Introduction
Chronic obstructive pulmonary disease (COPD) is a
complex respiratory disorder characterized by persistent airflow
limitation resulting from exposure to noxious particles and gases,
leading to an abnormal inflammatory response in the lungs (1). Acute exacerbations in COPD (AECOPD)
significantly contribute to the decline in lung function, diminish
the quality of life, increase the utilization of emergency
healthcare services and heighten the mortality rates associated
with COPD (2,3). Infectious pathogens, such as
bacteria, viruses and atypical pathogens, can trigger airway
inflammation in COPD and lead to acute exacerbations (4,5).
These pathogens are associated with acute exacerbations in up to
80% of patients with COPD, with bacteria potentially contributing
to 50% of these exacerbations (6).
Delayed diagnosis or delayed initiation of
antimicrobial therapy in patients with AECOPD was reported to be
associated with increased mortality (1,2,7).
Currently, the diagnosis primarily relies on pathogen cultures;
however, challenges such as the selection of appropriate specimens,
the conditions in which cultures are grown and the proficiency of
operators significantly hinder the prompt acquisition of results
from cultures and drug sensitivity tests (8). Therefore, early identification of
bacterial infections in patients with AECOPD remains challenging
(9). Consequently, clinicians are
actively seeking a simple and rapid indicator that can aid the
diagnosis of bacterial infections among individuals with
AECOPD.
Recently conducted studies have demonstrated that
several factors can augment the risk of developing bacterial
infections in patients with AECOPD (10,11).
The current diagnostic strategy for bacterial infections involves a
comprehensive assessment of risk factors, clinical manifestations
and laboratory test results (2,4,11).
Unveiling the complete spectrum of risk factors associated with
bacterial infections in patients with AECOPD is crucial to aid
clinicians in identifying such infections (7). Identifying one or more predisposing
conditions would be pivotal in initiating further diagnostic
investigations, thereby facilitating early diagnosis and treatment
(5,6).
The presence of multiple potential risk factors
poses a challenge for clinicians in assessing patient risk and
there is a dearth of risk-predictive scoring models for bacterial
infections in the existing literature. Consequently, there is an
urgent need to develop predictive models that can facilitate
clinical decision-making. To establish a more precise diagnostic
prediction model for bacterial infections in patients with AECOPD,
it is essential to incorporate more reliable predictors and include
patients from a broader cohort. The primary objective of the
present study was to develop and validate a risk prediction model
aimed at promptly identifying patients with severe AECOPD who
require immediate empirical antibacterial treatment, particularly
in healthcare facilities with limited resources.
Patients and methods
Data source
The present retrospective study was carried out
using clinical data from patients with AECOPD at Pingxiang People
Hospital (Pingxiang, China), from January 2023 to December 2023.
Patients who met the criteria of the study design were divided into
two groups as follows: 70% were randomly allocated to the training
set and the remaining 30% constituted the internal validation
set.
The present study complied with the Declaration of
Helsinki and was approved by the Ethics Committee of Pingxiang
People's Hospital (approval no. PK2023Z67-HS02).
The inclusion criteria for patients with AECOPD were
as follows: The diagnosis of AECOPD was established following the
guidelines set out by the Global Initiative for Chronic Obstructive
Lung Disease (1). AECOPD is
characterized by a sudden worsening of symptoms, such as
breathlessness, coughing and sputum production, which worsen over
<14 days. It is characterized by the presence of at least two of
the following symptoms: Increased sputum volume, altered sputum
color and exacerbated dyspnea.
Patients were excluded from the present study if
they lacked a spirometry report, had been admitted to the hospital
within one month before the study, were undergoing treatment with
immunosuppressants or had been diagnosed with malignant disease.
Patients with chronic respiratory conditions, such as asthma,
interstitial lung disease, active tuberculosis and bronchiectasis
were also excluded from the study.
Definition of bacterial infection
The diagnosis of bacterial infection was established
based on a combination of clinical manifestations, laboratory
findings, radiographic imaging, PCR detection and the patient's
response to antibiotic treatment. Bacterial infection was confirmed
through the isolation of causative agents. Spontaneous sputum
samples were incubated on sheep blood, chocolate and MacConkey agar
plates (Antu Biological) at 35˚C for 48 h in an atmosphere
containing 5% CO2. A sputum culture was deemed positive
if the cultured microorganisms showed potential pathogenicity,
exhibited high growth density (semi-quantitative), contained <25
squamous epithelial cells and had >15 leukocytes per high-power
microscopic field (magnification, x100) in the Gram-stained sputum
sample. The Gram staining involves five steps, all performed at
room temperature. First, a sputum smear on a microscope slide was
prepared and heat-fixed. Next, crystal violet stain was applied for
1 min, followed by rinsing with water. Iodine solution was then
added for 1 min to form a complex with the crystal violet. After
another rinse, the sample was decolorized with 95% ethanol for
10-30 sec and rinsed immediately. Finally, counterstaining was
performed with safranin for 30 sec and the slide was rinsed and
allowed to dry, followed by examination under a microscope.
Risk factors
The predictors utilized for constructing the
nomogram model were selected based on prior research studies
(10,11). All risk factors, encompassing
demographic data, comorbidities, pulmonary function,
pharmacological history, status of mechanical ventilation upon
admission and history of previous acute exacerbations, were readily
accessible from comprehensive early, current and historical medical
records. The detailed data are presented in Table I.
 | Table ICharacteristics of AECOPD in patients
in the training and validation sets. |
Table I
Characteristics of AECOPD in patients
in the training and validation sets.
| Variable | Total (n=544) | Validation cohort
(n=160) | Training cohort
(n=384) | P-value |
|---|
| Age, years | 59.75±10.91 | 59.51±10.94 | 59.86±10.90 | 0.733 |
| Sex | | | | 0.135 |
|
Male | 430 (79.0) | 120(75) | 310 (80.8) | |
|
Female | 114 (21.0) | 40(25) | 74 (19.2) | |
| Albumin, g/l | 38.30±12.92 | 39.30±14.94 | 37.87±11.97 | 0.240 |
| PaCO2,
mmHg | 44.84±10.45 | 44.48±9.62 | 44.99±10.78 | 0.607 |
| PaO2,
mmHg | 73.20±14.92 | 71.94±12.30 | 73.73±15.87 | 0.203 |
| Neutrophils, % | 67.24±12.85 | 67.12±12.58 | 67.28±12.98 | 0.891 |
| ESR, mm/h | 23.46±3.17 | 23.58±3.22 | 23.41±3.15 | 0.566 |
| WBC,
x109/l | 10.28±5.75 | 9.99±5.51 | 10.40±5.84 | 0.449 |
| NE, µg/ml | 20.51±22.06 | 20.31±21.66 | 20.59±22.25 | 0.892 |
| CRP, mg/l | 15.29±10.81 | 14.66±8.51 | 15.56±11.64 | 0.380 |
| D-D dimer,
mg/l | 0.36±0.85 | 0.39±1.02 | 0.35±0.78 | 0.594 |
| LDH, U/l | 208.27±76.02 | 195.45±63.99 | 203.61±79.97 | 0.051 |
| PCT, ng/l | 0.39±0.47 | 0.38±0.35 | 0.40±0.51 | 0.639 |
| Drinking | | | | 0.905 |
|
No | 372 (68.4) | 110 (68.8) | 262 (68.2) | |
|
Yes | 172 (31.6) | 50 (31.2) | 122 (31.8) | |
| GOLD | | | | 0.501 |
|
No | 366 (67.3) | 111 (69.4) | 255 (66.4) | |
|
Yes | 178 (32.7) | 49 (30.6) | 129 (33.6) | |
| Smoking | | | | 0.051 |
|
No | 350 (64.3) | 93 (58.1) | 257 (66.9) | |
|
Yes | 194 (35.7) | 67 (41.9) | 127 (33.1) | |
| Diabetes | | | | 0.310 |
|
No | 485 (89.2) | 146 (91.2) | 339 (88.3) | |
|
Yes | 59 (10.8) | 14 (8.8) | 45 (11.7) | |
| CAD | | | | 0.301 |
|
No | 430 (79.0) | 122 (76.2) | 308 (80.2) | |
|
Yes | 114 (21.0) | 38 (23.8) | 76 (19.8) | |
| Hypertension | | | | 0.658 |
|
No | 264 (48.5) | 80 (50.0) | 184 (47.9) | |
|
Yes | 280 (51.5) | 80 (50.0) | 200 (52.1) | |
| Cancer | | | | 0.722 |
|
No | 292 (53.7) | 84 (52.5) | 208 (54.2) | |
|
Yes | 252 (46.3) | 76 (47.5) | 176 (45.8) | |
| Anemia | | | | 0.152 |
|
No | 311 (57.2) | 99 (61.9) | 212 (55.2) | |
|
Yes | 233 (42.8) | 61 (38.1) | 172 (44.8) | |
| Eosinophils
≥2% | | | | 0.859 |
|
No | 245 (45.0) | 73 (45.6) | 172 (44.8) | |
|
Yes | 299 (55.0) | 87 (54.4) | 212 (55.2) | |
Statistical analysis
Statistical analyses were conducted using the
statistical software SPSS (version 25.0; IBM Corp.) and R software
(version 3.5.2). Continuous variables were presented as the mean ±
standard deviation and group comparisons were carried out using the
t-test or Mann-Whitney U-test. Categorical variables, presented as
numbers and percentages, were compared using the chi-square test or
Fisher's exact test, as appropriate. Multivariate analysis included
the aggregation of potential biomarkers that were identified as
significant using univariate logistic regression analysis. Only
variables with P<0.05 in the univariate analysis in the training
set were included in the multivariate analysis.
Subsequently, the nomogram was created utilizing the
‘rms’ package in R software, leveraging the outcomes of the
multivariate logistic regression analysis of the dependent
variable. The discriminatory power was evaluated by calculating the
area under the receiver operating characteristic curve (AUC). The
calibration of the model was evaluated by comparing the anticipated
and actual likelihood of bacterial infection. Furthermore, decision
curve analysis (DCA) was employed to assess the clinical utility of
the model. Unless explicitly mentioned, a two-tailed P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
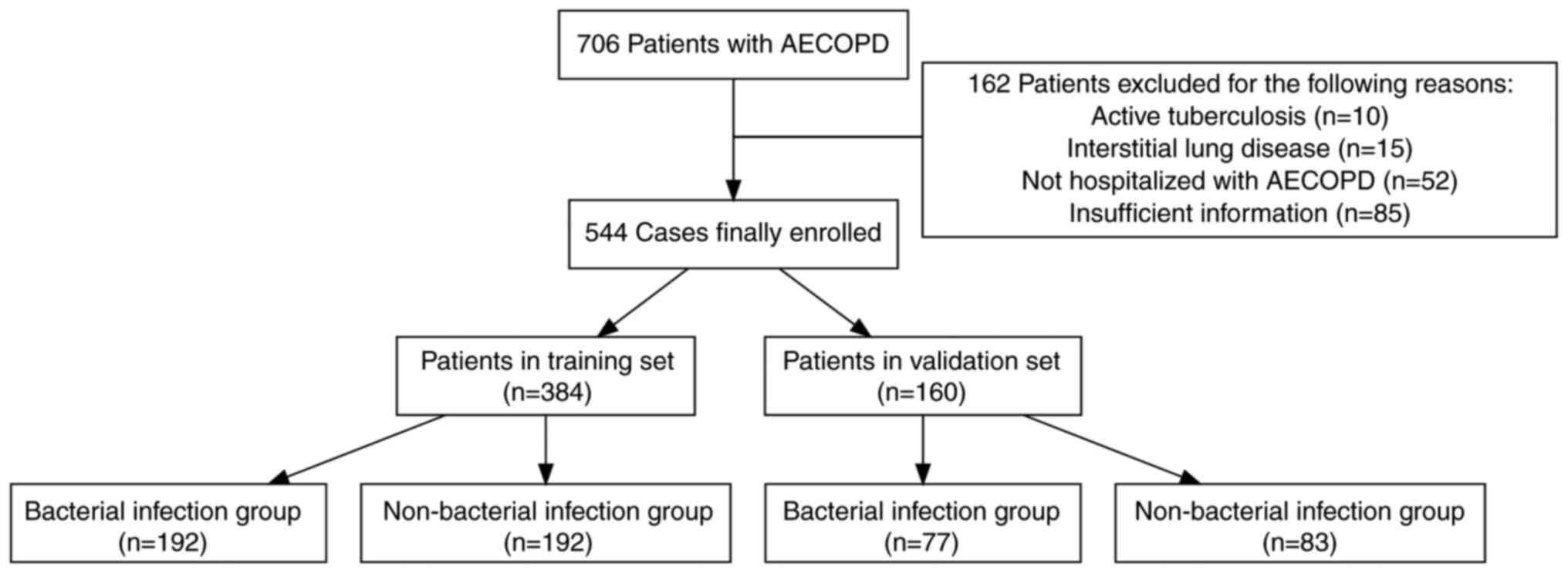
A total of 706 patients diagnosed with AECOPD were
screened, resulting in the exclusion of 162 patients based on
predefined exclusion criteria, ultimately yielding a final sample
size of 544 patients. The basic characteristics of the dataset are
summarized in Table I. The
training dataset comprised 384 patients, with 192 cases in the
bacterial infection group and 192 cases in the non-bacterial
infection group, which comprised 310 men and 74 women, aged between
41 and 97 years. The validation dataset comprised 160 patients,
with 77 cases in the bacterial infection group and 83 cases in the
non-bacterial infection group, which comprised 120 men and 40
women, aged between 44 and 92 years. The flowchart illustrates the
strategy for identifying bacterial infections in the AECOPD cohort
(Fig. 1). Table I presents the demographic and
clinical characteristics of patients in both the training and
validation sets.
Risk factors associated with bacterial
infections of AECOPD
The characteristics of the patients with AECOPD in
the training cohort, both with and without bacterial infection, are
compiled in Table II. A total of
21 clinical and basic indicators were included in the univariate
analysis to explore their association with bacterial infections.
The results revealed that procalcitonin (PCT), PaO2,
neutrophil elastase (NE), C-reactive protein (CRP), the neutrophil
percentage and eosinophil percentage ≥2% exhibited a significant
association with bacterial infection (P≤0.05; Table III). Subsequent multivariate
logistic regression analysis indicated that the following factors
were independently associated with bacterial infection: NE, CRP,
PCT and eosinophil percentage ≥2% (Table III).
 | Table IIComparison of AECOPD in patients with
and without bacterial infection in the training set. |
Table II
Comparison of AECOPD in patients with
and without bacterial infection in the training set.
| Variable | Total (n=384) | Non-bacterial
(n=192) | Bacterial
(n=192) | P-value |
|---|
| Age, years | 59.86±10.90 | 59.85±11.27 | 59.86±10.56 | 0.989 |
| Albumin, g/l | 37.87±11.97 | 38.86±13.98 | 36.89±9.48 | 0.106 |
| PaCO2,
mmHg | 44.99±10.78 | 46.00±12.31 | 43.98±8.92 | 0.066 |
| PaO2,
mmHg | 73.73±15.87 | 75.38±17.78 | 72.08±13.54 | 0.042 |
| Neutrophils, % | 67.28±12.98 | 64.89±12.65 | 69.68±12.89 | <0.001 |
| ESR, mm/h | 23.41±3.15 | 23.26±3.18 | 23.56±3.12 | 0.355 |
| WBC,
x109/l | 10.40±5.84 | 10.07±5.02 | 10.73±6.56 | 0.270 |
| NE, µg/ml | 20.59±22.25 | 12.81±10.93 | 28.38±27.42 | <0.001 |
| CRP, mg/l | 15.56±11.64 | 11.27±10.12 | 19.85±11.49 | <0.001 |
| D-D dimer,
mg/l | 0.35±0.78 | 0.41±0.90 | 0.29±0.63 | 0.126 |
| LDH, U/l | 213.61±79.97 | 213.37±67.82 | 213.86±90.69 | 0.953 |
| PCT, ng/l | 0.40±0.51 | 0.30±0.28 | 0.49±0.65 | <0.001 |
| Drinking | | | | >0.999 |
|
No | 262 (68.2) | 131 (68.2) | 131 (68.2) | |
|
Yes | 122 (31.8) | 61 (31.8) | 61 (31.8) | |
| GOLD | | | | 0.331 |
|
No | 255 (66.4) | 132 (68.8) | 123 (64.1) | |
|
Yes | 129 (33.6) | 60 (31.2) | 69 (35.9) | |
| Smoking | | | | 0.914 |
|
No | 257 (66.9) | 128 (66.7) | 129 (67.2) | |
|
Yes | 127 (33.1) | 64 (33.3) | 63 (32.8) | |
| Diabetes | | | | 0.874 |
|
No | 339 (88.3) | 169 (88.0) | 170 (88.5) | |
|
Yes | 45 (11.7) | 23 (12.0) | 22 (11.5) | |
| CAD | | | | 0.306 |
|
No | 308 (80.2) | 158 (82.3) | 150 (78.1) | |
|
Yes | 76 (19.8) | 34 (17.7) | 42 (21.9) | |
| Hypertension | | | | 0.307 |
|
No | 184 (47.9) | 87 (45.3) | 97 (50.5) | |
|
Yes | 200 (52.1) | 105 (54.7) | 95 (49.5) | |
| Cancer | | | | 0.539 |
|
No | 208 (54.2) | 107 (55.7) | 101 (52.6) | |
|
Yes | 176 (45.8) | 85 (44.3) | 91 (47.4) | |
| Anemia | | | | 0.412 |
|
No | 212 (55.2) | 110 (57.3) | 102 (53.1) | |
|
Yes | 172 (44.8) | 82 (42.7) | 90 (46.9) | |
| Eosinophils
≥2% | | | | <0.001 |
|
No | 172 (44.8) | 106 (55.2) | 66 (34.4) | |
|
Yes | 212 (55.2) | 86 (44.8) | 126 (65.6) | |
 | Table IIILogistic regression analysis of
predictors for bacterial infections of patients with AECOPD. |
Table III
Logistic regression analysis of
predictors for bacterial infections of patients with AECOPD.
| | Univariate | Multivariate |
|---|
| Variable | Coefficient | OR (95%CI) | P-value | Coefficient | OR (95%CI) | P-value |
|---|
| Age, years | 0.000 | 1.000 (0.982,
1.019) | 0.989 | | | |
| Drinking | -0.000 | 1.000 (0.651,
1.537) | >0.999 | | | |
| GOLD | 0.210 | 1.234 (0.807,
1.886) | 0.331 | | | |
|
Smoking | -0.024 | 0.977 (0.638,
1.494) | 0.914 | | | |
|
Diabetes | -0.050 | 0.951 (0.510,
1.771) | 0.874 | | | |
| CAD | 0.263 | 1.301 (0.786,
2.155) | 0.306 | | | |
| Hypertension | -0.209 | 0.811 (0.543,
1.212) | 0.307 | | | |
| Cancer | 0.126 | 1.134 (0.759,
1.695) | 0.539 | | | |
| Anemia | 0.169 | 1.184 (0.791,
1.770) | 0.412 | | | |
| Albumin | -0.017 | 0.983 (0.962,
1.005) | 0.124 | | | |
|
PaCO2 | -0.018 | 0.982 (0.964,
1.001) | 0.068 | | | |
|
PaO2 | -0.014 | 0.986 (0.973,
1.000) | 0.046 | 0.002 | 1.002 (0.986,
1.019) | 0.777 |
| Neutrophil
percent | 0.031 | 1.031 (1.014,
1.049) | <0.001 | 0.019 | 1.019 (1.000,
1.039) | 0.056 |
| ESR | 0.030 | 1.031 (0.967,
1.099) | 0.354 | | | |
| WBC | 0.019 | 1.020 (0.985,
1.056) | 0.270 | | | |
| NE | 0.042 | 1.042 (1.028,
1.057) | <0.001 | 0.042 | 1.043 (1.027,
1.059) | <0.001 |
| CRP | 0.098 | 1.103 (1.074,
1.134) | <0.001 | 0.082 | 1.085 (1.052,
1.119) | <0.001 |
| D-D dimer | -0.224 | 0.799 (0.591,
1.080) | 0.145 | | | |
| LDH | 0.000 | 1.000 (0.998,
1.003) | 0.953 | | | |
| PCT | 1.559 | 4.753 (2.373,
9.519) | <0.001 | 1.674 | 5.332 (2.263,
12.567) | <0.001 |
| Eosinophils
≥2% | 0.856 | 2.353 (1.559,
3.552) | <0.001 | 0.830 | 2.294 (1.403,
3.751) | 0.001 |
Nomogram development
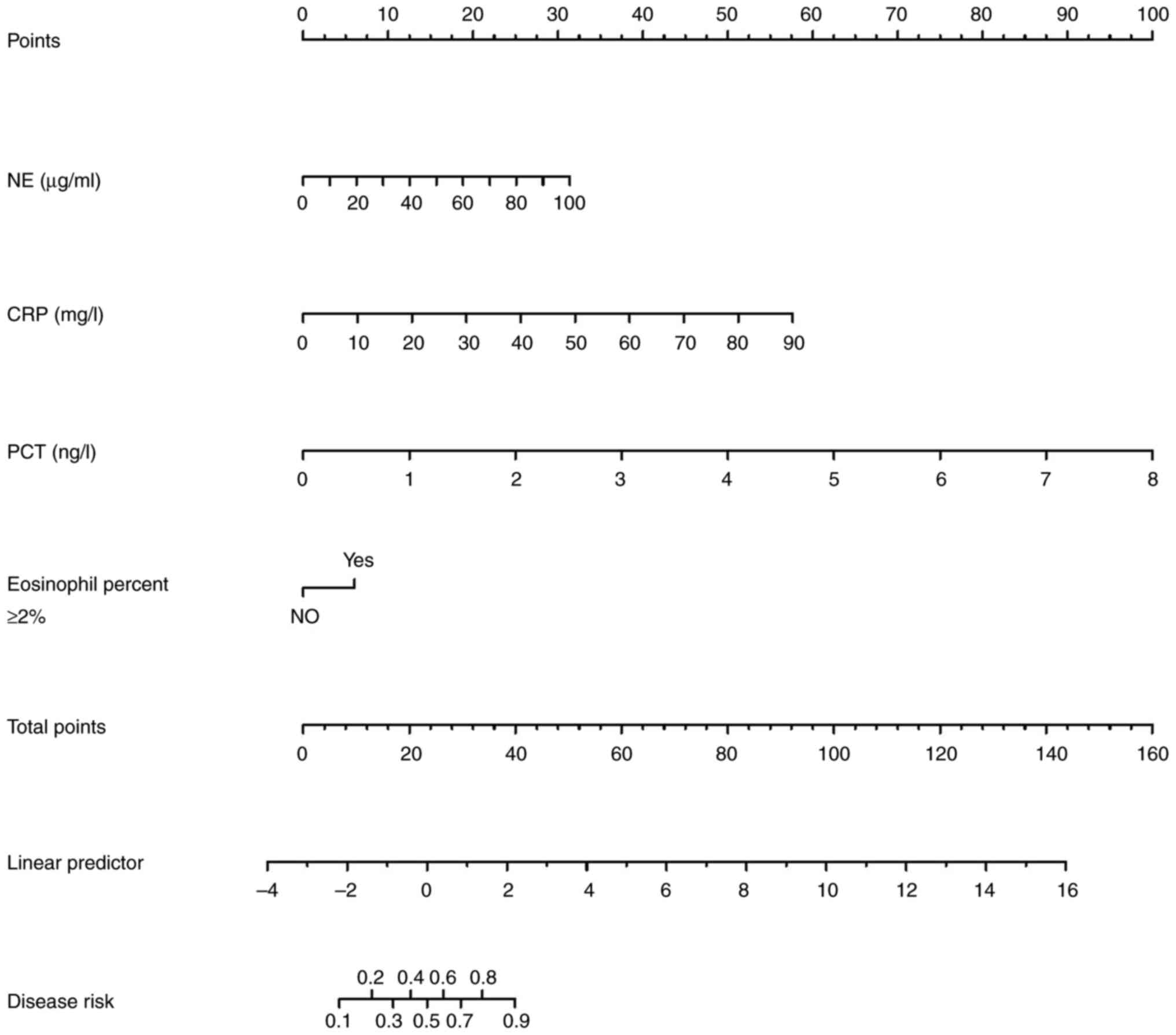
The logistic regression analysis identified four
independent risk factors for bacterial infection in the
multivariate analysis: Eosinophil percentage ≥2%, NE, PCT and CRP.
Based on the weights assigned to these factors from the training
set, a nomogram was generated using the results of the multivariate
regression to calculate the risk of bacterial infection (Fig. 2).
Discrimination and calibration
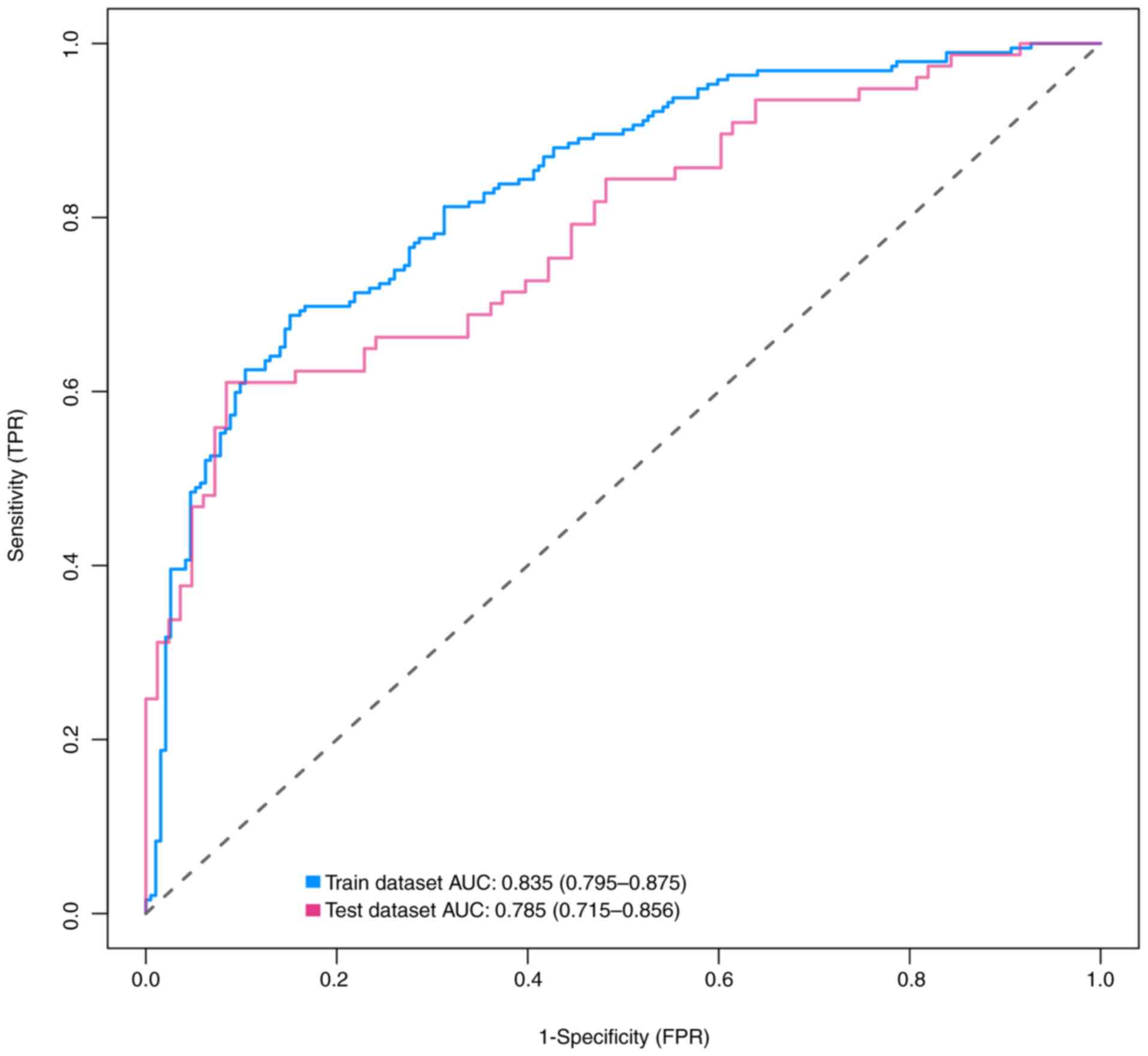
The AUC of the nomogram was calculated to be 0.835
[95% confidence interval (CI), 0.795-0.875] in the training
dataset, as demonstrated by the blue curve in Fig. 3. In the validation dataset, the AUC
was determined to be 0.785 (95% CI, 0.715-0.856), as indicated by
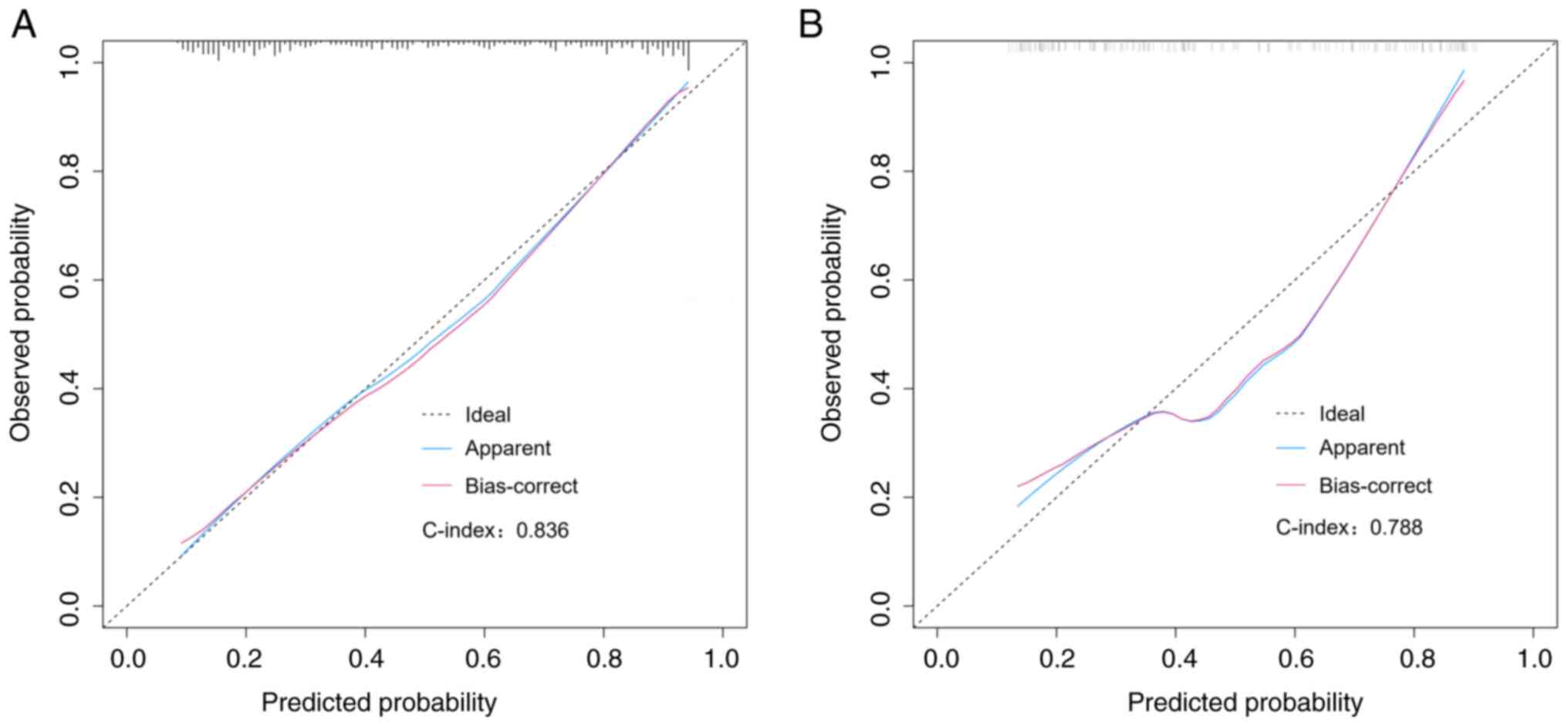
the red curve in the same figure. Further examination of the
calibration belt revealed strong concordance of the nomogram in
both the training and validation datasets, as illustrated in
Fig. 4A and B. The discrimination and overfitting bias
of the model were assessed through internal validation. The
findings revealed a C-index of 0.836 for the initial group and
0.788 for the subsequent validation set. The calibration plots
illustrated a robust agreement between the predictions of the
nomogram and the actual occurrence of bacterial infections in
patients. The model exhibited excellent goodness-of-fit, as
demonstrated by the Hosmer-Lemeshow test results in both the
training set and validation set, with P-values exceeding 0.05
(P=0.36 for the training set; P=0.12 for the validation set).
Meanwhile, our data (Fig. S1)
also showed that the nomogram model had a greater AUC than PCT
alone in the training dataset and in the validation dataset.
DCA
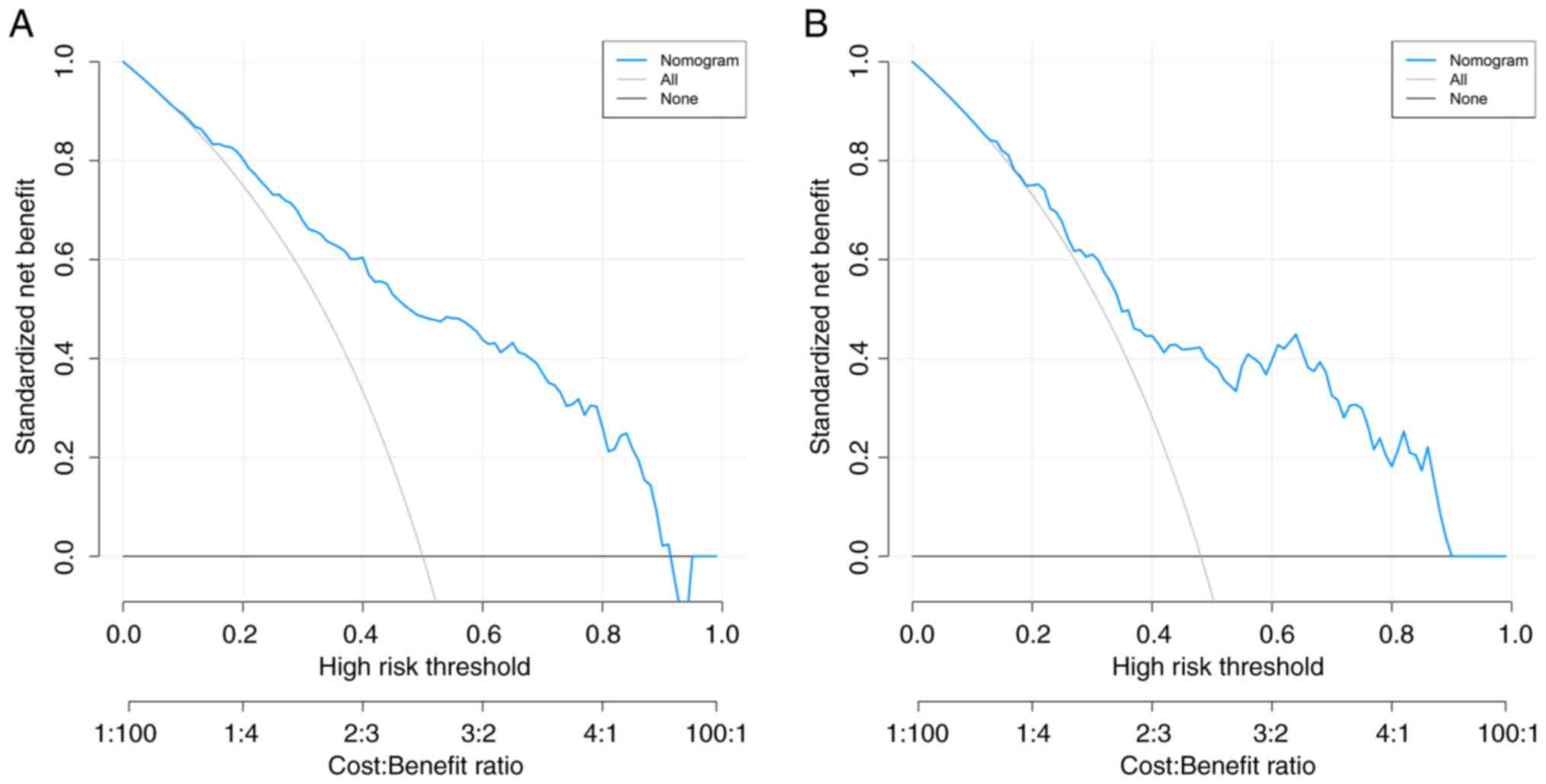
The results of the DCA for the risk nomogram in both
the training and validation sets were presented to determine the
optimal decision threshold for the nomogram (Fig. 5A and B). The nomogram model exhibited a
favorable net benefit across both datasets, encompassing predicted
risk thresholds ranging from 0-51%.
Discussion
AECOPD is a complex and heterogeneous disease
characterized by chronic airway inflammation. Over the past decade,
advancements in modern research techniques have significantly
enhanced the understanding of the role of bacterial infection in
AECOPD (8). Bacterial infections
contribute to up to 50% of acute exacerbations of COPD, leading to
increased morbidity and mortality (3,4,8,12).
Timely intervention is crucial for reducing the impact of bacterial
infections on the health outcomes of patients with AECOPD (7). Previous studies have aimed at
developing prediction models for identifying patients with
bacterial infections (13-15).
Therefore, it is imperative to prioritize this distinct patient
cohort and develop a concise yet precise prognostic model that
enables clinicians to accurately assess disease severity and
optimize therapeutic strategies for improved patient survival.
Utilizing a multiple logistic regression model, a
ranking chart was developed to predict bacterial infections in
patients with AECOPD, based on independently correlated and
identified risk factors. Each factor was assigned a weighted number
of points and the total points for each patient were calculated
using the nomogram, which yielded an estimated probability of
bacterial infections. Logistic regression analysis revealed that
CRP, PCT, eosinophil percentage and NE were significant predictors
of bacterial infections in patients with AECOPD. The nomogram
incorporating these predictors demonstrated exceptional
discriminatory and calibration abilities for predicting bacterial
infections in patients at high risk for developing AECOPD, thereby
offering valuable clinical guidance for the early identification
and initiation of empirical antibiotic treatment.
Consistent with previous studies, NE has been
identified as a robust predictor of the development of bacterial
infection in patients with AECOPD (14,16,17).
NE serves as a relatively specific biomarker for severe bacterial
infections and sepsis (14-16).
Research findings have indicated that bacterial infection can
trigger an immune response in the airways, leading to inflammation
(18,19). Of note, one study demonstrated
higher levels of neutrophilic inflammation during COPD-mediated
bacterial exacerbations compared to non-bacterial exacerbations
(18). NE can synergistically
impair tracheobronchial ciliary function when combined with
bacterial products (16,20). In AECOPD, there is a predominant
airway inflammation characterized by neutrophils, with sputum
samples from numerous patients showing neutrophil counts exceeding
60% (18,21). Neutrophils play various roles
within the innate immune system, including the defense against
invading microorganisms (18,22).
This demonstrates that NE serves as a highly sensitive and specific
predictor of bacterial-associated exacerbation, characterized by an
increased bacterial load or positive microbiological findings
during acute events.
The eosinophil count/percentage is a readily
available and straightforward test that can be utilized for
predicting bacterial infection (23,24).
However, relying solely on the eosinophil percentage is inadequate
to accurately predict bacterial infection, necessitating its
combination with other established biological markers to enhance
diagnostic accuracy. Thulborn et al (24) indicated that an eosinophil
percentage <2% demonstrated potential as an indicator of
bacterial infection in AECOPD events. Consequently, an eosinophil
percentage ≥2% could serve as a reference for guiding antibiotic
usage in the treatment of AECOPD.
Numerous previous literature reports have
consistently demonstrated that both PCT and CRP serve as common
clinical indicators for predicting the occurrence of bacterial
infection in AECOPD (25,26). The predictive capability of CRP
alone for bacterial infection is limited, necessitating the
integration of other established biological markers to enhance its
diagnostic accuracy in identifying bacterial infections (27). The present study further confirmed,
through univariate and multivariate analyses, that PCT is a robust
predictor of bacterial infection and an increasing concentration of
PCT was associated with a higher risk of infection. Consequently,
PCT was identified as a pivotal contributor in the current model.
The two most important biomarkers for detecting infection are PCT
and CRP, both of which are inflammatory markers.
Plasma levels of PCT are known to be increased in
individuals with sepsis or serious bacterial or fungal infections
(28). In contrast to these
observations, PCT levels tend to be lower in patients exhibiting
milder inflammatory reactions (29). Previous studies have demonstrated
that compared with CRP, PCT is a superior predictor of early
bacterial infection (26,30). It is interesting to know that,
owing to the presence of various types of infections in the current
model, CRP exhibited a comparatively lower diagnostic value
compared with PCT (27,29). It is worth noting that PCT may not
be effectively cleared by the kidneys in patients with severe renal
failure (25,29,31).
Consequently, relying solely on PCT as an indicator for determining
the optimal cut-off value to diagnose bacterial infection in
patients with AECOPD may not yield significantly different results
from those observed in the general population. Meanwhile, a
comparative analysis was conducted between the multivariate model
and a model using only PCT. The results showed that, while PCT is a
significant predictor, the incorporation of other parameters
enhances the model's overall predictive ability. Our findings
indicate that the nomogram model exhibits a significantly larger
AUC compared to the PCT for both the training and validation
datasets. Therefore, further research is warranted to fully
ascertain the predictive potential of PCT as a biomarker for
bacterial infection.
In the present study, a simplified nomogram was
pioneered for predicting the risk of bacterial infection in
patients with AECOPD. The prediction model was developed using
readily available clinical parameters, incorporating risk factors
such as NE, CRP, eosinophil percentage and PCT levels. The model's
discriminative and calibration abilities were proven to be
excellent through internal validation. The criteria for diagnosing
bacterial infections were proposed and the clinical algorithm for
patients with lower respiratory tract specimens exhibited favorable
validity in distinguishing colonization from true bacterial
infection. While previous research studies have identified certain
risk factors, such as antibiotic usage, plasma CRP and PCT, as
potential indicators of bacterial infection in patients with COPD
(26,27), the precise role and complex
interplay of these factors in predicting the probability of
bacterial infection have not been fully incorporated into current
diagnostic standards.
Nomograms are simple yet visually intuitive
predictive models that combine various indicators to aid in the
diagnosis or prediction of diseases. The nomogram developed in the
present study offers clinicians a valuable tool to assess the risk
of complications resulting from bacterial infections among patients
with COPD. An investigation of alternative methods for diagnosing
bacterial infections will be performed in future studies. A patient
with AECOPD who presents with two to three additional risk factors,
along with impaired lung function, may have a calculated predicted
risk of bacterial infection ranging from 8 to 52%, according to the
nomogram. In addition, the lack of a predictive model or serum
marker currently hinders the differentiation of bacterial
infections in patients with AECOPD, which is crucial for
determining appropriate antibiotic treatment strategies tailored to
specific bacteria types. Since culture and/or PCR methods are used
to confirm the infection, it would be reasonable to distinguish the
types of bacteria, as they may be strongly associated with disease
prognosis. Future research by our group will further investigate
the potential of developing models for identifying specific
bacterial pathogens, predicting clinical outcomes and customizing
treatment strategies for patients with AECOPD.
The present study exhibits certain limitations that
should be considered. First, the study was conducted at a single
center and was retrospective in nature. The evaluation of the
discrimination and calibration of the scoring model was limited to
internal validation. To generalize the results, conducting
multicenter prospective studies for external validation is
important. Furthermore, the simplistic binary classification
(positive/negative) utilized for categorizing COPD co-morbidities
does not accurately capture the true severity of the overall
disease burden faced by the patients. Incorporating a system for
classifying the severity of these comorbidities could significantly
enhance the quality and value of the findings.
The nomogram model integrates four distinct risk
factors for bacterial infection and has the potential to provide
clinicians and patients with advanced, precise insights into the
risk of bacterial infection in AECOPD cases. Additional research is
essential to confirm the effectiveness and validity of using the
nomogram in real-world clinical settings to enhance the prediction
of bacterial infections.
Supplementary Material
ROC curve indicating performance of
the prediction model and PCT using both the training set and
validation set. ROC, receiver operating characteristic; AUC, area
under the ROC curve; FPR, false-positive rate; TPR, true-positive
rate; PCT, procalcitonin.
Acknowledgements
Not applicable.
Funding
Funding: The study was supported by Jiangxi Provincial Health
Commission Science and Technology Project Plan (grant nos.
202311691 and 202311693).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XLC conceived and designed the study. XMW and WQY
collected the data and performed the literature search. DZ was
involved in writing the manuscript and performed the statistical
analyses. All authors have read and approved the final version of
the manuscript. XMW and XLC confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The Ethics Committee of Pingxiang People's Hospital
(Pingxiang, China; approval no. PK2023Z67-HS02) approved the
protocol of the current study and waived the requirement for
informed consent owing to the retrospective design of the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vestbo J, Hurd SS, Agusti AG, Jones PW,
Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ,
Nishimura M, et al: Global strategy for the diagnosis, management,
and prevention of chronic obstructive pulmonary disease: GOLD
executive summary. Am J Respir Crit Care Med. 187:347–365.
2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ouyang P, Zhou Z, Pan C, Tang P, Long S,
Liao X, Liu Q and Xie L: Acute exacerbation of chronic obstructive
pulmonary disease due to carbapenem-resistant klebsiella
pneumoniae-induced pneumonia: Clinical features and prognostic
factors. Int J Chron Obstruct Pulmon Dis. 19:683–693.
2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Abi Abdallah G, Diop S, Jamme M, Legriel S
and Ferre A: Respiratory infection triggering severe acute
exacerbations of chronic obstructive pulmonary disease. Int J Chron
Obstruct Pulmon Dis. 19:555–565. 2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Messous S, Elargoubi A, Pillet S,
Rajoharison A, Hoffmann J, Trabelsi I, Grissa MH, Boukef R,
Beltaief K, Mastouri M, et al: Bacterial and viral infection in
patients hospitalized for acute exacerbation of chronic obstructive
pulmonary disease: Implication for antimicrobial management and
clinical outcome. COPD. 18:53–61. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Martinez-Garcia MA, Faner R, Oscullo G, la
Rosa-Carrillo D, Soler-Cataluna JJ, Ballester M, Muriel A and
Agusti A: Chronic bronchial infection is associated with more rapid
lung function decline in chronic obstructive pulmonary disease. Ann
Am Thorac Soc. 19:1842–1847. 2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Martinez-Garcia MA and Alvar A: POINT: Is
chronic bacterial infection clinically relevant in COPD? Yes.
Chest. 162:970–972. 2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Brennan M, McDonnell MJ, Harrison MJ,
Duignan N, O'Regan A, Murphy DM, Ward C and Rutherford RM:
Antimicrobial therapies for prevention of recurrent acute
exacerbations of COPD (AECOPD): Beyond the guidelines. Respir Res.
23(58)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Moghoofei M, Azimzadeh Jamalkandi S, Moein
M, Salimian J and Ahmadi A: Bacterial infections in acute
exacerbation of chronic obstructive pulmonary disease: A systematic
review and meta-analysis. Infection. 48:19–35. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Smith D, Gill A, Hall L and Turner AM:
Prevalence, pattern, risks factors and consequences of antibiotic
resistance in COPD: A systematic review. COPD. 18:672–682.
2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pazmany P, Soos A, Hegyi P, Dohos D, Kiss
S, Szakacs Z, Parniczky A, Garami A, Peterfi Z and Molnar Z:
Inflammatory biomarkers are inaccurate indicators of bacterial
infection on admission in patients with acute exacerbation of
chronic obstructive pulmonary disease-A systematic review and
diagnostic accuracy network meta-analysis. Front Med (Lausanne).
8(639794)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Qian Y, Cai C, Sun M, Lv D and Zhao Y:
Analyses of factors associated with acute exacerbations of chronic
obstructive pulmonary disease: A review. Int J Chron Obstruct
Pulmon Dis. 18:2707–2723. 2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Duan T: Analysis of microbiological and
clinical characteristics of bacterial infection in patients with
pulmonary infection. Comput Intell Neurosci.
2022(5607358)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang Y, Zheng SP, Hou YF, Jie XY, Wang D,
Da HJ, Li HX, He J, Zhao HY, Liu JH, et al: A predictive model for
frequent exacerbator phenotype of acute exacerbations of chronic
obstructive pulmonary disease. J Thorac Dis. 15:6502–6514.
2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Thulborn SJ, Mistry V, Brightling CE,
Moffitt KL, Ribeiro D and Bafadhel M: Neutrophil elastase as a
biomarker for bacterial infection in COPD. Respir Res.
20(170)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hoult G, Gillespie D, Wilkinson TMA,
Thomas M and Francis NA: Biomarkers to guide the use of antibiotics
for acute exacerbations of COPD (AECOPD): A systematic review and
meta-analysis. BMC Pulm Med. 22(194)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zheng S, Kummarapurugu AB, Bulut GB, Syed
A, Kang L and Voynow JA: Neutrophil elastase activates the release
of extracellular traps from COPD blood monocyte-derived
macrophages. Clin Transl Sci. 16:2765–2778. 2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Boxio R, Wartelle J, Nawrocki-Raby B,
Lagrange B, Malleret L, Hirche T, Taggart C, Pacheco Y, Devouassoux
G and Bentaher A: Neutrophil elastase cleaves epithelial cadherin
in acutely injured lung epithelium. Respir Res.
17(129)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Benjamin JT, Plosa EJ, Sucre JM, van der
Meer R, Dave S, Gutor S, Nichols DS, Gulleman PM, Jetter CS, Han W,
et al: Neutrophilic inflammation during lung development disrupts
elastin assembly and predisposes adult mice to COPD. J Clin Invest.
131(e139481)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Park S, Lee SJ, Shin B, Lee SJ, Kim SH,
Kwon WC, Kim J and Lee MK: The association of delta neutrophil
index with the prognosis of acute exacerbation of chronic
obstructive pulmonary disease. BMC Pulm Med. 20(47)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Deng F, Zhong S, Yu C, Zhao H, Huang H,
Meng X, Lin C and Cai S: Abnormal neutrophil polarization in
chronic obstructive pulmonary disease and how cigarette smoke
extracts attract neutrophils. Ann Transl Med.
10(472)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gu L, Liu W, Huang JA, Zhu L, Hu X, Yue J
and Lin J: The role of neutrophil counts, infections and smoking in
mediating the effect of bronchiectasis on chronic obstructive
pulmonary disease: A mendelian randomization study. BMC Pulm Med.
24(144)2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Guan X, Yuan Y, Wang G, Zheng R, Zhang J,
Dong B, Ran N, Hsu AC, Wang C and Wang F: Ginsenoside Rg3
ameliorates acute exacerbation of COPD by suppressing neutrophil
migration. Int Immunopharmacol. 83(106449)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Choi J, Oh JY, Lee YS, Hur GY, Lee SY,
Shim JJ, Kang KH and Min KH: The association between blood
eosinophil percent and bacterial infection in acute exacerbation of
chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon
Dis. 14:953–959. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Thulborn SJ, Dilpazir M, Haldar K, Mistry
V, Brightling CE, Barer MR and Bafadhel M: Investigating the role
of pentraxin 3 as a biomarker for bacterial infection in subjects
with COPD. Int J Chron Obstruct Pulmon Dis. 12:1199–1205.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Titova E, Christensen A, Henriksen AH,
Steinshamn S and Asberg A: Comparison of procalcitonin, C-reactive
protein, white blood cell count and clinical status in diagnosing
pneumonia in patients hospitalized with acute exacerbations of
COPD: A prospective observational study. Chron Respir Dis.
16(1479972318769762)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Daniels JM, Schoorl M, Snijders D, Knol
DL, Lutter R, Jansen HM and Boersma WG: Procalcitonin vs C-reactive
protein as predictive markers of response to antibiotic therapy in
acute exacerbations of COPD. Chest. 138:1108–1115. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
An X, Zhang C, Weng X, Xiao W, Sun Z, Zeng
Z and Huang Q: C-reactive protein testing to guide antibiotic
prescribing for COPD exacerbations: A protocol for systematic
review and meta-analysis. Medicine (Baltimore).
99(e21152)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mathioudakis AG,
Chatzimavridou-Grigoriadou V, Corlateanu A and Vestbo J:
Procalcitonin to guide antibiotic administration in COPD
exacerbations: A meta-analysis. Eur Respir Rev.
26(160073)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang L, Wang J, Gu X, Sheng W, Wang Y and
Cao B: Procalcitonin-guided initiation of antibiotics in AECOPD
inpatients: Study protocol for a multicenter randomised controlled
trial. BMJ Open. 11(e049515)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tang L, Shi S, Wang B, Liu L, Yang Y, Sun
X, Ni Z and Wang X: Effect of urban air pollution on CRP and
coagulation: A study on inpatients with acute exacerbation of
chronic obstructive pulmonary disease. BMC Pulm Med.
21(296)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mou S, Zhang W, Deng Y, Tang Z and Jiang
D: Comparison of CRP, procalcitonin, neutrophil counts, eosinophil
counts, sTREM-1, and OPN between pneumonic and nonpneumonic
exacerbations in COPD patients. Can Respir J.
2022(7609083)2022.PubMed/NCBI View Article : Google Scholar
|