSGB-BIO; OriGene
Technologies, Inc.), mouse anti-human CD3 monoclonal (ready to use;
cat. no. ZM-0417; ZSGB-BIO; OriGene Technologies, Inc.), mouse
antihuman CD4 monoclonal (ready to use; cat. no. ZM-0418; ZSGB-BIO;
OriGene Technologies, Inc.), rabbit anti-human CD8 monoclonal
(ready to use; cat. no. ZM-0508 ZSGB-BIO; OriGene Technologies,
Inc.) and mouse anti-human CD56 monoclonal (ready to use; cat. no.
ZM-0057; ZSGB-BIO; OriGene Technologies, Inc.) were used for
staining overnight at 4˚C. HRP-labeled goat anti-mouse secondary
antibody (1:1,000; cat. no. ab6728; Abcam) and HRP-labeled goat
anti-rabbit IgG (1:1,000; cat. no. Ab6721; Abcam) were used at room
temperature for 30 min. Sections were incubated with DAB (cat. no.
DA1010; Beijing Solarbio Science & Technology Co., Ltd.) for
~10 min, counterstained with hematoxylin (room temperature, 20-30
sec) and sealed with gelatin glycerol. PBS served as a negative
control. Staining results were double-blinded and analyzed by two
senior pathologists (XYR and SZ).
Cells were light imaged through an optical
microscope (BX51, OLYMPUS) of immunohistochemistry staining were
defined as brownish-yellow or brownish-brown nuclei (ELF1) and
cytoplasm (MMP9) staining of GC tumor cells, nuclei (ELF1) of the
lymphocytes in mesenchyme, and membrane (CD19, CD3, CD4, CD8 and
CD56) of infiltrated immune cells. The intensity was scored as
follows: i) 0, negative; ii) 1, weak intensity; iii) 2, moderate
intensity; and iv) 3, strong intensity. Percentage of positive
cells was scored as follows: i) 0, negative; ii) 1, 1-25% positive;
iii) 2, 26-50% positive; iv) 3, 51-75% positive; and v) 4, 76-100%
positive). The multiplication of the two aforementioned scores was
used as the final score, where 0-6 would be deemed no or low
expression (-) and 7-12 was considered high expression (+)
(28).
Bioinformatics analysis
Data on 375 primary GC tissues and 32 adjacent
normal tissues were sourced from The Cancer Genome Atlas (TCGA)
database (portal.gdc.cancer.gov/), where ‘STAD’ was searched to
download RNA-Seq expression and clinical data for GC patients.
Cases with gene expression of ‘0’, lost-visit cases, and cases with
incomplete clinical information were excluded by R language
(version 4.2.1). Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/) was used
to compare ELF1 mRNA expression levels in GC and normal tissues.
GEPIA is an interactive web server that can analyze RNA sequencing
expression data from TCGA and GTEx for tumor and normal samples and
can be used for numerous analyses, such as differential analysis,
characterization based on cancer type or pathological stage,
survival analysis, correlation analysis and downscaling analysis.
The specific settings in ‘Expression DIY’ (Boxplot) were as
follows: i) Gene symbol, ELF1; ii) Datasets Selection (Cancer
name), stomach adenocarcinoma (STAD); iii) |Log2fold
change (FC)| Cutoff, 1; iv) P-value cut-off, 0.01; and v) Matched
Normal data, Match TCGA normal and GTEx data. The Strawberry Perl
software (version 5.38.2, perl.org/get.html) and R language (version 4.2.1) with
Limma package (Log2FC filter >1; adjusted P-value
filter=0.05; Wilcox test) were used to obtain the ELF1 mRNA
expression differentiation. Strawberry Perl software was used to
convert probe names from transcriptome files to gene names, and
‘Limma’ was used for analyzing chip data.
GEPIA was also utilized for various correlation
analyses, including the analysis of correlations between ELF1 and
expressions of GC molecular typing-related genes, including MutL
homolog 1 (MLH1), erythroblastic leukemia viral oncogene homolog 2
(ERBB2), PI3K subunit α (PIK3CA) and p53(29). EMT-related molecules, including
MMP9, Cadherin 1 (CDH1) and TIMP1; and angiogenic indices,
including vascular endothelial growth factor A (VEGFA) and kinase
insert domain receptor (KDR). ‘Correlation Analysis’ was chosen on
the database. Then ELF1 was designated as ‘Gene A’ for the x-axis,
whilst other genes to be studied served as ‘Gene B’ for the y-axis.
‘Pearson’ was selected as ‘Correlation Coefficient’. STAD tumor was
selected from ‘TCGA Tumor’ dialog box to ‘Used Expression
Datasets’.
Tumor Immune Estimation Resource 2.0 (TIMER)
(https://cistrome.shinyapps.io/timer/), a web server
for the comprehensive analysis of infiltrated immune cells, is a
resource for the systematic analysis of immune infiltrates among
the different cancer types. The abundance of six infiltrated immune
cell types (B cells, CD4+ T cells, CD8+ T cells, neutrophils,
macrophages and dendritic cells) can be estimated using the TIMER
algorithm (30). Users can
conveniently access tumor immunological, clinical and genomic
features by inputting function-specific parameters. ‘Gene’ module
was chosen to visualize the scatter plots visualizing the
correlation between ELF1 expression and different levels of
infiltration by immune cell types in GC. ‘Spearman’ was selected as
the ‘Correlation Coefficient’.
Cell-type Identification By Estimating Relative
Subsets Of RNA Transcript (CIBERSORT; version 2023.1.2, cibersort.stanford.edu/), a software for the
deconvolution of transcriptome expression matrices to estimate the
composition and abundance of immune components in mixed cells on
the basis of linear support vector regression principles, was
further applied to study the correlation of 22 immune cell scores
with ELF1 expression levels (31).
CIBERSORT was used to analyze immune cell content of each sample,
those with a CIBERSORT output of P<0.05 were considered accurate
and enrolled for further construction of the immune landscape,
otherwise, samples were eliminated. Subsequently the correlation of
ELF1 with these immune cells was analyzed.
Statistical analysis
Epidata 3.1 software (epidata.dk;
Epidata Association from Denmark, was utilized to manage clinical
and laboratory data. SPSS 21.0 (IBM Corp.) and GraphPad 5.0
software (Dotmatics) were used for data analysis. Wilcoxon
signed-rank non-parametric test was used to analyze the
experimental data from RT-qPCR, as it was non-normally distributed.
MedCalc software (version 18.2.1, MedCalc Software Ltd.) was used
for receiver operating characteristic (ROC) curve analysis to
estimate the diagnostic value of ELF1. Categorical variables in IHC
were compared by the χ2 or Fisher exact tests, as
appropriate. Survival curves were plotted by using the Kaplan-Meier
method and tested through the log-rank test. Cox regression
provided the hazard ratio (HR) and 95% CI in all prognostic
analyses. P<0.05 was considered to indicate a statistically
significant difference, association or correlation.
Results
Expression level of ELF1 mRNA in GC
tissues
Examination in GEPIA revealed increased ELF1 mRNA
expression in GC compared with normal gastric tissue (P<0.05;
Fig. 1A). This trend was supported
by results reported by the R4.2.1 software based on data from TCGA
database (Fig. 1B and C). RT-qPCR assay was next performed to
measure ELF1 mRNA expression in GC tissues, with an amplification
efficiency of 99.33%. ELF1 mRNA expression in GC tissues was found
to be significantly higher compared with that in adjacent normal
tissues (P<0.05; Fig. 1D).
Increased ELF1 mRNA expression was also revealed to be a reliable
indicator for distinguishing GC tissues from normal adjacent
tissues, as evidenced by an area under the curve of 0.8078 and a
95% CI of 0.7015-0.9141. The sensitivity and specificity of this
differentiation were 87.5 and 77.5%, respectively (P<0.05;
Fig. 1E).
Expression levels of ELF1 and MMP9
proteins in GC tissues
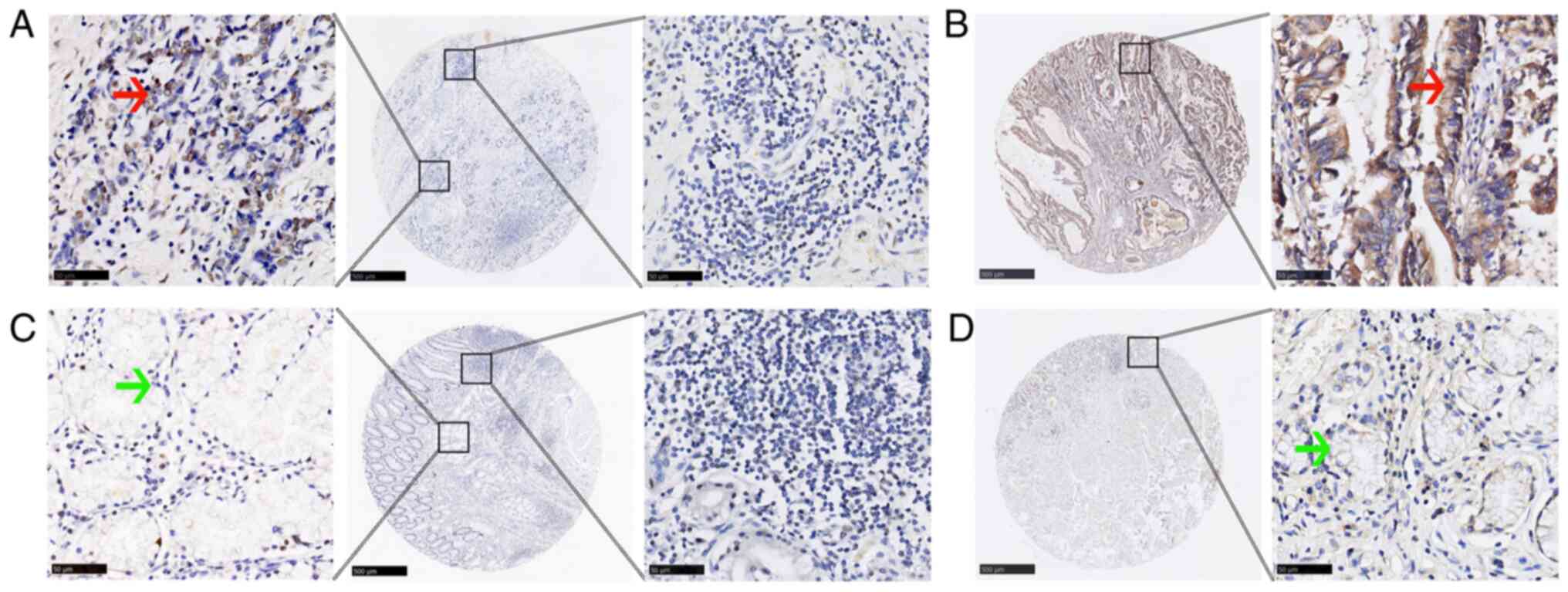
TMA-IHC revealed that the incidence of ELF1 (mainly
localized to the nucleus) and MMP9 (mainly localized to the
cytoplasm) positivity were statistically significantly higher in GC
tissues compared with that in adjacent normal tissues (both
P<0.001; Fig. 2 and Table I). However, no significant
difference in the incidence of positive ELF1 expression in
lymphocytes in the mesenchyme of GC tissues and adjacent normal
tissues could be found (Fig. 2;
Table I).
 | Table IExpression of ELF1 and MMP9 by
immunohistochemistry in GC and adjacent normal tissues (N=355). |
Table I
Expression of ELF1 and MMP9 by
immunohistochemistry in GC and adjacent normal tissues (N=355).
| Characteristic | Negative | Positive | P-value |
|---|
| GC | 103 (29.0) | 252 (71.0) | <0.001 |
| Adjacent
normal | 245 (69.0) | 110 (31.0) | |
| Characteristic | Negative | Positive | P-value |
| GC | 129 (36.3) | 226 (63.7) | <0.001 |
| Adjacent
normal | 257 (72.4) | 98 (27.6) | |
| Characteristic | Negative | Positive | P-value |
| GC | 241 (67.9) | 114 (32.1) | 0.099 |
| Adjacent
normal | 140 (62.0) | 215 (38.0) | |
Correlations of ELF1 and MMP9 protein
expression levels, their associations and clinical characteristics
in GC
TMA-IHC revealed a positive association between the
incidence of ELF1 and MMP9 positivity in GC tissues (P<0.001;
Table II). ELF1 and MMP9 protein
expression levels were also found to associate with T (both
P<0.001), N (both P<0.001), M (ELF1, P=0.047; MMP9, P=0.001),
TNM staging (both P<0.001), MVI (ELF1, P=0.040; MMP9, P=0.008),
lymphatic invasion (ELF1, P=0.015; MMP9, P=0.017) and blood serum
CEA levels (ELF1, P=0.002; MMP9, P<0.001; Table III). However, ELF1 expression in
lymphocytes in the GC mesenchyme was not associated found to be
with any of the clinicopathological parameters of patients with GC
(Table III).
 | Table IIAssociation between ELF1 and MMP9
protein expression positivity in tissue microarray according to
immunohistochemistry in patients with gastric cancer. |
Table II
Association between ELF1 and MMP9
protein expression positivity in tissue microarray according to
immunohistochemistry in patients with gastric cancer.
| | MMP9 | |
|---|
| ELF1 | - | + | P-value |
|---|
| - | 90 | 13 | <0.001 |
| + | 39 | 213 | |
 | Table IIIAssociation of ELF1 and MMP9
expression by tissue microarray-immunohistochemistry with clinical
characteristics in patients with GC. |
Table III
Association of ELF1 and MMP9
expression by tissue microarray-immunohistochemistry with clinical
characteristics in patients with GC.
| A, ELF1
expression |
|---|
| Characteristic | N | Negative | Positive | P-value |
|---|
| Total | 355 | 103 (29.0) | 252 (71.0) | |
| Sex | | | | 0.204 |
|
Male | 241 | 75 (31.1) | 166 (68.9) | |
|
Female | 114 | 28 (24.6) | 86 (75.4) | |
| Age | | | | 0.680 |
|
≤60 | 109 | 30 (27.5) | 79 (72.5) | |
|
>60 | 246 | 73 (29.7) | 173 (70.3) | |
| Histological
type | | | | 0.212 |
|
Tubular | 246 | 71 (28.9) | 175 (71.1) | |
|
Papillary | 7 | 2 (28.6) | 5 (71.4) | |
|
Mucinous | 25 | 3 (12.0) | 22 (88.0) | |
|
Mixed
(tubular and mucinous) | 5 | 1 (20.0) | 4 (80.0) | |
|
Signet ring
cell | 72 | 26 (36.1) | 46 (63.9) | |
|
Differentiation | | | | 0.158 |
|
Well | 55 | 14 (25.5) | 41 (74.5) | |
|
Middle | 166 | 50 (30.1) | 116 (69.9) | |
|
Poor | 104 | 35 (33.7) | 69 (66.3) | |
|
Others | 30 | 4 (13.3) | 26 (86.7) | |
| T | | | | <0.001 |
|
Tis | 6 | 5 (83.3) | 31 (16.7) | |
|
1 | 31 | 19 (61.3) | 12 (38.7) | |
|
2 | 60 | 23 (38.3) | 37 (61.7) | |
|
3 | 238 | 55 (23.1) | 183 (76.9) | |
|
4 | 20 | 1 (5.0) | 19 (95.0) | |
| N | | | | <0.001 |
|
0 | 145 | 58 (40.0) | 87 (60.0) | |
|
1 | 55 | 16 (29.1) | 39 (70.9) | |
|
2 | 84 | 19 (22.6) | 65 (77.4) | |
|
3 | 71 | 10 (14.1) | 61 (85.9) | |
| M | | | | 0.047 |
|
0 | 339 | 102 (30.1) | 237 (69.9) | |
|
1 | 16 | 1 (6.3) | 15 (93.8) | |
| TNM stage | | | | <0.001 |
|
0+1 | 42 | 22 (52.4) | 20 (47.6) | |
|
2 | 115 | 52 (45.2) | 63 (54.8) | |
|
3 | 182 | 28 (15.4) | 154 (84.6) | |
|
4 | 16 | 1 (6.3) | 15 (93.7) | |
| MVI | | | | 0.040 |
|
No | 201 | 67 (33.3) | 134 (66.7) | |
|
Yes | 154 | 36 (23.4) | 118 (76.6) | |
| Lymphatic
invasion | | | | 0.015 |
|
No | 269 | 87 (32.3) | 182 (67.7) | |
|
Yes | 86 | 16 (18.6) | 70 (81.4) | |
| Perineural
invasion | | | | 0.628 |
|
No | 318 | 91 (28.6) | 277 (71.4) | |
|
Yes | 37 | 12 (32.4) | 25 (67.6) | |
| CEA (ng/ml) | | | | 0.002 |
|
≤5 | 122 | 48 (39.3) | 74 (60.7) | |
|
>5 | 219 | 52 (23.7) | 167 (76.3) | |
|
Unknown | 14 | 3 (21.4) | 11 (78.6) | |
| CA 19-9 (U/ml) | | | | 0.232 |
|
≤37 | 191 | 61 (31.9) | 130 (68.1) | |
|
>37 | 150 | 39 (26.0) | 111 (74.0) | |
|
Unknown | 14 | 3 (21.4) | 11 (78.6) | |
| Laurén
classification | | | | 0.704 |
|
Intestinal
type | 260 | 74 (28.5) | 186 (71.5) | |
|
Diffuse
type | 95 | 29 (30.5) | 66 (69.5) | |
| B, MMP9
expression |
| Characteristic | N | Negative, N
(%) | Positive, N
(%) | P-value |
| Total | 355 | 129 (36.3) | 226 (63.7) | |
| Sex | | | | 0.418 |
|
Male | 241 | 91 (37.8) | 150 (62.2) | |
|
Female | 114 | 38 (33.3) | 76 (66.7) | |
| Age | | | | 0.884 |
|
≤60 | 109 | 39 (35.8) | 70 (64.2) | |
|
>60 | 246 | 90 (36.6) | 156 (63.4) | |
| Histological
type | | | | 0.097 |
|
Tubular | 246 | 85 (34.6) | 161 (65.4) | |
|
Papillary | 7 | 4 (57.1) | 3 (42.9) | |
|
Mucinous | 25 | 5 (20.0) | 20 (80.0) | |
|
Mixed
(tubular and mucinous) | 5 | 2 (40.0) | 3 (60.0) | |
|
Signet ring
cell | 72 | 33 (45.8) | 39 (54.2) | |
|
Differentiation | | | | 0.200 |
|
Well | 55 | 20 (36.4) | 35 (63.6) | |
|
Middle | 166 | 57 (34.3) | 109 (65.7) | |
|
Poor | 104 | 45 (43.3) | 59 (56.7) | |
|
Others | 30 | 7 (23.3) | 23 (76.7) | |
| T | | | | <0.001 |
|
Tis | 6 | 5 (83.3) | 1 (16.7) | |
|
1 | 31 | 23 (74.2) | 8 (25.8) | |
|
2 | 60 | 33 (55.0) | 27 (45.0) | |
|
3 | 238 | 66 (27.7) | 172 (72.3) | |
|
4 | 20 | 2 (10.0) | 18 (90.0) | |
| N | | | | <0.001 |
|
0 | 145 | 73 (50.3) | 72 (49.7) | |
|
1 | 55 | 19 (34.5) | 36 (65.5) | |
|
2 | 84 | 22 (26.2) | 62 (73.8) | |
|
3 | 71 | 15 (21.1) | 56 (78.9) | |
| M | | | | 0.001 |
|
0 | 339 | 129 (38.1) | 210 (61.9) | |
|
1 | 16 | 0 (0.0) | 16 (100.0) | |
| TNM stage | | | | <0.001 |
|
0+1 | 42 | 27 (64.3) | 15 (35.7) | |
|
2 | 115 | 61 (53.0) | 54 (47.0) | |
|
3 | 182 | 41 (22.5) | 141 (77.5) | |
|
4 | 16 | 0 (0.0) | 16 (100.0) | |
| MVI | | | | 0.008 |
|
No | 201 | 85 (42.3) | 116 (57.7) | |
|
Yes | 154 | 44 (28.6) | 110 (71.4) | |
| Lymphatic
invasion | | | | 0.017 |
|
No | 269 | 107 (39.8) | 162 (60.2) | |
|
Yes | 86 | 22 (25.6) | 64 (74.4) | |
| Perineural
invasion | | | | 0.108 |
|
No | 318 | 120 (37.7) | 198 (62.3) | |
|
Yes | 37 | 9 (24.3) | 28 (75.7) | |
| CEA (ng/ml) | | | | <0.001 |
|
≤5 | 122 | 60 (49.2) | 62 (50.8) | |
|
>5 | 219 | 64 (29.2) | 155 (70.8) | |
|
Unknown | 14 | 5 (35.7) | 9 (64.3) | |
| CA 19-9 (U/ml) | | | | 0.017 |
|
≤37 | 191 | 80 (41.9) | 111 (58.1) | |
|
>37 | 150 | 44 (29.3) | 106 (70.7) | |
|
Unknown | 14 | 5 (35.7) | 9 (64.3) | |
| Laurén
classification | | | | 0.386 |
|
Intestinal
type | 260 | 91 (35.0) | 169 (65.0) | |
|
Diffuse
type | 95 | 38 (40.0) | 57 (60.0) | |
| C, ELF1 expression
in lymphocytes |
| Characteristic | N | Negative | Positive | P-value |
| Total | 355 | 241 (67.9) | 114 (32.1) | |
| Sex | | | | 0.525 |
|
Male | 241 | 161 (66.8) | 80 (33.2) | |
|
Female | 114 | 80 (70.2) | 34 (29.8) | |
| Age | | | | 0.459 |
|
≤60 | 109 | 77 (70.6) | 32 (29.4) | |
|
>60 | 246 | 164 (66.7) | 82 (33.3) | |
| Histological
type | | | | 718 |
|
Tubular | 246 | 165 (67.1) | 81 (32.9) | |
|
Papillary | 7 | 5 (71.4) | 2 (28.6) | |
|
Mucinous | 25 | 20 (80.0) | 5 (20.0) | |
|
Mixed
(tubular and mucinous) | 5 | 4 (80.0) | 1 (20.0) | |
|
Signet ring
cell | 72 | 47 (65.3) | 25 (34.7) | |
|
Differentiation | | | | 0.305 |
|
Well | 55 | 40 (72.7) | 15 (27.3) | |
|
Middle | 166 | 111 (66.9) | 55 (33.1) | |
|
Poor | 104 | 66 (63.5) | 38 (36.5) | |
|
Others | 30 | 24 (80.0) | 6 (20.0) | |
| T | | | | 0.988 |
|
Tis | 6 | 4 (66.7) | 2 (33.3) | |
|
1 | 31 | 22 (71.0) | 9 (29.0) | |
|
2 | 60 | 40 (66.7) | 20 (33.3) | |
|
3 | 238 | 162 (68.1) | 76 (31.9) | |
|
4 | 20 | 13 (65.0) | 7 (35.0) | |
| N | | | | 0.699 |
|
0 | 145 | 102 (70.3) | 43 (29.7) | |
|
1 | 55 | 34 (61.8) | 21 (38.2) | |
|
2 | 84 | 56 (66.7) | 28 (33.3) | |
|
3 | 71 | 49 (69.0) | 22 (31.0) | |
| M | | | | 0.104 |
|
0 | 339 | 227 (67.0) | 112 (33.0) | |
|
1 | 16 | 14 (87.5) | 2 (12.5) | |
| TNM stage | | | | 0.296 |
|
0+1 | 42 | 29 (69.0) | 13 (31.0) | |
|
2 | 115 | 80 (69.6) | 35 (30.4) | |
|
3 | 182 | 118 (64.8) | 64 (35.2) | |
|
4 | 16 | 14 (87.5) | 2 (12.5) | |
| MVI | | | | 0.723 |
|
No | 201 | 138 (68.7) | 63 (31.3) | |
|
Yes | 154 | 103 (66.9) | 51 (33.1) | |
| Lymphatic
invasion | | | | 0.487 |
|
No | 269 | 180 (66.9) | 89 (33.1) | |
|
Yes | 86 | 61 (70.9) | 25 (29.1) | |
| Perineural
invasion | | | | 0.743 |
|
No | 318 | 215 (67.6) | 103 (32.4) | |
|
Yes | 37 | 26 (70.3) | 11 (29.7) | |
| CEA (ng/ml) | | | | 0.756 |
|
≤5 | 122 | 81 (66.4) | 41 (33.6) | |
|
>5 | 219 | 149 (68.0) | 70 (32.0) | |
|
Unknown | 14 | 11 (78.6) | 3 (21.4) | |
| CA 19-9 (U/ml) | | | | 0.175 |
|
≤37 | 191 | 123 (64.4) | 68 (35.6) | |
|
>37 | 150 | 107 (71.3) | 43 (28.7) | |
|
Unknown | 14 | 11 (78.6) | 3 (21.4) | |
| Laurén
classification | | | | 0.896 |
|
Intestinal
type | 260 | 176 (67.7) | 84 (32.3) | |
|
Diffuse
type | 95 | 65 (68.4) | 30 (31.6) | |
Association of ELF1 and MMP9
expression with the prognosis of patients with GC
Kaplan-Meier survival analysis revealed that
patients with GC and elevated levels of ELF1 and MMP9 protein
expression both faced significantly inferior overall survival (OS)
compared with that in patients with lower expression levels of ELF1
and MMP9 proteins (P<0.001 for both; Fig. 3A and B). Specifically, OS stood at 17.1 and
11.5% in patients with high ELF1 and MMP9 expression levels,
respectively. This is in contrast to that in patients with lower
expression levels of ELF1 and MMP9 proteins, where the OS was
observed to be 72.8% for ELF1 and 71.3% for MMP9 (Fig. 3A and B). The lowest OS, which was 10.8%, was
observed in patients with simultaneously high expression levels of
ELF1 and MMP9 (Fig. 3C). In
addition, OS decreased with increasing TNM stage (P<0.001;
Fig. 3D). Further prognostic
analysis using the Cox regression model showed that high ELF1
expression (HR, 2.555; 95% CI, 1.546-4.224; P=0.002), high MMP9
expression (HR, 3.813; 95% CI, 2.406-6.041; P<0.001), advanced
TNM stage (P=0.001) and advanced N stage (P=0.011) were independent
prognostic factors for patients with GC (Table IV). However, there was no
significant association between ELF1 expression in lymphocytes in
the GC mesenchyme and the prognosis of patients with GC (Table IV).
 | Table IVCox regression analysis of prognostic
factors for 5-year survival from gastric cancer. |
Table IV
Cox regression analysis of prognostic
factors for 5-year survival from gastric cancer.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Parameter | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| ELF1 expression,
high vs. low + none | 5.519
(3.707-8.217) | <0.001 | 2.555
(1.546-4.224) | 0.002 |
| MMP9 expression,
high vs. low + none | 6.125
(4.291-8.744) | <0.001 | 3.813
(2.406-6.041) | <0.001 |
| Age, ≤60 vs. >60
years | 0.868
(0.662-1.138) | 0.306 | | |
| Sex, male vs.
female | 0.978
(0.745-1.283) | 0.870 | | |
| Histological
type | | 0.043 | | 0.803 |
|
Tubular vs.
papillary | 0.533
(0.170-1.669) | 0.280 | 1.030
(0.315-3.365) | 0.961 |
|
Tubular vs.
mucinous | 1.365
(0.849-2.194) | 0.199 | 1.108
(0.672-1.828) | 0.687 |
|
Tubular vs.
mixed (tubular + mucinous) | 2.023
(0.829-4.936) | 0.121 | 0.900
(0.322-2.517) | 0.841 |
|
Tubular vs.
signet ring cells | 0.716
(0.509-1.007) | 0.055 | 0.811
(0.561-1.172) | 0.266 |
|
Differentiation | | 0.064 | | |
|
Well vs.
middle | 1.075
(0.744-1.553) | 0.699 | | |
|
Well vs.
poor | 0.828
(0.553-1.241) | 0.361 | | |
|
Well vs.
others | 1.557
(0.934-2.596) | 0.090 | | |
| TNM stage | | <0.001 | | 0.001 |
|
0+1 vs.
2 | 2.992
(1.579-5.667) | 0.001 | 4.440
(1.966-10.029) | <0.001 |
|
0+1 vs.
3 | 5.380
(2.908-9.956) | <0.001 | 4.983
(2.022-12.276 | <0.001 |
|
0+1 vs.
4 | 12.030
(5.494-26.345) | <0.001 | 8.796
(3.031-25.521) | <0.001 |
| T trend | | <0.001 | | 0.179 |
|
Tis vs.
1 | 0.550
(0.146-2.072) | 0.377 | 0.290
(0.070-1.197) | 0.087 |
|
Tis vs.
2 | 1.406
(0.431-4.585) | 0.572 | 0.365
(0.102-1.302) | 0.120 |
|
Tis vs.
3 | 2.403
(0.767-7.525) | 0.132 | 0.255
(0.069-0.935) | 0.039 |
|
Tis vs.
4 | 3.243
(0.944-11.143) | 0.062 | 0.219
(0.053-0.914) | 0.037 |
| N trend | | <0.001 | | 0.011 |
|
0 vs. 1 | 2.200
(1.512-3.201) | <0.001 | 1.452
(0.958-2.203) | 0.079 |
|
0 vs. 2 | 2.750
(1.968-3.842) | <0.001 | 1.909
(1.275-2.859) | 0.002 |
|
0 vs. 3 | 2.236
(1.562-3.200) | <0.001 | 1.273
(0.821-1.971) | 0.280 |
| M, 0 vs. 1 | 3.159
(1.866-5.348) | <0.001 | 0.919
(0.481-1.757) | 0.799 |
| MVI, no vs.
yes | 1.346
(1.043-1.737) | 0.023 | 0.946
(0.718-1.246) | 0.692 |
| Lymphatic invasion,
no vs. yes | 1.275
(0.953-1.707) | 0.102 | | |
| Perineural
invasion, no vs. yes | 1.316
(0.875-1.978) | 0.187 | | |
| Carcinoembryonic
antigen level, ≤5 vs. >5 ng/ml | 1.645
(1.234-2.193) | 0.001 | 0.690
(0.460-1.037) | 0.074 |
| Carbohydrate
antigen 19-9 level, ≤37 vs. >37 U/ml | 1.268
(0.977-1.646) | 0.074 | | |
| Laurén
classification, intestinal type vs. diffuse type | 0.836
(0.622-1.125) | 0.238 | | |
| ELF1 expression in
lymphocytes, high vs. low and none | 1.015
(0.772-1.334) | 0.918 | | |
Gene correlation analysis
Investigations using the GEPIA database revealed
notable positive associations between ELF1 expression and several
crucial genes involved in GC occurrence (32). ELF1 expression showed a mild
positive association with MLH1 (r=0.35; P<0.001), PIK3CA (r=0.4;
P<0.001) and CDH1 (r=0.39; P<0.001; Fig. 4A-C), but not with ERBB2 (r=0.16),
TP53 (r=0.1), MMP9 (r=0.12), TIMP1 (r=-0.13), VEGFA (r=0.19) and
KDR (r=0.16; Fig. 4D-I). These
findings suggest an association between ELF1 expression and the GC
molecular subtypes, implying roles in various processes, such as
EMT and angiogenesis.
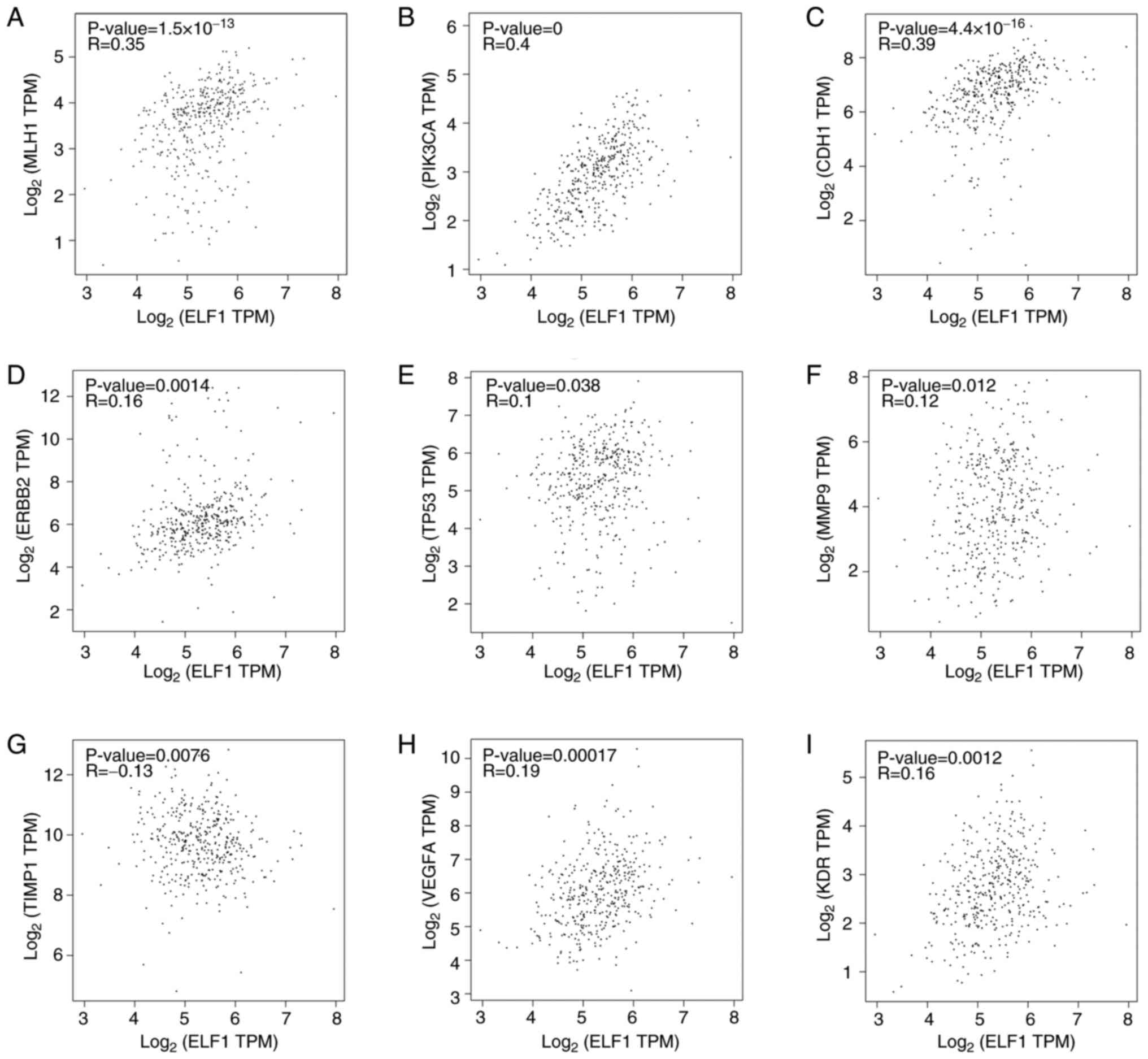 | Figure 4Correlation between ELF1 expression
and that of MLH1, ERBB2, PIK3CA, TP53, MMP9, CDH1, TIMP1, VEGFA and
KDR in GC tissues according to the GEPIA database. Correlation
between ELF1 and (A) MLH1, (B) PIK3CA, (C) CDH1, (D) ERBB2, (E)
TP53, (F) MMP9, (G) TIMP1, (H) VEGFA and (I) KDR. TPM, transcripts
per kilobase of exon model per million mapped reads; ELF1, E74-like
Factor 1; MLH1, MutL homolog 1; ERBB2, erythroblastic leukemia
viral oncogene homolog 2; PIK3CA, PI3K subunit α; CDH1, cadherin 1;
TIMP1, tissue inhibitors of metalloproteinases; KDR, kinase insert
domain receptor. |
Association between ELF1 expression
and immune cell infiltration into GC
Infiltrating immune cell types into tumor tissues
mainly include B-cells (CD19+), T-cells (CD3+, CD4+ and CD8+) and
natural killer cells (NKs, CD56+) (33,34).
The association between ELF1 expression and the infiltrating immune
cells in GC was examined. TIMER database was used as an initial
analysis to assess the correlation between the degree of
infiltration by various immune cell types and ELF1 expression
levels. This examination revealed no correlations between ELF1
expression and the levels of infiltration by B cells (r=0.187),
CD8+ T cells (r=-0.017), CD4+ T cells (r=0.169), Macrophage
(r=0.058), neutrophil (r=0.039) or dendritic cells (r=0.067;
Fig. 5A). Additionally, by using
the CIBERSORT software, the relationship between ELF1 expression
and infiltration by 24 subsets of different immune cell types
integral to tumor immunity was evaluated. The outcomes revealed
marked variations in several immune cell types across groups with
differing ELF1 expression levels. In particular, ELF1 had a
positive association with naive B cells (P=0.028), memory-activated
CD4+ T cells (P=0.009), γδ T cells (P=0.029) and activated NK cells
(P=0.001), but no relationship with others. (Fig. 5B).
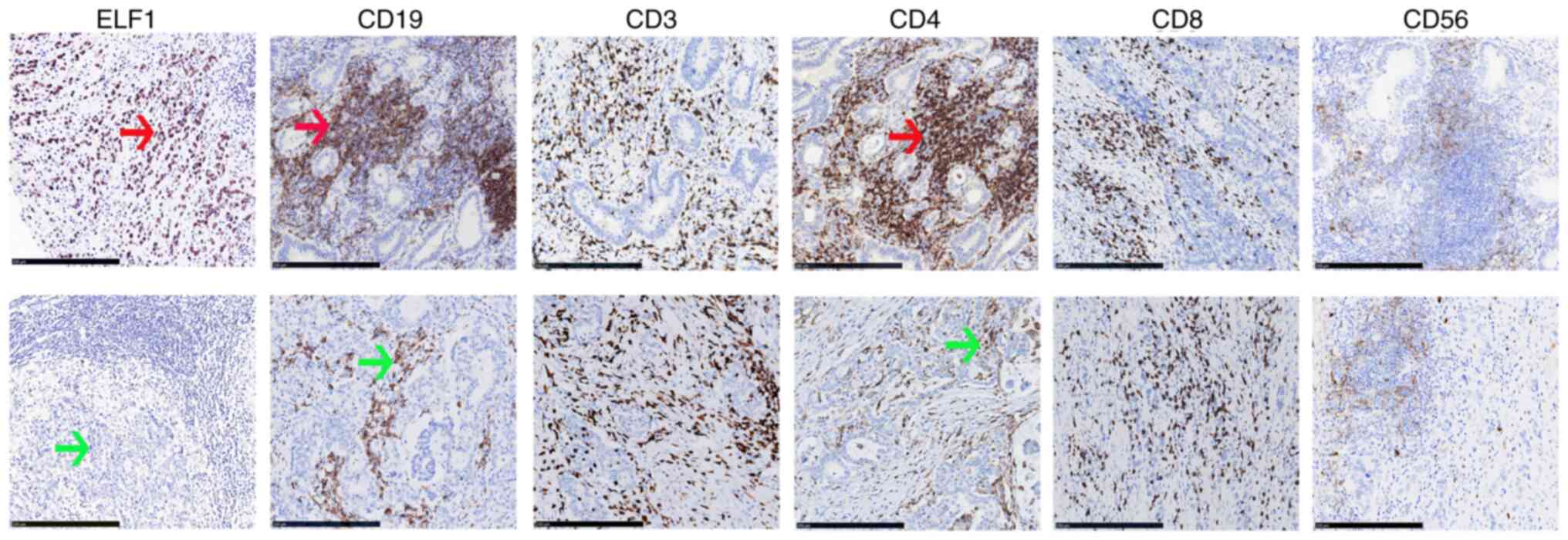
IHC was next used to further validate the
relationship between ELF1 expression and infiltrating immune cells
in GC. The results revealed that ELF1 had associations with CD19
(B-cells; P<0.001) and CD4 (CD4+ T cells; P=0.002), but not with
CD3, CD8 or CD56 (Table V;
Fig. 6).
 | Table VAssociation between ELF1 expression
by tissue microarray-immunohistochemistry and that of different
immune cell markers in GC. |
Table V
Association between ELF1 expression
by tissue microarray-immunohistochemistry and that of different
immune cell markers in GC.
| | CD19 | CD3 | CD4 | CD8 | CD56 |
|---|
| ELF1 | - | + | - | + | - | + | - | + | - | + |
|---|
| - | 93 | 10 | 70 | 33 | 69 | 34 | 62 | 41 | 71 | 32 |
| + | 113 | 139 | 167 | 85 | 124 | 128 | 160 | 92 | 169 | 83 |
| P-value | <0.001 | | 0.759 | | 0.002 | | 0.560 | | 0.733 | |
Discussion
GC is a common malignant tumor worldwide and is
characterized by high incidence and mortality rates, with
multifactorial associations encompassing dietary habits (including
high-fat intake, elevated salt consumption, and smoking),
infections (such as Helicobacter pylori and Epstein-Barr
virus infections) and genetic predispositions (35,36).
Familial clustering is discernible in GC development, with ~25% of
autosomal dominant diffuse GC-prone families harboring E-cadherin
mutations (37). The aggressive
and metastatic attributes of GC are closely associated with the
poor patient prognosis, with a median survival of <12 months
(38). Although significant
improvements have been made in identifying biomarkers and molecular
targeted agents (trastuzumab, Bevacizumab, Panitumumab, Everolimus,
etc.) for improving patient prognosis, such as epidermal growth
factor receptor (EGFR), vascular endothelial growth factor (VEGF)
(39), mTOR and PI3K (40), predictive biomarkers and
corresponding targeted agents specifically addressing GC invasion
and metastasis remain scarce.
Ets genes, known for their erythroblast
transformation specificity, constitute a class of highly conserved
oncogenes that serve important roles in regulating tumor
infiltration and metastasis (41).
Among the genes in the Ets transcription factor family, ELF1 is one
of the most prominent members (42). ELF1 is a T cell-specific
transcription factor that was originally cloned from a human T-cell
library through hybridization to a probe encoding the DNA-binding
structural domain (Ets structural domain) of human Ets-1 cDNA. This
transcription factor mainly exerts its influence by binding to gene
promoters or enhancers (42) and
can function to either activate and repress the expression of
target genes. In addition, it is unique in being subject to
glycosylation and phosphorylation (10). Elevated levels of ELF1 expression
have been previously associated with the initiation and progression
of various malignancies, such as pancreatic and colon cancer
(13,43). The high expression of ELF1 has been
associated with poor prognosis in patients with endometrial
(44) and ovarian cancer (45). ELF1 can also promote the malignant
progression of glioma (46). In
addition, ELF1 has been found to promote the proliferation of oral
squamous cell carcinoma cells by increasing the expression of
β-catenin mRNA (47). By contrast,
ELF1 can activate the doublecortin-like kinase 1/Janus kinase/STAT
signaling pathway, thereby promoting the malignant progression of
pancreatic cancer (13). Another
recent study has elucidated ELF1 effect on the biological behavior
of colon cancer, which was by modulating the transcriptional
activation of serine peptidase inhibitor kazal type 4, thereby
promoting colon cancer progression (43). However, conflicting reports exist
that also suggest ELF1 to serve an inhibitory role in certain
cancers, such as prostate cancer (48) and Hodgkin's lymphoma (49).
The extracellular matrix (ECM) is divided into
basement membrane and interstitial matrix, and is composed of
various proteins such as type IV collagen, laminin, elastin, and
hyaluronic acid, which provide structural support for cells and are
also involved in the development of epithelial cells. The
composition and stability of the ECM is closely related to the
protein-protein and polysaccharide-protein binding in the
extracellular matrix, which is one of the main barriers to prevent
tumor metastasis. Degradation of ECM facilitates tumor cell
invasion and metastasis and is a key trigger of EMT, where MMPs
serve a key role because they can degrade almost all ECM components
(50). The MMP family includes 26
different members, which can then be divided into various subtypes,
including collagenases, gelatinases, matrix proteases and
membrane-type MMPs (22). Previous
studies have shown that MMP9 (a member of the gelatinase family),
which can degrade the ECM and basement membranes, in addition to
being associated with cancer cell adhesion and migration, is an
important marker for predicting poor tumor prognosis in colorectal
cancer (51) and endometrial
carcinoma (44). In addition, MMP9
has been reported to positively correlate with the infiltration of
a diverse range of immune cell types, including Th1 cells,
neutrophils and macrophages, and regulates their transport in uveal
melanoma and clear Cell Renal Cell Carcinoma, which is essential
for establishing and coordinating the tumor immune environment
(52,53). The influence of MMP9 has been
documented to extend to angiogenic and lymphangiogenic factors,
such as VEGF, TGF-β, tumor necrosis factor-α, ΙL-8 and EGFR
(54). Elevated levels of MMP9
mRNA and protein expression have been frequently found in various
cancer types, including ovarian (55), colorectal (22), breast (56) and lung cancers (57). High MMP9 expression is also
consistently associated with advanced tumor stages and adverse
clinical outcomes, underscoring its potential as a prognostic
indicator for patients with cancer of uveal melanoma and breast
cancer (54,56).
Ets family proteins enhance the expression of the
urokinase-type plasminogen activator (uPA) gene by binding to its
inducible enhancer region. uPA in turn activates a variety of MMPs
(MMP1, MMP2 and MMP9) whilst promoting protein hydrolysis in the
ECM (44). ELF1 serves a pivotal
role in tumor angiogenesis, ECM remodeling and metastasis, by
regulating various stages of neovascularization (which encompasses
the generation of proangiogenic factors, MMPs and protease
inhibitors) (58). In addition,
ELF1 overexpression has been documented to contribute to a critical
facet of tumor infiltration (pancreatic and colon cancer), namely
ECM penetration. This was mediated through the transcriptional
control of genes encoding enzymes involved in ECM degradation, such
as MMP9(59). The aforementioned
findings suggest that ELF1 may regulate tumorigenesis and
progression through the TME, which is associated with EMT,
angiogenesis and immune infiltration. The present study revealed
the expression profiles of ELF1 in GC, the possible mechanistic
interplay between ELF1 and MMP9, in addition to their collective
effect on patient survival and prognosis.
Initial analysis using the GEPIA database and R
4.2.1 software indicated elevated ELF1 expression in GC.
Subsequently, RT-qPCR analysis corroborated these findings,
suggesting the diagnostic value of ELF1 expression in
distinguishing GC from adjacent normal tissues. Therefore, it
became possible to initially distinguish GC tissues from normal
tissues based on ELF1 expression. Recent in vitro
experiments showed that ELF1 overexpression directly activated MMP9
promoter activity, which identified an ELF1-triggered
transcriptional mechanism by which neurotrauma upregulated MMP9
expression in the dorsal root ganglion (59). ELF1 has also been shown to activate
forkhead box D3-antisense 1 to promote the migration, invasion and
EMT of osteosarcoma cells by sponging microRNA-296-5p, preventing
the inhibition of zinc finger CCHC-type containing 3(60). Transcription factor ELF1 activates
MEIS1 transcription and promotes MEIS1 expression. Overexpression
of MEIS1 increases growth factor independent protein 1) expression
by activating the GFI1 enhancer, but decreases FBW7 expression and
thus promoting glioma cell proliferation (decreased PCNA),
migration and invasive ability (decreased MMP-9), and reducing
apoptosis (increased capase-3) promotes glioma development
(46). ELF1 is involved in
prostate cancer tumor cell migration and EMT by interfering with
the oncogenic ETS function of ETS/Activator protein-1
cis-regulatory motifs (48). To
elucidate the association between ELF1 and EMT in GC, TMA-IHC was
performed. The results revealed increased levels of ELF1 and MMP9
proteins in GC tissues compared with adjacent normal tissues, where
both exhibited positive associations with key clinical parameters,
including T, N and M stages, TNM staging, MVI, lymphatic invasion
and blood CEA levels. Survival and prognostic analyses also
revealed that patients with high expression levels of ELF1 and MMP9
proteins and advanced TNM staging had shorter survival and poorer
prognoses. The findings of IHC revealed a positive correlation
between the expression levels of ELF1 and MMP9. These findings
suggest that high ELF1 expression may serve an important role in
the pathogenesis and malignant progression of GC, which can be
exploited to predict the survival and prognosis of patients with
GC. However, the specific mechanistic interactions between ELF1 and
MMP9 remain unclear, which require further study.
Based on heterogeneity of gastric cancer and the
rapid development of molecular biology, TCGA proposed a molecular
classification system for gastric cancer by analyzing data from
multiple platforms, consisting of four distinct subtypes, including
Epstein-Barr virus+ (accompanied by PIK3CA and ERBB2
amplification), microsatellite instability (usually accompanied by
MLH1 silence, high-frequency mutation of ERBB2 and PIK3CA),
genomically stable (accompanied by CDH1 gene mutation) and
chromosome instability (accompanied by high mutation rates of TP53
and ERBB2 amplification (30).
TIMP1, an endogenous inhibitor of MMP-9, executes a role not only
in impeding the matrix degradation activity of MMP-9 whilst also
activating pivotal cytokines, including VEGF, TGF-α,TGF-β
(transforming growth factor-β) and CAM (cell adhesion molecule),
binding cell surface protein CD63, activating FAK-PI3K/AKT (focal
adhesion kinase-phosphatidylinositol 3 kinase/protein kinase B)
signaling pathway and MAPK) signaling pathway, leading to tumor
cell proliferation (61). VEGFA
and KDR are involved in tumor angiogenesis in addition to molecular
subtype identification of GC (62,63).
The CDH1 gene encodes the vital cell adhesion molecule E-cadherin,
which is crucial for maintaining epithelial tissue integrity. E-CAD
reduction is associated with EMT), generation of stem-cell, and
metastasis (64,65). Analysis of the GEPIA database in
the present study revealed associations of ELF1 with MLH1, PIK3CA
and CDH1. ELF1 has been previously reported to serve an
indispensable role in the survival, differentiation and maturation
of T and B lymphocyte cells, especially NK/T cells, where the
absence of which results in T cell apoptosis (66). Findings from the TIMER database,
CIBERSORT and IHC in the present study unveiled associations
between ELF1 expression and B cells and CD4+ T cells.
CD4+ and CD8+ T cells contribute to tumor growth and
became effective targets for cancer survival, prognosis and
treatment in lung cancer (67). B
cells can produce cytokines (e.g., IL-10) that inhibit the
antitumor response of T cells (inhibit CTL-mediated tumor
clearance) and promote the production of immune complexes
(generated by antitumor antibodies), inducing tumorigenesis. Breg
(regulatory B) cells exhibit pro-tumorigenic activity due to a lack
of response to CTLA4, resulting in a shortened overall survival in
cutaneous melanoma, and produce adenosine, which is involved in the
inhibition of T cell activation and/or inactivation influencing
prognosis (such as urothelial bladder and gastric cancer) (68,69).
CD4+ T cells can produce tumor cytokines (such as IL-4), associate
with other cell types (myeloid-derived suppressor cells and
tumor-associated macrophages) and transform into regulatory T
cells, which ultimately exert pro-tumorigenic functions (69,70).
Based on the aforementioned results, we hypothesized that ELF1
appeared to be associated with GC development, which are
intricately intertwined with the TME including EMT, angiogenesis
and immune infiltration.
However, the present study has several limitations.
All the specimens used for this study were selected randomly, and
the size and quality of specimens could not be controlled. The
detection of protein expression using IHC may be affected by tumor
heterogeneity and subjective scoring system analysis. The selected
tumor location may not accurately represent the entire tumor due to
intratumor heterogeneity. In addition, the r-value of the
correlation analysis based on data from the GEPIA database was low,
which require confirmation through further detailed exploration in
subsequent studies. The majority of the studies on the involvement
of ELF1 in EMT and angiogenesis in GC were based on bioinformatics,
where the exact mechanism and related signaling pathways requires
further experimental verification. However, the results may provide
guidance for future prospective clinical trials. Any future studies
should conduct comprehensive and in-depth studies at the
cytological and molecular levels to provide a solid theoretical
foundation for the study of GC molecular targets.
Supplementary Material
Clinical characteristics for the 40
patients with gastric cancer.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Nantong Science
and Technology Program (grant no. JCZ19093).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YW designed the study. Data analysis was performed
by XZ and XR. XZ drafted the article. XZ performed the experiments.
Critical revision of article was done by SZ and YW. All authors
approved the final version of the article. YW and XZ confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The present study complies with the ethical
standards of the Declaration of Helsinki (as revised in 2013) and
was approved by the Ethics Committee of Affiliated Hospital of
Nantong University (approval no. 2018-L042). Written informed
consent was obtained by each patient or his or her family prior to
this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lei ZN, Teng QX, Tian Q, Chen W, Xie Y, Wu
K, Zeng Q, Zeng L, Pan Y, Chen ZS and He Y: Signaling pathways and
therapeutic interventions in gastric cancer. Signal Transduct
Target Ther. 7(358)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yeoh KG and Tan P: Mapping the genomic
diaspora of gastric cancer. Nat Rev Cancer. 22:71–84.
2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sexton RE, Al Hallak MN, Diab M and Azmi
AS: Gastric cancer: A comprehensive review of current and future
treatment strategies. Cancer Metastasis Rev. 39:1179–1203.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cai H, Li M, Deng R, Wang M and Shi Y:
Advances in molecular biomarkers research and clinical application
progress for gastric cancer immunotherapy. Biomark Res.
10(67)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zang YS, Dai C, Xu X, Cai X, Wang G, Wei
J, Wu A, Sun W, Jiao S and Xu Q: Comprehensive analysis of
potential immunotherapy genomic biomarkers in 1000 Chinese patients
with cancer. Cancer Med. 8:4699–4708. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ajani JA, D'Amico TA, Bentrem DJ, Chao J,
Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, et al:
Gastric cancer, version 2.2022, NCCN clinical practice guidelines
in oncology. J Natl Compr Canc Netw. 20:167–192. 2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Thompson CB, Wang CY, Ho IC, Bohjanen PR,
Petryniak B, June CH, Miesfeldt S, Zhang L, Nabel GJ, Karpinski B,
et al: cis-acting sequences required for inducible interleukin-2
enhancer function bind a novel Ets-related protein, Elf-1. Mol Cell
Biol. 12:1043–1053. 1992.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tsokos GC, Nambiar MP and Juang YT:
Activation of the Ets transcription factor Elf-1 requires
phosphorylation and glycosylation: Defective expression of
activated Elf-1 is involved in the decreased TCR zeta chain gene
expression in patients with systemic lupus erythematosus. Ann NY
Acad Sci. 987:240–245. 2003.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Andrews PGP, Kennedy MW, Popadiuk CM and
Kao KR: Oncogenic activation of the human Pygopus2 promoter by
E74-like factor-1. Mol Cancer Res. 6:259–266. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hu M, Li H, Xie H, Fan M, Wang J, Zhang N,
Ma J and Che S: ELF1 transcription factor enhances the progression
of glioma via ATF5 promoter. ACS Chem Neurosci. 12:1252–1261.
2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yang B, Shen F, Zhu Y, Lu W and Cai H:
E74-like ETS transcription factor 1 promotes the progression of
pancreatic cancer by regulating doublecortin-like kinase 1/Janus
kinase/signal transducer and activator of transcription pathway. Am
J Cancer Res. 14:616–629. 2024.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang J, Chen L, Wei W and Mao F: Long
non-coding RNA signature for predicting gastric cancer survival
based on genomic instability. Aging (Albany NY). 15:15114–15133.
2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Xu TP, Wang YF, Xiong WL, Ma P, Wang WY,
Chen WM, Huang MD, Xia R, Wang R, Zhang EB, et al: E2F1 induces
TINCR transcriptional activity and accelerates gastric cancer
progression via activation of TINCR/STAU1/CDKN2B signaling axis.
Cell Death Dis. 8(e2837)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
He DX, Zhang GY, Gu XT, Mao AQ, Lu CX, Jin
J, Liu DQ and Ma X: Genome-wide profiling of long non-coding RNA
expression patterns in anthracycline-resistant breast cancer cells.
Int J Oncol. 49:1695–1703. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zeltz C, Primac I, Erusappan P, Alam J,
Noel A and Gullberg D: Cancer-associated fibroblasts in
desmoplastic tumors: Emerging role of integrins. Semin Cancer Biol.
62:166–181. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Harrison JD and Fielding JW: Prognostic
factors for gastric cancer influencing clinical practice. World J
Surg. 19:496–500. 1995.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Grzesiak M, Kaminska K, Knapczyk-Stwora K
and Hrabia A: The expression and localization of selected matrix
metalloproteinases (MMP-2, -7 and -9) and their tissue inhibitors
(TIMP-2 and -3) in follicular cysts of sows. Theriogenology.
185:109–120. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang Y, Chuang CY, Hawkins CL and Davies
MJ: Activation and inhibition of human matrix metalloproteinase-9
(MMP9) by HOCl, myeloperoxidase and chloramines. Antioxidants
(Basel). 11(1616)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Farina AR and Mackay AR: Gelatinase
B/MMP-9 in tumour pathogenesis and progression. Cancers (Basel).
6:240–296. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Buttacavoli M, Di Cara G, Roz E,
Pucci-Minafra I, Feo S and Cancemi P: Integrated multi-omics
investigations of metalloproteinases in colon cancer: Focus on MMP2
and MMP9. Int J Mol Sci. 22(12389)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Teng Z, Wang S, Yuan H, Wang H, Li J,
Chang X, Zhang Y, Han Z and Wang Y: MMP-9 gene polymorphisms on
cancer risk: An updated systematic review and meta-analysis.
Nucleosides Nucleotides Nucleic Acids. 3:1–24. 2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Fu CK, Chang WS, Tsai CW, Wang YC, Yang
MD, Hsu HS, Chao CY, Yu CC, Chen JC, Pei JS and Bau DT: The
association of MMP9 promoter Rs3918242 genotype with gastric
cancer. Anticancer Res. 41:3309–3315. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Costache S, Sajin M, Wedden S and D'Arrigo
C: A consolidated working classification of gastric cancer for
histopathologists (review). Biomed Rep. 19(58)2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu JY, Peng CW, Yang XJ, Huang CQ and Li
Y: The prognosis role of AJCC/UICC 8th edition staging system in
gastric cancer, a retrospective analysis. Am J Transl Res.
10:292–303. 2018.PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liao Q and Xiong J: YTHDF1 regulates
immune cell infiltration in gastric cancer via interaction with
p53. Exp Ther Med. 27(255)2024.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cancer Genome Atlas Research Network.
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48 (W1):W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Usui G, Matsusaka K, Mano Y, Urabe M,
Funata S, Fukayama M, Ushiku T and Kaneda A: DNA methylation and
genetic aberrations in gastric cancer. Digestion. 102:25–32.
2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Quan Q, Guo L, Huang L, Liu Z, Guo T, Shen
Y, Ding S, Liu C and Cao L: Expression and clinical significance of
PD-L1 and infiltrated immune cells in the gastric adenocarcinoma
microenvironment. Medicine (Baltimore). 102(e36323)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu D, Heij LR, Czigany Z, Dahl E, Lang
SA, Ulmer TF, Luedde T, Neumann UP and Bednarsch J: The role of
tumor-infiltrating lymphocytes in cholangiocarcinoma. J Exp Clin
Cancer Res. 41(127)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Alhalabi MM, Alsayd SA and Albattah ME:
Advanced diffuse gastric adenocarcinoma in young Syrian woman. A
case report. Ann Med Surg (Lond). 78(103728)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tuei VC, Maiyoh GK and Ndombera FT: The
role of infections in the causation of cancer in Kenya. Cancer
Causes Control. 33:1391–1400. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chang ZW, Dong L, Qin YR, Song M, Guo HY
and Zhu QL: Correlations between gastric cancer family history and
ROBO2 and RASSF2A gene methylations. J Cancer Res Ther. 12:597–600.
2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tan AC, Chan DL, Faisal W and Pavlakis N:
New drug developments in metastatic gastric cancer. Therap Adv
Gastroenterol. 11(1756284818808072)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li Z, Zhao Z, Wang C, Wang D, Mao H, Liu
F, Yang Y, Tao F and Lu Z: Association between DCE-MRI perfusion
histogram parameters and EGFR and VEGF expressions in different
lauren classifications of advanced gastric cancer. Pathol Oncol
Res. 27(1610001)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
He Y and Wang X: Identification of
molecular features correlating with tumor immunity in gastric
cancer by multi-omics data analysis. Ann Transl Med.
8(1050)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hsing M, Wang Y, Rennie PS, Cox ME and
Cherkasov A: ETS transcription factors as emerging drug targets in
cancer. Med Res Rev. 40:413–430. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Oettgen P, Akbarali Y, Boltax J, Best J,
Kunsch C and Libermann TA: Characterization of NERF, a novel
transcription factor related to the Ets factor ELF-1. Mol Cell
Biol. 16:5091–5106. 1996.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li T, Jia Z, Liu J, Xu X, Wang H, Li D and
Qiu Z: Transcription activation of SPINK4 by ELF-1 augments
progression of colon cancer by regulating biological behaviors.
Tissue Cell. 84(102190)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Takai N, Miyazaki T, Nishida M, Shang S,
Nasu K and Miyakawa I: Clinical relevance of Elf-1 overexpression
in endometrial carcinoma. Gynecol Oncol. 89:408–413.
2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Takai N, Miyazaki T, Nishida M, Nasu K and
Miyakawa I: The significance of Elf-1 expression in epithelial
ovarian carcinoma. Int J Mol Med. 12:349–354. 2003.PubMed/NCBI
|
|
46
|
Cheng M, Zeng Y, Zhang T, Xu M, Li Z and
Wu Y: Transcription factor ELF1 activates MEIS1 transcription and
then regulates the GFI1/FBW7 axis to promote the development of
glioma. Mol Ther Nucleic Acids. 23:418–430. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Qiao C, Qiao T, Yang S, Liu L and Zheng M:
SNHG17/miR-384/ELF1 axis promotes cell growth by transcriptional
regulation of CTNNB1 to activate Wnt/β-catenin pathway in oral
squamous cell carcinoma. Cancer Gene Ther. 29:122–132.
2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Budka JA, Ferris MW, Capone MJ and
Hollenhorst PC: Common ELF1 deletion in prostate cancer bolsters
oncogenic ETS function, inhibits senescence and promotes docetaxel
resistance. Genes Cancer. 9:198–214. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Paczkowska J, Soloch N, Bodnar M, Kiwerska
K, Janiszewska J, Vogt J, Domanowska E, Martin-Subero JI, Ammerpohl
O, Klapper W, et al: Expression of ELF1, a lymphoid ETS
domain-containing transcription factor, is recurrently lost in
classical Hodgkin lymphoma. Br J Haematol. 185:79–88.
2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Karamanos NK, Theocharis AD, Piperigkou Z,
Manou D, Passi A, Skandalis SS, Vynios DH, Orian-Rousseau V,
Ricard-Blum S, Schmelzer CEH, et al: A guide to the composition and
functions of the extracellular matrix. FEBS J. 288:6850–6912.
2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Peltonen R, Hagström J, Tervahartiala T,
Sorsa T, Haglund C and Isoniemi H: High expression of MMP-9 in
primary tumors and high preoperative MPO in serum predict improved
prognosis in colorectal cancer with operable liver metastases.
Oncology. 99:144–160. 2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wang T, Zhang Y, Bai J, Xue Y and Peng Q:
MMP1 and MMP9 are potential prognostic biomarkers and targets for
uveal melanoma. BMC Cancer. 21(1068)2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Xu T, Gao S, Liu J, Huang Y, Chen K and
Zhang X: MMP9 and IGFBP1 regulate tumor immune and drive tumor
progression in clear cell renal cell carcinoma. J Cancer.
12:2243–2257. 2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Quintero-Fabián S, Arreola R,
Becerril-Villanueva E, Torres Romero JC, Arana-Argáez V,
Lara-Riegos J, Ramírez Camacho MA and Alvarez-Sánchez ME: Role of
matrix metalloproteinases in angiogenesis and cancer. Front Oncol.
9(1370)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Liu C, Shen Y and Tan Q: Diagnostic and
prognostic values of MMP-9 expression in ovarian cancer: A study
based on bioinformatics analysis and meta-analysis. Int J Biol
Markers. 38:15–24. 2023.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Jiang H and Li H: Prognostic values of
tumoral MMP2 and MMP9 overexpression in breast cancer: A systematic
review and meta-analysis. BMC Cancer. 21(149)2021.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Shao W, Wang W, Xiong XG, Cao C, Yan TD,
Chen G, Chen H, Yin W, Liu J, Gu Y, et al: Prognostic impact of
MMP-2 and MMP-9 expression in pathologic stage IA non-small cell
lung cancer. J Surg Oncol. 104:841–846. 2011.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Lin Z, Liu Y, Sun Y and He X: Expression
of Ets-1, Ang-2 and maspin in ovarian cancer and their role in
tumor angiogenesis. J Exp Clin Cancer Res. 30(31)2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zhang L, Li X, Feng X, Berkman T, Ma R, Du
S, Wu S, Huang C, Amponsah A, Bekker A and Tao YX: E74-like factor
1 contributes to nerve trauma-induced nociceptive hypersensitivity
through transcriptionally activating matrix metalloprotein-9 in
dorsal root ganglion neurons. Pain. 164:119–131. 2023.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wang L: ELF1-activated FOXD3-AS1 promotes
the migration, invasion and EMT of osteosarcoma cells via sponging
miR-296-5p to upregulate ZCCHC3. J Bone Oncol.
26(100335)2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Liu Y, Ma R, Juan D, Yuan Z, Sun J, Wang
M, Li Y, Bao Y and Jin H: Adipose-derived mesenchymal stem
cell-loaded β-chitin nanofiber hydrogel activates the AldoA/HIF-1α
pathway to promote diabetic wound healing. Am J Stem Cells.
12:1–11. 2023.PubMed/NCBI
|
|
62
|
Wang Y, Hu C, Kwok T, Bain CA, Xue X,
Gasser RB, Webb GI, Boussioutas A, Shen X, Daly RJ and Song J:
DEMoS: a deep learning-based ensemble approach for predicting the
molecular subtypes of gastric adenocarcinomas from
histopathological images. Bioinformatics. 38:4206–4213.
2022.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Bonsang B, Maksimovic L, Maille P, Martin
N, Laurendeau I, Pasmant E, Bièche I, Deschamps J, Wolkenstein P
and Ortonne N: VEGF and VEGFR family members are expressed by
neoplastic cells of NF1-associated tumors and may play an oncogenic
role in malignant peripheral nerve sheath tumor growth through an
autocrine loop. Ann Diagn Pathol. 60(151997)2022.PubMed/NCBI View Article : Google Scholar
|
|
64
|
De Re V, Alessandrini L, Brisotto G,
Caggiari L, De Zorzi M, Casarotto M, Miolo G, Puglisi F, Garattini
SK, Lonardi S, et al: HER2-CDH1 interaction via Wnt/B-catenin is
associated with patients' survival in HER2-positive metastatic
gastric adenocarcinoma. Cancers (Basel). 14(1266)2022.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Qin X, Chen Y, Ma S, Shen L and Ju S:
Immune-related gene TM4SF18 could promote the metastasis of gastric
cancer cells and predict the prognosis of gastric cancer patients.
Mol Oncol. 16:4043–4059. 2022.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Liu C, Omilusik K, Toma C, Kurd NS, Chang
JT, Goldrath AW and Wang W: Systems-level identification of key
transcription factors in immune cell specification. PLoS Comput
Biol. 18(e1010116)2022.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Klugman M, Fazzari M, Xue X, Ginsberg M,
Rohan TE, Halmos B, Hanna DB, Shuter J and Hosgood HD III: The
associations of CD4 count, CD4/CD8 ratio, and HIV viral load with
survival from non-small cell lung cancer in persons living with
HIV. AIDS Care. 34:1014–1021. 2022.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Fridman WH, Petitprez F, Meylan M, Chen
TWW, Sun CM, Roumenina LT and Sautès-Fridman C: B cells and cancer:
To B or not to B? J Exp Me. 218(e20200851)2021.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Peña-Romero AC and Orenes-Piñero E: Dual
effect of immune cells within tumour microenvironment: Pro- and
anti-tumour effects and their triggers. Cancers (Basel).
14(1681)2022.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Dou A and Fang J: Heterogeneous myeloid
cells in tumors. Cancers (Basel). 13(3772)2021.PubMed/NCBI View Article : Google Scholar
|