Introduction
Ulcerative colitis (UC) is a chronic inflammatory
bowel disease characterized by inflammation and ulcers in the
lining of the colon and rectum, and the incidence rate of UC is
steadily increasing globally, with the prevalence of UC estimated
to be 5 million cases worldwide by 2023(1). It typically affects the innermost
lining of the colon and causes symptoms such as abdominal pain,
diarrhea, rectal bleeding and weight loss (2). The etiology of this disease is
believed to be multifaceted, with a growing emphasis on aberrant
immune reactions, gut dysbiosis, genetic predisposition and
environmental influences (3).
Treatment of UC usually involves medication to reduce inflammation,
manage symptoms and maintain remission (3). Although sulfasalazine,
5-aminosalicylates, corticosteroids, thiopurines and methotrexate
have traditionally served as the primary and initial treatment
options for patients with UC, treatment with biological agents is
now advisable for those experiencing moderate to severe symptoms
(4,5). In severe cases, surgery to remove the
affected portion of the colon is necessary (6). However, none of the currently
available treatment methods are deemed entirely satisfactory due to
the side effects caused by long-term medication use and the high
recurrence rate, and the identification of the ideal treatment
remains elusive.
Multiple studies have reported that UC activates the
colonic Wnt/β-catenin signaling pathway, which is intricately
linked to the upregulation of inflammatory cytokines (7-9).
Furthermore, chronic inflammation in UC increases the risk of
developing colorectal cancer, which is closely related to the
excessive activation of the Wnt/β-catenin signaling pathway
(10,11). Furthermore, inhibiting the
Wnt/β-catenin signaling pathway has been reported to be
advantageous in the treatment of UC (12,13).
Among the inhibitors of the Wnt/β-catenin signaling pathway, XAV939
exhibits suppressive effects on the inflammatory response triggered
by lipopolysaccharide (LPS) (14,15).
Consequently, the utilization of XAV939 to block the Wnt/β-catenin
signaling pathway presents a feasible approach to effectively
manage the inflammation associated with UC.
The Wnt/β-catenin signaling pathway serves a
critical role in regulating the differentiation of intestinal stem
cells (ISCs) (16). By modulating
the expression of Wnt/β-catenin and its downstream effector SOX9,
it regulates the process of ISC differentiation into secretory cell
progenitor cells (17). It is
suggested that there is upregulated activity of the Wnt/β-catenin
signaling pathway in UC and this is believed to contribute to the
abnormal expansion of ISCs observed in UC (18). There is an increased tendency for
ISCs to differentiate into secretory progenitor cells in the colon
of patients with UC (19).
However, this disruption hinders the normal healing process of the
intestinal mucosa and compromises the functionality of the
intestinal barrier and its absorption capabilities. Understanding
the mechanisms underlying ISCs and their potential as therapeutic
targets could provide invaluable insights and novel approaches for
the management of UC.
The present study aimed to validate the therapeutic
effects of the Wnt/β-catenin inhibitor XAV939 on dextran sulfate
sodium (DSS)-induced UC and to assess its impact on ISC
differentiation.
Materials and methods
Animals
All animal procedures were performed in accordance
with the Guidelines for Care and Use of Laboratory Animals of
Guangdong Medical University and the experiments were approved by
the Ethics Committee of Shunde Women and Children's Hospital of
Guangdong Medical University (approval no. 2023054; Foshan,
China).
Animal welfare
An enriched environment, nutritious diet and humane
handling were provided to the animals. All the mice were provided
by the Guangdong Medical Laboratory Animal Center and housed in a
specific pathogen-free facility (temperature, 23±1˚C; 12/12 h
light/dark cycle; humidity, 50-60%). A single mouse was used as the
experimental unit and the placement of cages was random throughout
the space used. Measures were taken to ensure the welfare of the
animals and to minimize discomfort for all animals involved in the
present study. Animals were fed daily with a fresh diet to maintain
body weight and normal growth. All mice were allowed ad
libitum access to food and water. The criteria used to
determine when animals should be euthanized included, but were not
limited to, loss of body weight of 20% compared with the body
weight before the study began, severe behavioral abnormalities or
physiological distress. In the present study, a maximum weight loss
of 19.78% was observed. The mice were euthanized by intraperitoneal
injection of 100 mg/kg pentobarbital sodium. Death was verified by
checking for the cessation of the heartbeat and pupil dilation.
Experimental design
The experiments were performed during the 12 h light
cycle. For RNA-sequencing (RNA-seq) experiment, a total of 6
healthy male C57BL/6 mice (body weight, 18-22 g; age, 8 weeks) were
randomly divided into two groups (n=3/group). The control group
received distilled water, while the treatment group received 3.5%
DSS for 7 days. For the intervention study, a total of 18 healthy
male C57BL/6 mice (body weight, 18-22 g; age, 8 weeks) were
randomly divided into three groups (n=6/group): i) Control group;
ii) DSS group (3.5% DSS) and iii) DSS + XAV939 group [DSS + XAV,
3.5% DSS + 10 mg/kg body weight (BW) XAV939]. In our previous
study, we did not observe colitis-related changes when utilizing
low concentrations of DSS to induce the UC model (unpublished
data). Subsequently, findings from other literature were
cross-referenced and the experimental model of DSS administration
was adjusted accordingly (20,21).
Throughout the experiment, there were no instances of animal
mortality observed prior to its conclusion. The dose of XAV939 was
determined in accordance with the report conducted by Distler et
al (22). DSS was dissolved in
drinking water and XAV939 was dissolved in DMSO (≥99.7%; cat. no.
D2650; Sigma-Aldrich, Inc.) and 0.4 ml was administered by
intraperitoneal injection for 7 days. The control and DSS groups
were injected with 0.4 ml PBS mixed with the same concentration of
DMSO (Fig. S1). All mice were
euthanized and the entire colon was collected. The length of
colons, average daily weight gain and weight of the colon per unit
length were examined to measure the treatment outcome. DSS was
purchased from Dalian Meilun Biology Technology Co., Ltd. and
XAV939 was purchased from MedChemExpress (cat. no. HY-15147).
Analysis of colonic crypt RNA-seq
transcriptomic data using bioinformatics
RNA extraction and purification from colonic crypts
were carried out in accordance with the manufacturer's instructions
(cat. no. 74104; Qiagen GmbH). The total RNA of colonic crypts from
the control and DSS groups were sequenced by Guangdong Magigene
Biotechnology Co., Ltd. The Agilent 4200 Bioanalyzer (Agilent
Technologies, Inc.) was used for quality control of samples. All
samples had RNA integrity numbers >8. Subsequently, 150 bp
paired-end reads were generated using the Illumina NextSeq
sequencing platform (Illumina, Inc.).
Raw reads were first trimmed with Trimmomatic
(version 0.36; http://www.usadellab.org/cms/index.php?page=trimmomatic)
to acquire the clean reads, which were then mapped to National
Center for Biotechnology Information Rfam databases (version 14.10;
https://rfam.org) and the rRNA sequences removed using
Bowtie2 (version 2.33; https://github.com/BenLangmead/bowtie2). The reads
were mapped to the mouse reference genome
(Mus_musculus_GCF_000001635.27) using Hisat2 (version 2.1.0;
https://github.com/infphilo/hisat2)
(23,24). HTSeq-count (version 0.9.1;
http://htseq.readthedocs.io/en/release_0.9.1/) was
used to obtain the read count and function information of each
gene. The count tables were normalized based on their library size
using trimmed mean of M-values normalization implemented in
R/Bioconductor EdgeR (version 3.34.0; http://www.bioconductor.org/packages/release/bioc/html/edgeR.html)
(25,26). Normalized read counts were fitted
to a negative binomial distribution with a quasi-likelihood F-test.
Principal component analysis (PCA; https://www.r-project.org) was performed for the
regularized log transform of the normalized counts using plotPCA
tools with default parameters (27,28).
Differential gene expression analysis was further carried out using
EdgeR. The log2 fold change <-1 or >1 and a P-value <0.05
were used to examine differentially expressed genes (DEGs). The
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology
(GO) analysis was also used to calculate DEG enrichment, which
could be used to obtain information on the fold changes of
expressed genes at the molecular level. To determine the metabolic
and signaling pathways, the DEGs at various KEGG pathway levels
were counted. The log2 fold change <-1 or >1 and a P-value
<0.05 were used to examine differentially for KEGG and GO
analyses.
Histological examination of tissues
and immunohistochemical (IHC) staining
The colons were collected and fixed in 4%
paraformaldehyde, with fixation typically conducted at room
temperature (20-25˚C) for 24 h. Following fixation, the colons were
paraffin embedded and sectioned at a thickness of 5 µm for
histological analysis and IHC. Hematoxylin and eosin (H&E)
staining was used to assess morphological changes in the intestine,
and was imaged (Evos XL Core; Thermo Fisher Scientific, Inc.).
Tissue sections were dewaxed and rehydrated, and then stained with
hematoxylin for 5 min to highlight nuclear structures. Next, the
sections were rinsed and counterstained with eosin for 1 min to
color the cytoplasm and extracellular matrix. The staining was
carried out at room temperature (20-25˚C). Finally, the sections
were dehydrated, cleared and mounted onto slides for microscopic
examination. Antigen repair was performed on paraffin sections
using EDTA antigen repair solution (cat. no. ZLI-9067; ZSBG-BIO) in
boiling water for 10 min, then blocked with fetal bovine serum at
room temperature for 1 h. IHC staining for IL-1β (cat. no.
ab234437; Abcam), SOX9 (cat. no. ab185230; Abcam), β-catenin (cat.
no. R22820; Zenbio), Villin (cat. no. SC58897; Santa Cruz
Biotechnology, Inc.) and PPAR-γ (cat. no. 16643-1-AP; Wuhan Sanying
Biotechnology) was performed. The dilution ratio of primary
antibody was 1:100, and the incubations for primary antibodies were
conducted at 4˚C for a duration of 16 h. The samples were then
washed with PBS three times and incubated with Cy3 (cat. no. K1209;
APeXBIO Technology LLC) and FITC (cat. no. IF-0091; Beijing Dingguo
Changsheng Biotechnology Co., Ltd.)-labeled fluorescent secondary
antibodies (1:500) for 1 h at room temperature. The dilutions for
all antibodies used were prepared using a universal antibody
dilution buffer (cat. no. WB500D; New Cell & Molecular Biotech
Co., Ltd.). The nuclei were stained with DAPI (cat. no. C55-141215;
Biosharp Life Sciences) (1:1,000 dilution) for 10 min at room
temperature. Images were obtained using a confocal microscope (LSM
900; Carl Zeiss AG). The fluorescence signal intensity was measured
with ImageJ software (ImageJ2x 2.1.4.7; National Institutes of
Health), and the minimum value in the control group was set to
1.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
The RNA was extracted from cultured intestinal
organoids. RNA extraction was performed was performed using a
FastPure cell/tissue RNA isolation kit V2 (cat. no. RC112-01;
Vazyme Biotech Co., Ltd.), according to the manufacturer's
instructions. The reverse transcription process followed RNA
extraction using a reverse transcription kit (HiScript II Q RT
SuperMix for qPCR; cat. no. R222-01; Vazyme Biotech Co., Ltd.),
which involved incubating the reaction mixture at 50˚C for 15 min
and subsequently at 85˚C for 30 sec. The primers were purchased
from Sangon Biotech Co., Ltd. (Table
I). qPCR of TNF-α, IL-1β, Villin, peroxisome
proliferator-activated receptor γ (PPAR-γ), SOX9 and β-catenin was
performed. To normalize gene expression, a GAPDH endogenous control
was used. The ChamQ Universal SYBR qPCR Master Mix (cat. no.
Q711-02; Vazyme Biotech Co., Ltd.) and qPCR was employed to verify
the mRNA expression using a platform from Applied Biosystems;
Thermo Fisher Scientific, Inc. The reaction conditions set were as
follows: An initial denaturation at 94˚C for 5 min; followed by 36
cycles of (denaturation at 94˚C for 30 sec, annealing at 60˚C for
30 sec, and extension at 72˚C for 30 sec); a final extension at
72˚C for 5 min; and a subsequent incubation at 60˚C for 30 sec
followed by 95˚C for 30 sec. PCR amplification was analyzed using
the comparative 2-ΔΔCq method (29).
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5'-3') |
|---|
| TNF-α | F:
CTGGGACAGTGACCTGGACT |
| | R:
GCACCTCAGGGAAGAGTCTG |
| IL-1β | F:
TTCAGGCAGGCAGTATCACTCATT |
| | R:
TTGTTCATCTCGGAGCCTGTAGTG |
| SOX9 | F:
GGGGCTTGTCTCCTTCAGAG |
| | R:
TGGTAATGAGTCATACACAGTAC |
| Villin | F:
TATCATCGTGGTGAAGCAGGGACA |
| | R:
GGGCTCATAACCTCGTCAGCAATCT |
| β-catenin | F:
TGTACTGTTCTACGCCATCACGA |
| | R:
CTAGAGCAGACAGACAGCACCTTC |
| Peroxisome
proliferator activated receptor-γ | F:
TTTTCAAGGGTGCCAGTTTCG |
| | R:
GGGCTTCCGCAGGCTTTT |
| GAPDH | F:
TGTTTGTGATGGGTGTGAACC |
| | R:
GCAGTGATGGCATGGACTGTG |
Crypt culture and treatment of
intestinal organoids
The crypts were isolated by taking ~5 centimeters of
colon and splitting it longitudinally. The intestinal contents were
then cleaned using PBS and transferred to a 15 ml centrifuge tube
and 5 ml of PBS was added. After gently shaking by hand for 5 min,
the solution was replaced with fresh PBS and the shaking process
was repeated six times. The colon segments were then cut into 1 cm
pieces. The PBS was discarded, and a solution containing 30 mmol/l
EDTA-2Na and 1.5 mmol/l dithiothreitol in PBS was added. The
solution was gently shaken by hand for 5 min, then the separation
solution was replaced and the shaking process was repeated once.
Fresh PBS was added, and the suspension was observed for crypt
units and debris impurities. When there were more crypt units and
less debris impurities, fresh PBS was added and the sample was
shaken vigorously by hand for 1 min, incubated on ice for 5 min and
the crypts were collected. The quality of the isolated crypts
served a crucial role in the success of organoid culture. The
crypts obtained exhibited intact structures and a high purity
(>90% of them retained the intact structure of the crypt base),
ensuring the quality of the resulting organoids (Fig. S2). Crypts were embedded in
Matrigel and cultured using a mouse colonic organoid kit (cat. no.
K2204-MC; Biogenous), and the seeding density of the crypts was 5/1
µl of matrix gel, with 8 µl per well (40 crypts in total). The
organoid was cultured in a CO2 incubator at 37˚C. At 24
h post-crypt seeding, a gradual transformation of the crypts into
rounded and transparent structures was observed using a Evos XL
Core microscope (Thermo Fisher Scientific, Inc.), which is a
characteristic feature of organoid formation. The intestinal
organoid was induced with TNF-α to establish an inflammatory model.
TNF-α and XAV939 treatment was performed after 48 h of seeding, and
the sample in each well served as a replicate and was collected
after 24 h of treatment. The intestinal organoids were divided into
three groups: i) Control group; ii) TNF-α group treated with 100
ng/ml TNF-α (cat. no. 10602-R101-F; Sino Biological, Inc.); and
iii) TNF-α+XAV939 group treated with 100 ng/ml TNF-α and 10 µM
XAV939. The dose of XAV939 used was determined in accordance with
the previous report by Liang et al (30). The organoid formation efficiency
was calculated by dividing the number of colonies formed by the
number of crypts seeded and expressing this as a percentage. The
surface area was measured using ImageJ software (ImageJ2X 2.1.4.7,
National Institutes of Health).
Western blotting
The colon samples collected from various groups in
the XAV939 experiment were used to prepare protein samples. The
colon was lysed by RIPA (cat. no. WB3100; New Cell & Molecular
Biotech Co., Ltd.) and prepared into protein samples of the same
concentration by the BCA kit (cat. no. P0011; Beyotime Institute of
Biotechnology). The sample loading amount per lane was 10 µg.
Protein samples were separated on 10% SDS-PAGE gels and transferred
to PVDF membranes. The membranes were blocked with 5% bovine serum
albumin at room temperature for 1 h. The membranes were incubated
with primary antibodies at 4˚C for 12-16 h. The selection and use
of these primary antibodies were consistent with the description
for IHC: IL-1β (cat. no. ab234437; Abcam), SOX9 (cat. no. ab185230;
Abcam), β-catenin (cat. no. R22820; Zenbio), Villin (cat. no.
SC58897; Santa Cruz Biotechnology, Inc.) and PPAR-γ (cat. no.
16643-1-AP; Wuhan Sanying Biotechnology). The dilution ratio of
primary antibody was 1:1,000, and the dilutions for all antibodies
used were prepared using a universal antibody dilution buffer (cat.
no. WB500D; New Cell & Molecular Biotech Co., Ltd.). Samples
were washed using TBS with Tween-20 (0.1%) three times and
incubated with a HRP-goat anti-rabbit secondary antibody (cat. no.
RGAR001; Wuhan Sanying Biotechnology) and a HRP-goat anti-mouse
secondary antibody (cat. no. SA00001-1; Wuhan Sanying
Biotechnology) for 1 h at room temperature. The enhanced
chemiluminescence (KF8001; Affinity Biosciences) signals were
scanned using a ChemiDOC XRS+ (Bio-Rad Laboratories, Inc.), and the
band densities were analyzed using ImageJ software.
Statistical analysis
GraphPad Prism (version 8.0; Dotmatics) software was
utilized for statistical analysis. All data were presented as the
mean ± SEM. Multiple comparisons were performed using a one-way
ANOVA and Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
RNA-seq demonstrated changes in ISC
differentiation in the UC model
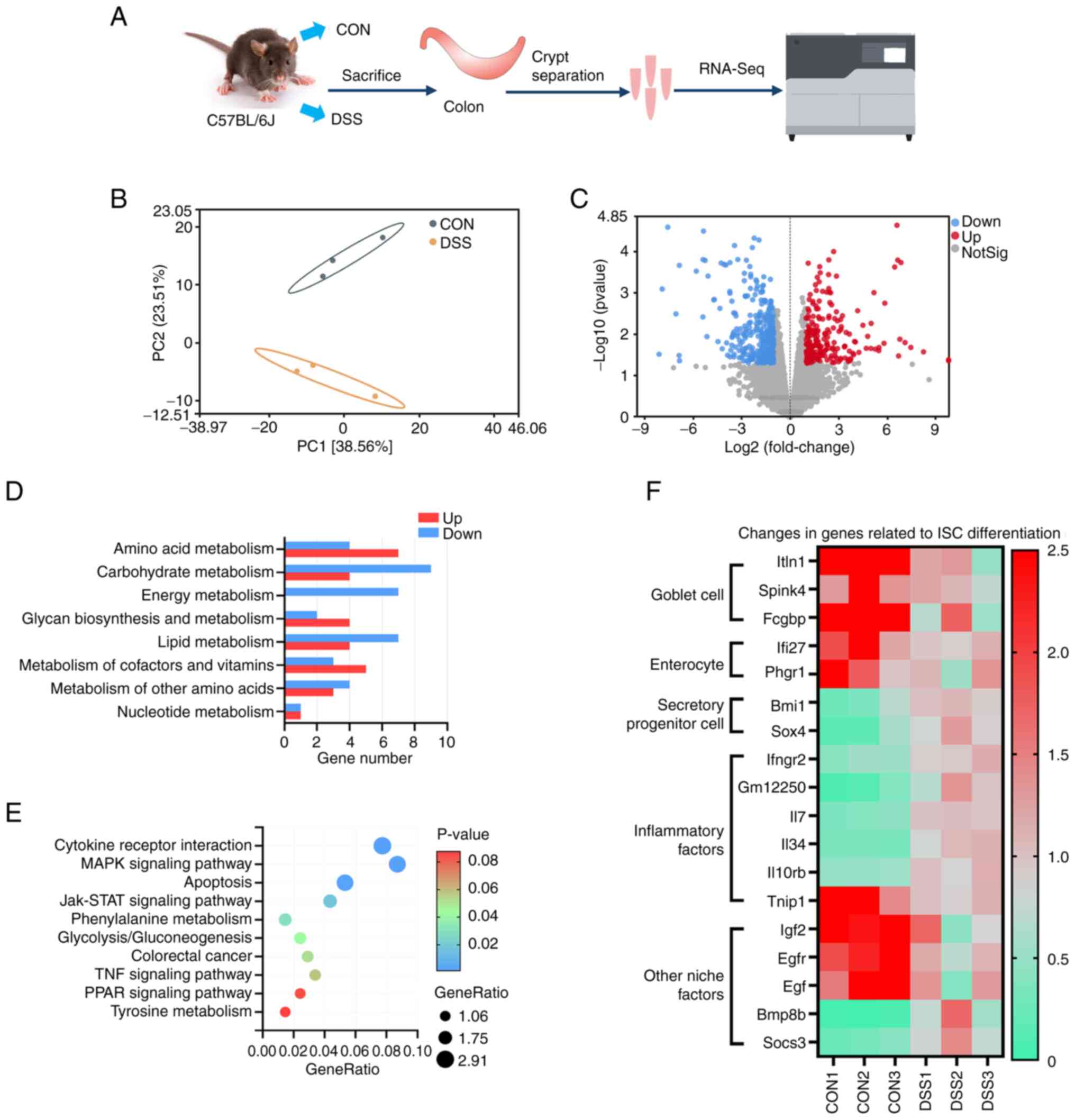
The colonic crypts were isolated for RNA-seq
(Fig. 1A). PCA results showed a
significant difference between the control group compared with the
DSS group (Fig. 1B). A total of
212 genes were upregulated and 315 genes were downregulated after
DSS treatment (Fig. 1C). KEGG
analysis demonstrated that DSS treatment resulted in the
differentially expressed genes related to various types of
metabolic processes (Fig. 1D).
Additionally, KEGG analysis showed significant changes in
inflammatory cytokine receptor interactions and MAPK signaling
pathways (Fig. 1E). The DSS group
showed significant down-regulation of intestinal goblet cell and
enterocyte marker genes, and a significant increase in secreting
cell progenitor marker genes, which indicated a potential disorder
in ISC differentiation (Fig. 1F).
Furthermore, DSS significantly altered the gene abundance of
inflammatory factors and other niche factors, which is crucial for
the microenvironmental homeostasis of ISCs (Fig. 1F).
Effect of XAV939 on the DSS-induced UC
model in mice
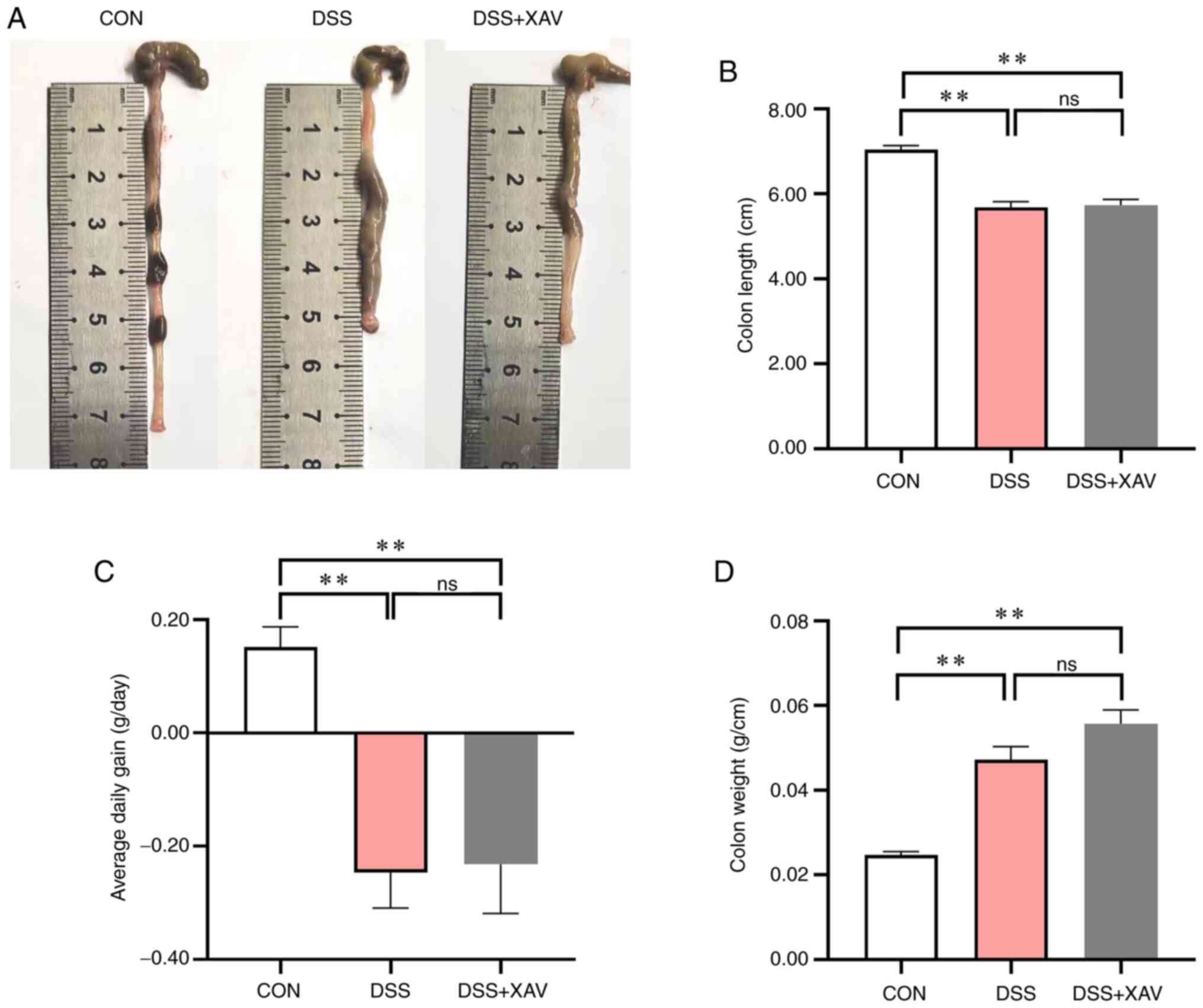
To confirm the therapeutic effect of XAV939 on
DSS-induced UC in mice, the length of the colon was measured
(Fig. 2A). Compared with the
control group, DSS caused significant shortening of the colon
(Fig. 2B, P<0.01) and a
significant reduction in average daily weight gain (Fig. 2C, P<0.01). However, the results
showed that treatment with XAV939 had no significant effect on the
colon length compared with the DSS group (Fig. 2B, P=0.95). Additionally, XAV939 did
not significantly improve the average daily weight gain compared
with the DSS group (Fig. 2C,
P=0.98). XAV939 treatment caused an increase in the weight of the
colon per unit length compared with the DSS group (Fig. 2D, P=0.09), which indicated that the
colon injury treated by DSS was not improved. Therefore, XAV939 may
not be beneficial in mitigating the colonic changes associated with
DSS-induced UC in mice.
XAV939 did not improve the DSS-induced
inflammation
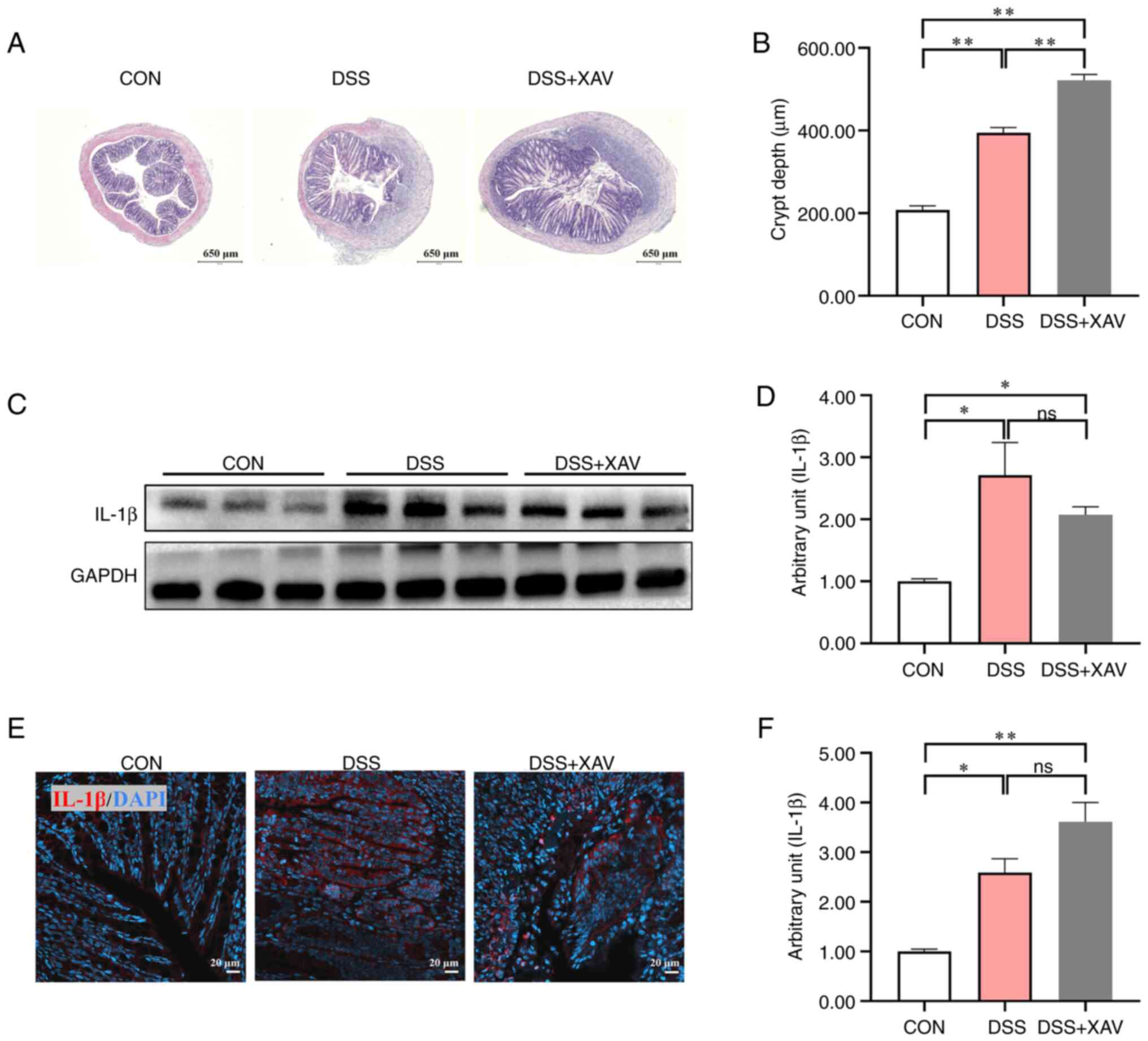
The results of H&E staining showed that XAV939
treatment caused no significant difference in colonic epithelial
structure and inflammatory infiltration compared with the DSS group
(Fig. 3A). The increase in crypt
depth, which can be considered a marker of intestinal damage, was
significantly more pronounced after XAV939 treatment compared with
the DSS group, which indicated that there was improvement in colon
injury (Fig. 3B, P<0.01).
Furthermore, the results of western blotting demonstrated that,
compared with the control group, the DSS group exhibited a
significant increase in IL-1β protein expression levels (Fig. 3C and D, P<0.05). The non-specific bands in
the GAPDH blot may have been due to the high concentration of
antibodies used. The results of IHC also demonstrated that the DSS
induced a significant increase in the expression level of IL-1β
(Fig. 3E and F, P<0.05). Notably, supplementation
with XAV939 did not lead to significant changes in IL-1β protein
expression levels (Fig. 3D,
P=0.38; Fig. 3F, P=0.09). These
findings collectively suggested that XAV939 had no beneficial
effect on DSS-induced colon injury and inflammation.
Effect of XAV939 on the mouse
intestinal organoid treated with TNF-α
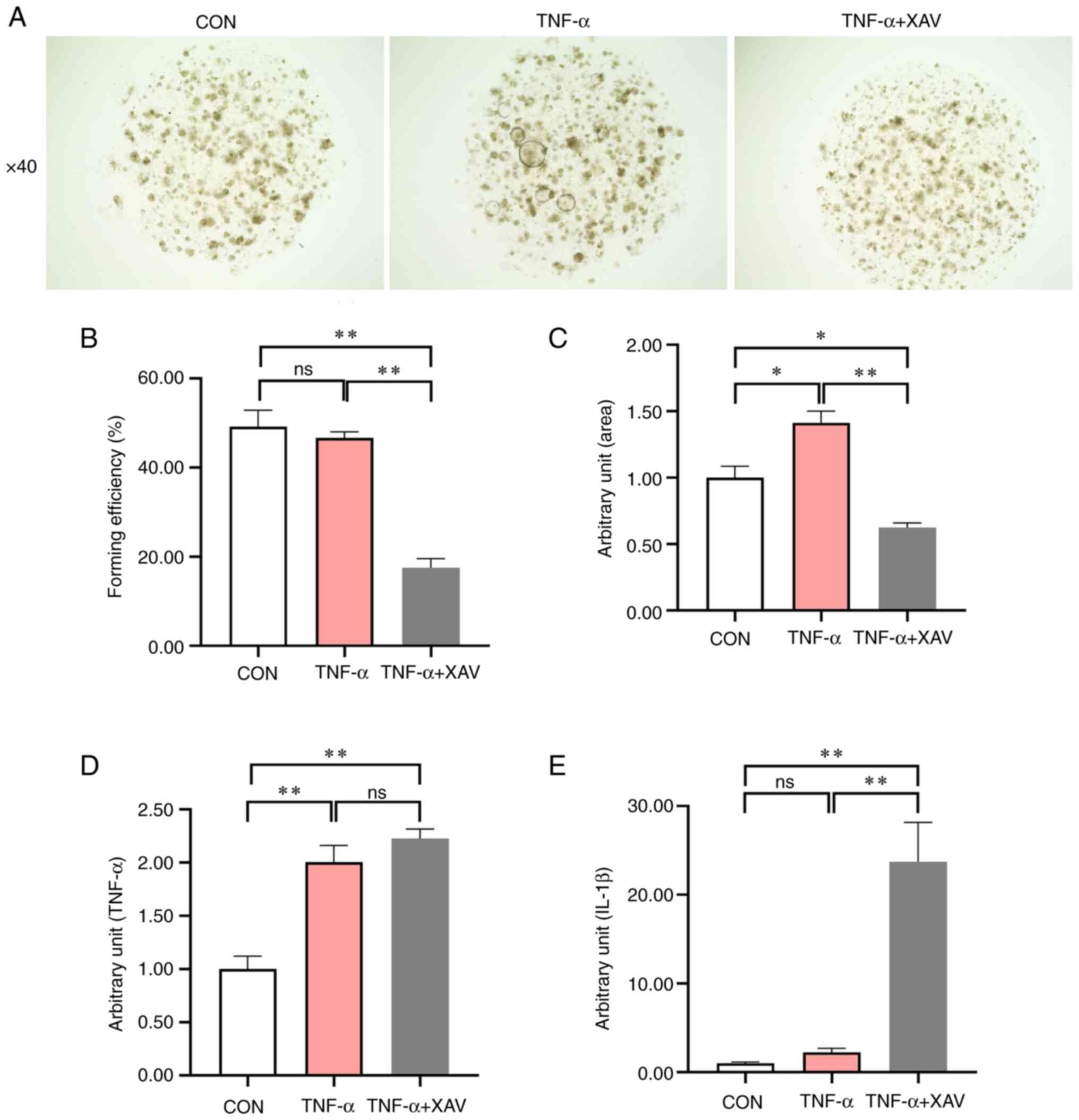
Due to the severe damage caused by DSS to the
intestinal epithelium and the key role of ISCs in promoting the
repair process of the epithelium, the intestinal organoid model was
used to verify the anti-inflammatory effect of XAV939 (Fig. 4A). These results demonstrated that
XAV939 significantly reduced the forming efficiency of organoids
compared with the TNF-α group (Fig.
4B, P<0.01), and significantly alleviated the TNF-α-induced
increase in organoid area (Fig.
4C, P<0.01). TNF-α did not affect the forming efficiency of
the organoid, but the stimulation was successful because TNF-α
treatment led to a significant increase in TNF-α (Fig. 4D, P<0.01) expression level, and
an increase in IL-1β (Fig. 4E,
P=0.81) expression level. Nevertheless, the addition of XAV939 did
not significantly reduce the expression levels of IL-1β and TNF-α
induced by TNF-α treatment (Fig.
4D, P=0.47). Notably, the addition of XAV939 contributed to an
increase in IL-1β expression levels (Fig. 4E, P<0.01), which indicated that
XAV939 did not significantly relieve inflammation.
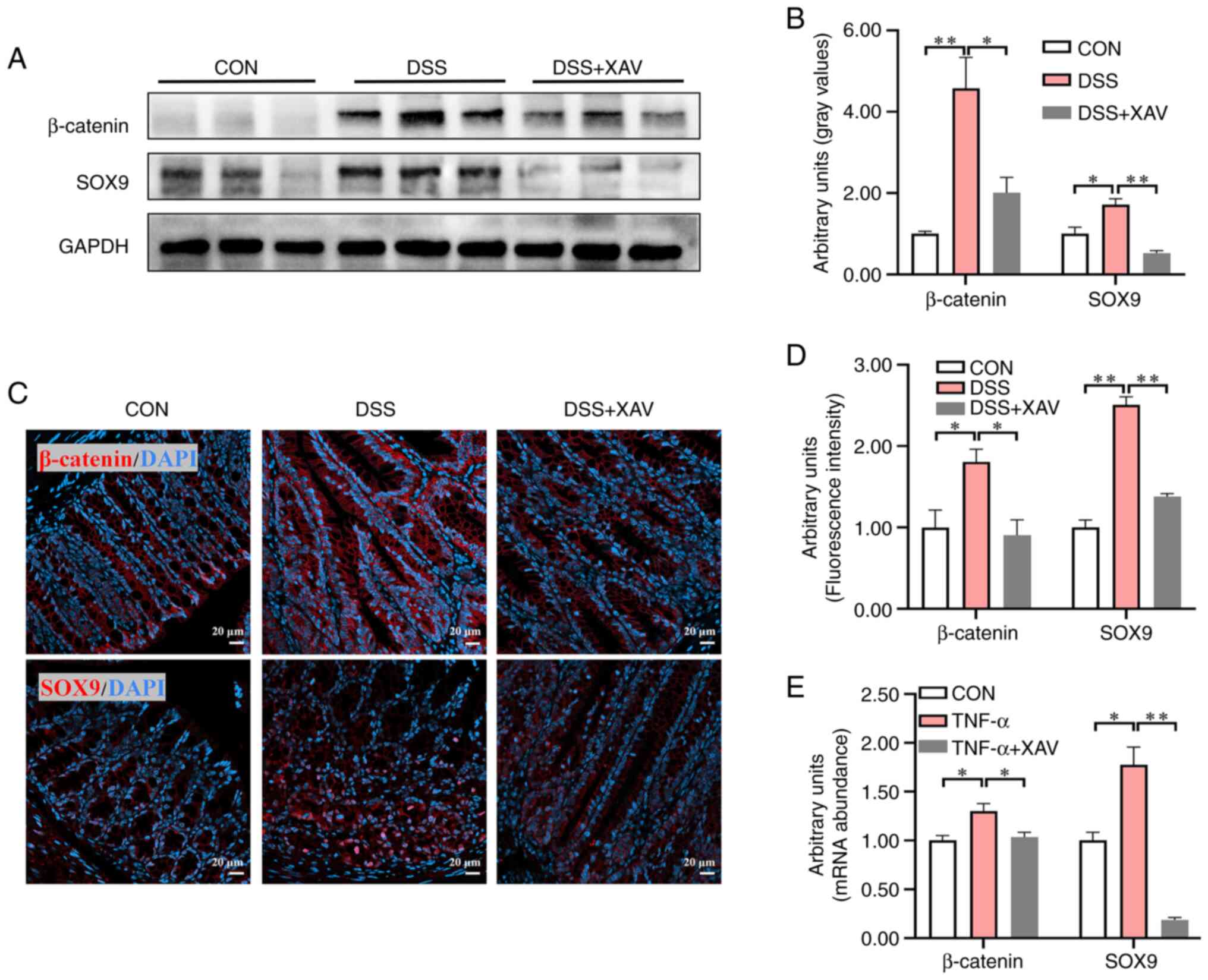
XAV939 reduced the expression levels
of β-catenin and SOX9
As the differentiation of ISCs is regulated by the
Wnt/β-catenin signaling pathway, the protein expression levels of
β-catenin and its downstream target SOX9 were examined (Fig. 5A). DSS significantly increased
β-catenin (Fig. 5B, P<0.01) and
SOX9 (Fig. 5B, P<0.05) protein
expression levels, while XAV939 significantly reduced the protein
expression levels of β-catenin (Fig.
5B, P<0.05) and SOX9 (Fig.
5B, P<0.01) compared with the DSS group. Simultaneously,
β-catenin and SOX9 protein expression was determined by IHC
(Fig. 5C). DSS significantly
increased the fluorescence intensity of β-catenin (Fig. 5D, P<0.05) and SOX9 (Fig. 5D, P<0.01), while XAV939
significantly reduced the fluorescence intensity of β-catenin
(Fig. 5D, P<0.05) and SOX9
(Fig. 5D, P<0.01). In the
intestinal organoid experiment, XAV939 treatment significantly
reduced the TNF-α-induced increase in the expression levels of
β-catenin (Fig. 5E, P<0.05) and
SOX9 (Fig. 5E, P<0.01). These
results indicated that XAV939 could reverse the excessive
activation of Wnt/β-catenin induced by DSS and TNF-α.
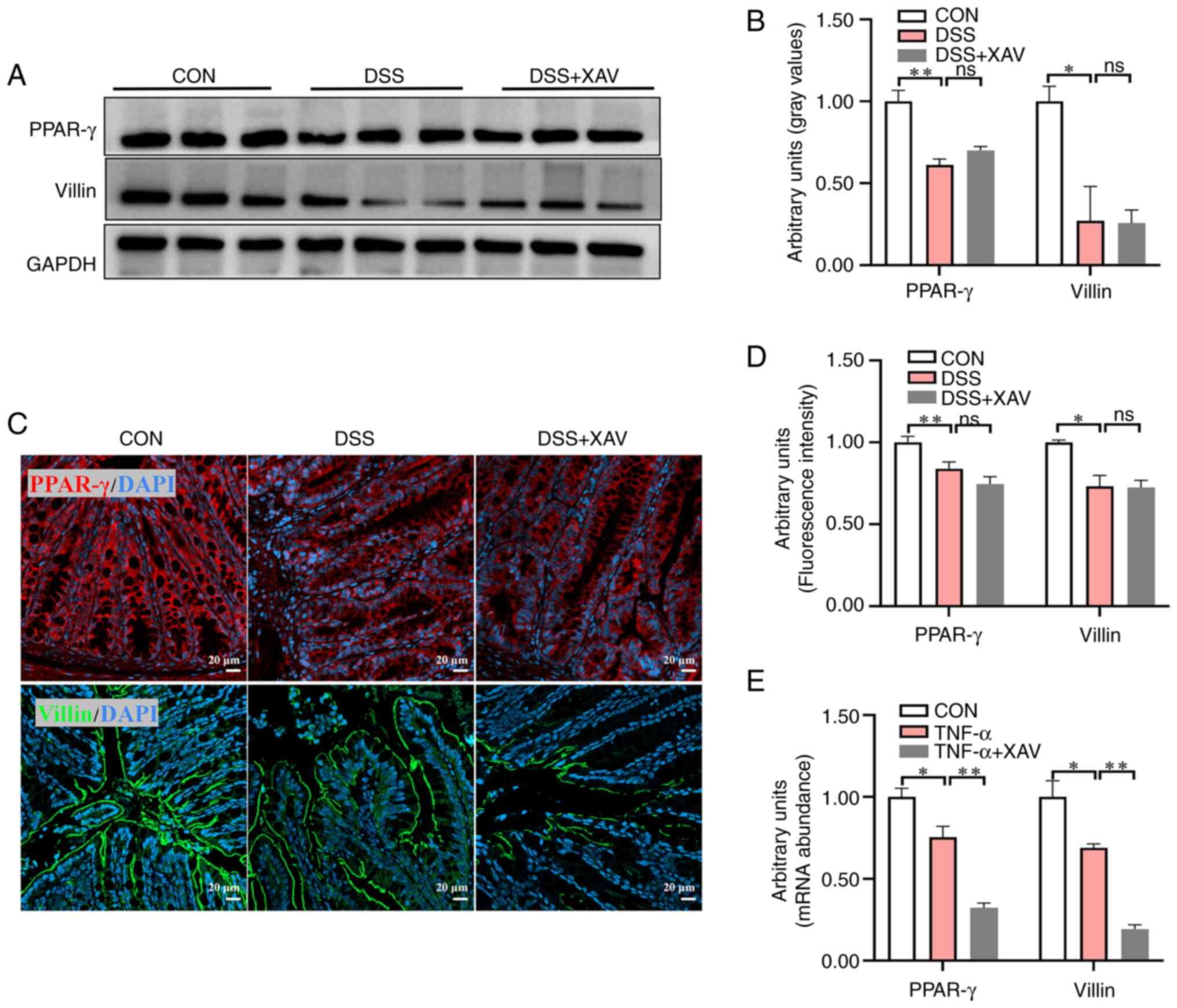
XAV939 failed to reverse the reduction
of PPAR-γ and Villin
Previous studies have suggested that the PPAR-γ
signaling pathway serves a role in enterocyte differentiation, and
Villin serves as a marker for enterocytes in the intestine
(31,32). To further investigate the
regulatory effect of XAV939 on enterocyte differentiation, western
blotting was used to assess the protein expression levels of PPAR-γ
and Villin (Fig. 6A). The results
demonstrated that DSS treatment led to decreased protein expression
levels of PPAR-γ (Fig. 6B,
P<0.01) and Villin (Fig. 6B,
P<0.05). However, XAV939 did not significantly reverse this
decreased expression (Fig. 6B).
Simultaneously, PPAR-γ and Villin protein expression was determined
by IHC (Fig. 6C). DSS
significantly decreased the fluorescence intensity of PPAR-γ
(Fig. 6D, P<0.01) and Villin
(Fig. 6D, P<0.05), while XAV939
did not significantly reverse the reduction of the fluorescence
intensity of PPAR-γ (Fig. 6D,
P=0.31) and Villin (Fig. 6D,
P=0.99). In the intestinal organoid model, XAV939 significantly
reduced the expression levels of PPAR-γ (Fig. 6E, P<0.01) and Villin (Fig. 6E, P<0.01) compared with the
TNF-α group, which indicated that the organoid was more sensitive
to the inhibitory effects of XAV939 compared with the ISC in
vivo.
Discussion
The present study provided insights into the effect
of XAV939 on Wnt/β-catenin signaling in a mouse model of
DSS-induced UC and suggested its potential association with the
differentiation of secretory progenitor cells. Nevertheless, the
present findings showed that XAV939 did not increase the length of
the colon, enhance epithelial structure or reduce inflammatory
cytokines. These results indicated that XAV939 may not be efficient
in the treatment of UC. The present study further demonstrated that
XAV939 effectively suppressed SOX9, a downstream target of the
Wnt/β-catenin signaling pathway, in DSS-induced UC. SOX9 serves a
pivotal role in regulating the differentiation of ISCs into
secretory progenitor cells (33).
Therefore, XAV939 may show potential to reverse the excessive
differentiation of secretory progenitor cells in the DSS-induced UC
mouse model. However, the present study demonstrated that the use
of XAV939 did not mitigate inflammation. XAV939 regulated the
differentiation of secretory cell progenitor cells via the
Wnt/β-catenin signaling pathway. Unraveling the complex mechanisms
underlying ISC differentiation and its dysregulation in UC is
crucial for developing effective therapeutic strategies.
The mechanism by which DSS induces UC is not fully
understood, but it may include several aspects, such as damaging
colonic epithelial cells, activating immune inflammatory responses
and disrupting the intestinal flora balance (34). XAV939 may exert therapeutic effects
by regulating ISC differentiation and inhibiting inflammatory
responses. However, the present study demonstrated that XAV939 did
not exert a significant inhibitory effect on the inflammatory
response induced by TNF-α, therefore, it was ineffective at
suppressing the inflammatory storm caused by DSS. In the context of
UC treatment, controlling inflammation is crucial for restoring the
intestinal epithelium. The purpose of using XAV939 was to affect
the Wnt/β-catenin signaling pathway and to simultaneously modulate
the inflammatory response. A previous study reported that XAV939
exhibits suppressive effects on the inflammatory response induced
by LPS (35). However, in the
present study, XAV939 treatment did not lead to a decrease in the
expression levels of inflammatory cytokines in the TNF-α-induced
intestinal organoid model. This may be due to LPS and TNF-α
inducing inflammation through distinct molecular mechanisms. LPS
primarily acts through Toll-like receptor 4 (TLR4), while TNF-α
signals through the tumor necrosis factor receptor superfamily
(36,37). This suggests that the
anti-inflammatory effects of XAV939 are mechanism-specific and may
not be broadly applicable to all inflammatory stimuli.
Additionally, the TLR4 and NF-κB signaling pathways serve crucial
roles in the pathogenesis of UC. XAV939 can inhibit the activity of
TLR4, which may be achieved by blocking the binding of TLR4 to its
ligand or interfering with downstream signaling molecules of
TLR4(38). XAV939 does not have a
direct inhibitory effect on NF-κB (39). As the activation of NF-κB is also
regulated by other signaling pathways, the inhibitory effect of
XAV939 on TLR4 may not be sufficient to completely block the
regulatory effects of these signaling pathways on the inflammatory
response, which results in its inability to effectively inhibit
inflammation caused by DSS. The Wnt/β-catenin signaling pathway
exhibits both stimulatory and inhibitory effects on NF-κB-mediated
inflammation, and the underlying molecular mechanisms involved are
complex and multifaceted (40,41).
Notably, the impact of β-catenin on NF-κB may vary depending on the
specific genetic context or cell type examined. Further research is
required to gain a deeper understanding into how the Wnt/β-catenin
and NF-κB signaling pathways interact in UC. Such insights could
ultimately contribute to the development of more effective
therapeutic interventions for the treatment of UC.
PPAR-γ serves a crucial role in regulating lipid and
glucose metabolism, as well as inflammatory responses (42,43).
Notably, in patients with UC, there is a negative association
between PPAR-γ expression and disease severity (44). By activating PPAR-γ, inflammatory
factors can be inhibited to reduce the inflammatory response in the
colonic mucosa, thereby improving the condition of patients with UC
(45). On the other hand, a
previous study reported that PPAR-γ functions as a brake on the
Wnt/β-catenin signaling pathway (46). Consequently, the activation of
PPAR-γ effectively inhibits the Wnt/β-catenin signaling pathway
(47). Whether downregulation of
the Wnt/β-catenin signaling pathway also leads to the activation of
the PPAR-γ signaling pathway remains to be investigated. However,
the present study demonstrated that XAV939 treatment did not
increase the expression levels of PPAR-γ, suggesting that PPAR-γ
potentially resides upstream of β-catenin. This finding underscores
the importance of PPAR-γ as a potential therapeutic target in
modulating both inflammatory processes and the Wnt/β-catenin
signaling cascade in UC. Further exploration of the intricate
interplay between these pathways and their respective regulators
may reveal novel strategies for the management of symptoms and
progression of UC.
Within the intestinal tract, enterocytes serve as a
vital barrier, as they are responsible for nutrient absorption and
defense against harmful microbial invasion (48). A number of studies have reported
that UC is associated with the loss of colonic crypt structures,
and disruption of enterocytes impacts intestinal absorption and
compromises the barrier function of the intestine (49,50).
Through the use of single-cell sequencing, research has shown a
notable decrease in the population of enterocytes and their
progenitor cells within the intestines of patients with UC
(51). Villin serves as a marker
for enterocytes, and the present in vivo and in vitro
experiments demonstrated that XAV939 could not reverse the
differentiation of enterocytes. Notably, the differentiation of
enterocytes is regulated by both the Wnt/β-catenin signaling
pathway and the Notch signaling pathway. XAV939 cannot promote the
differentiation of enterocytes by inhibiting Wnt/β-catenin alone
(52). The present results
demonstrated an association between PPAR-γ and Villin expression,
implying that PPAR-γ may serve a regulatory role in the
differentiation of enterocytes. A study examining the fruit fly
intestine showed that the activation of the PPAR-γ homolog,
ecdysone-induced protein 75B, stimulated the differentiation of
ISCs into absorptive intestinal epithelial cell lineages (53). Furthermore, it has been documented
that the UC risk gene, organic cation/carnitine transporter 2
(OCTN2), is under the regulatory influence of PPAR-γ, and that
OCTN2 is predominantly expressed in enterocytes (54-56).
OCTN2 participates in the transport of carnitine, which is a
crucial step in fatty acid oxidation (FAO). Repairing the impeded
FAO improved ISC function and ameliorated DSS-induced colitis in
mice (57). Research has shown
that improving oxidative phosphorylation metabolism enhanced
mitochondrial function in intestinal epithelial cells and reduced
DSS-induced intestinal inflammation (58). Thereby, continuous and in-depth
research on the dysregulation of energy metabolism in enterocytes,
as well as the regulatory role of PPAR-γ, may offer valuable
insights into the underlying mechanisms of UC.
The results of the present in vivo
experiments on the effects of XAV939 on PPAR-γ and Villin were
inconsistent with those obtained from intestinal organoid cultures.
The potential reasons include the effect of microenvironment and
the impact of Wnt/β-catenin signaling on ISCs expansion. In the
crypt, the ISCs are protected by surrounding niche cells (59). However, under ex vivo
conditions, ISCs are directly exposed to the XAV939. The activation
of the Wnt/β-catenin signaling pathway ex vivo mainly
originates from Wnt ligands and enhancers in the culture medium,
while Wnt signals in the crypt mainly come from niche cells, which
are more stable (60).
Additionally, a certain level of Wnt signaling is necessary for the
maintenance and expansion of ISCs. The loss of other sources of Wnt
activation ex vivo hinders the expansion of stem cells,
which partly explained why XAV939 intervention ex vivo led
to a further decrease in PPAR-γ and Villin expression.
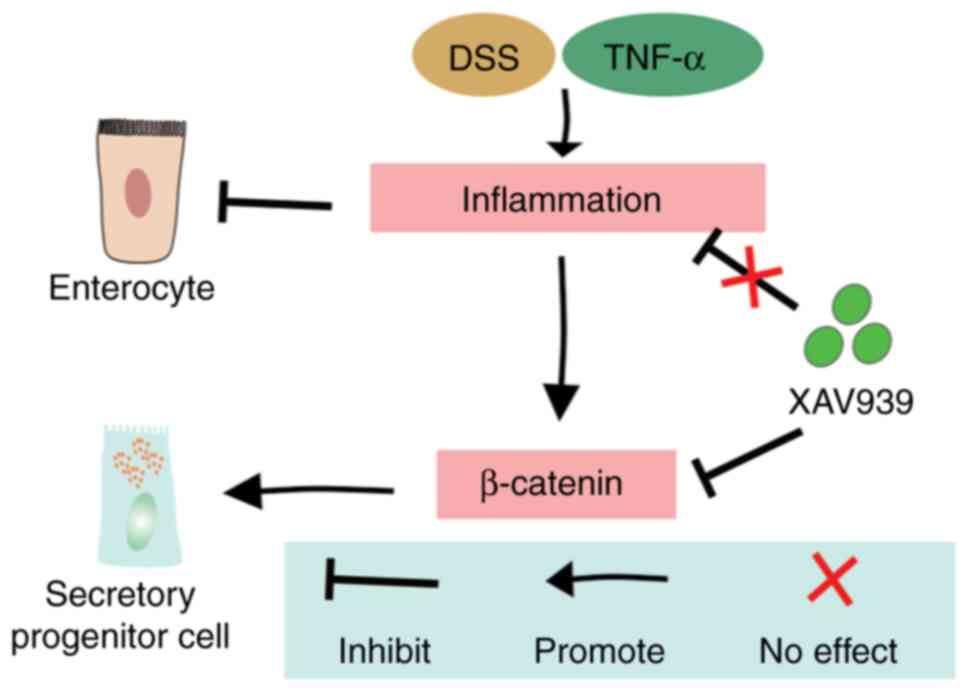
In conclusion, the present study showed that XAV939
did not improve inflammation or intestinal morphology in
DSS-induced UC. However, the present study elucidated the role of
XAV939 in regulating ISC for DSS-induced intestinal injury through
inhibition of the Wnt/β-catenin pathway (Fig. 7). Considering the significance of
PPAR-γ and the differentiation of enterocytes, it is necessary to
further investigate their interaction and understand the underlying
mechanisms. Furthermore, given the complexity of UC and the
involvement of multiple inflammatory pathways, a single agent such
as XAV939 may not be sufficient to address all aspects of the
disease. Consequently, a combination of therapeutic strategies
targeting different inflammatory pathways and ISC functions may be
necessary for effective UC treatment.
Supplementary Material
Experimental design outlining the
grouping and treatment of mice. The DSS and DSS + XAV groups
received 3.5% DSS for 7 days. XAV was dissolved in DMSO and
administered by intraperitoneal injection for 7 days. The control
and DSS groups were injected with PBS mixed with the same DMSO
concentration. CON, control; XAV, XAV939; DSS, dextran sulfate
sodium; ns, not significant; D, day.
Crypts obtained exhibited intact
structures and high purity, which ensured the quality of the
resulting organoids. Left magnification, x40; right magnification,
x100.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Guangdong Basic and
Applied Basic Research Foundation (grant no. 2023A1515110203) and
the Postdoctoral Startup Fund of Shunde Women and Children's
Hospital of Guangdong Medical University (grant no.
2022BSHQD002).
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus database under accession number
GSE275191 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE275191.
Authors' contributions
LWX and DJZ conceived the research and critically
revised the manuscript. SJL, DBM and KW performed the experiments
and prepared the manuscript. SJL and KW confirm the authenticity of
all the raw data. All authors read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
All animal procedures were performed in accordance
with the Guidelines for Care and Use of Laboratory Animals of
Guangdong Medical University and experiments were approved by
Ethics Committee of Shunde Maternal and Children's Hospital of
Guangdong Medical University (approval no. 2023054).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Le Berre C, Honap S and Peyrin-Biroulet L:
Ulcerative colitis. Lancet. 402:571–584. 2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang M, Fu R, Xu D, Chen Y, Yue S, Zhang S
and Tang Y: Traditional Chinese Medicine: A promising strategy to
regulate the imbalance of bacterial flora, impaired intestinal
barrier and immune function attributed to ulcerative colitis
through intestinal microecology. J Ethnopharmacol.
318(116879)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hassan SA, Kapur N, Sheikh F, Fahad A and
Jamal S: Disease clearance in ulcerative colitis: A new therapeutic
target for the future. World J Gastroenterol. 30:1801–1809.
2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Le Berre C, Roda G, Nedeljkovic Protic M,
Danese S and Peyrin-Biroulet L: Modern use of 5-aminosalicylic acid
compounds for ulcerative colitis. Expert Opin Biol Ther.
20:363–378. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
De Deo D, Dal Buono A, Gabbiadini R,
Spaggiari P, Busacca A, Masoni B, Ferretti S, Bezzio C and Armuzzi
A: Management of proctitis in ulcerative colitis and the place of
biological therapies. Expert Opin Biol Ther. 24:443–453.
2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Heimann TM, Swaminathan S, Slater GI and
Kurtz RJ: Perianal fistula after ileoanal pouch in patients with
ulcerative colitis: A review of 475 patients operated on at a major
IBD center. Dis Colon Rectum. 65:76–82. 2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hou Q, Huang J, Ayansola H, Masatoshi H
and Zhang B: Intestinal stem cells and immune cell relationships:
Potential therapeutic targets for inflammatory bowel diseases.
Front Immunol. 11(623691)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Quandt J, Arnovitz S, Haghi L, Woehlk J,
Mohsin A, Okoreeh M, Mathur PS, Emmanuel AO, Osman A, Krishnan M,
et al: Wnt-β-catenin activation epigenetically reprograms
Treg cells in inflammatory bowel disease and dysplastic
progression. Nat Immunol. 22:471–484. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Swafford D, Shanmugam A, Ranganathan P,
Manoharan I, Hussein MS, Patel N, Sifuentes H, Koni PA, Prasad PD,
Thangaraju M and Manicassamy S: The Wnt-β-catenin-IL-10 signaling
axis in intestinal APCs protects mice from colitis-associated colon
cancer in response to gut microbiota. J Immunol. 205:2265–2275.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chang M, Chang L, Chang HM and Chang F:
Intestinal and extraintestinal cancers associated with inflammatory
bowel disease. Clin Colorectal Cancer. 17:e29–e37. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hirano T, Hirayama D, Wagatsuma K,
Yamakawa T, Yokoyama Y and Nakase H: Immunological mechanisms in
inflammation-associated colon carcinogenesis. Int J Mol Sci.
21(3062)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li F, Yan H, Jiang L, Zhao J, Lei X and
Ming J: Cherry polyphenol extract ameliorated dextran sodium
sulfate-induced ulcerative colitis in mice by suppressing
Wnt/β-catenin signaling pathway. Foods. 11(49)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Dong LN, Wang M, Guo J and Wang JP:
Influences of probiotics combined with sulfasalazine on rats with
ulcerative colitis via the Wnt/β-catenin signaling pathway. Eur Rev
Med Pharmacol Sci. 23:6371–6378. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jang J, Jung Y, Chae S, Bae T, Kim SM,
Shim YJ, Chung SI and Yoon Y: XAV939, a Wnt/β-catenin pathway
modulator, has inhibitory effects on LPS-induced inflammatory
response. Immunopharmacol Immunotoxicol. 41:394–402.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yao YY, Bian LG, Yang P, Sui Y, Li R, Chen
YL, Sun L, Ai QL, Zhong LM and Lu D: Gastrodin attenuates
proliferation and inflammatory responses in activated microglia
through Wnt/β-catenin signaling pathway. Brain Res. 1717:190–203.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Takahashi T: Roles of nAChR and Wnt
signaling in intestinal stem cell function and inflammation. Int
Immunopharmacol. 81(106260)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hiramatsu Y, Fukuda A, Ogawa S, Goto N,
Ikuta K, Tsuda M, Matsumoto Y, Kimura Y, Yoshioka T, Takada Y, et
al: Arid1a is essential for intestinal stem cells through Sox9
regulation. Proc Natl Acad Sci USA. 116:1704–1713. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Moparthi L and Koch S: Wnt signaling in
intestinal inflammation. Differentiation. 108:24–32.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Parikh K, Antanaviciute A, Fawkner-Corbett
D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen
HH, Alham NK, et al: Colonic epithelial cell diversity in health
and inflammatory bowel disease. Nature. 567:49–55. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wei P, He Q, Liu T, Zhang J, Shi K, Zhang
J and Liu S: Baitouweng decoction alleviates dextran sulfate
sodium-induced ulcerative colitis by suppressing leucine-related
mTORC1 signaling and reducing oxidative stress. J Ethnopharmacol.
304(116095)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang H, Lang W, Li S, Xu C, Wang X, Li Y,
Zhang Z, Wu T and Feng M: Corynoline ameliorates dextran sulfate
sodium-induced colitis in mice by modulating Nrf2/NF-κB pathway.
Immunopharmacol Immunotoxicol. 45:26–34. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Distler A, Deloch L, Huang J, Dees C, Lin
NY, Palumbo-Zerr K, Beyer C, Weidemann A, Distler O, Schett G and
Distler JH: Inactivation of tankyrases reduces experimental
fibrosis by inhibiting canonical Wnt signalling. Ann Rheum Dis.
72:1575–1580. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lachmann A, Clarke DJB, Torre D, Xie Z and
Ma'ayan A: Interoperable RNA-Seq analysis in the cloud. Biochim
Biophys Acta Gene Regul Mech. 1863(194521)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: a Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
McDermaid A, Monier B, Zhao J, Liu B and
Ma Q: Interpretation of differential gene expression results of
RNA-seq data: Review and integration. Brief Bioinform.
20:2044–2054. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mi JX, Zhang YN, Lai Z, Li W, Zhou L and
Zhong F: Principal component analysis based on nuclear norm
minimization. Neural Netw. 118:1–16. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yoo YE, Lee S, Kim W, Kim H, Chung C, Ha
S, Park J, Chung Y, Kang H and Kim E: Early chronic memantine
treatment-induced transcriptomic changes in wild-type and
Shank2-mutant mice. Front Mol Neurosci. 14(712576)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liang B, Jiang Y, Song S, Jing W, Yang H,
Zhao L, Chen Y, Tang Q, Li X, Zhang L, et al: ASPP2 suppresses
tumour growth and stemness characteristics in HCC by inhibiting
Warburg effect via WNT/β-catenin/HK2 axis. J Cell Mol Med.
27:659–671. 2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ming Z, Vining B, Bagheri-Fam S and Harley
V: SOX9 in organogenesis: Shared and unique transcriptional
functions. Cell Mol Life Sci. 79(522)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kitamura S, Miyazaki Y, Shinomura Y, Kondo
S, Kanayama S and Matsuzawa Y: Peroxisome proliferator-activated
receptor gamma induces growth arrest and differentiation markers of
human colon cancer cells. Jpn J Cancer Res. 90:75–80.
1999.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Centonze M, Berenschot EJW, Serrati S,
Susarrey-Arce A and Krol S: The fast track for intestinal tumor
cell differentiation and in vitro intestinal models by inorganic
topographic surfaces. Pharmaceutics. 14(218)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yang C and Merlin D: Unveiling colitis: A
journey through the dextran sodium sulfate-induced model. Inflamm
Bowel Dis. 30:844–853. 2024.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhong Y, Wang K, Zhang Y, Yin Q, Li S,
Wang J, Zhang X, Han H and Yao K: Ocular Wnt/β-catenin pathway
inhibitor XAV939-loaded liposomes for treating alkali-burned
corneal wound and neovascularization. Front Bioeng Biotechnol.
9(753879)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu J, Kang R and Tang D:
Lipopolysaccharide delivery systems in innate immunity. Trends
Immunol. 45:274–287. 2024.PubMed/NCBI View Article : Google Scholar
|
|
37
|
So T and Ishii N: The TNF-TNFR family of
co-signal molecules. Adv Exp Med Biol. 1189:53–84. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Shi J, Ma C, Hao X, Luo H and Li M:
Reserve of Wnt/β-catenin signaling alleviates mycoplasma pneumoniae
P1-C-induced Inflammation in airway epithelial cells and lungs of
mice. Mol Immunol. 153:60–74. 2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chen T, Zhou R, Chen Y, Fu W, Wei X, Ma G,
Hu W and Lu C: Curcumin ameliorates IL-1β-induced apoptosis by
activating autophagy and inhibiting the NF-κB signaling pathway in
rat primary articular chondrocytes. Cell Biol Int. 45:976–988.
2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhou J, Wu H, Hou J, Wang J, Wang J, Li M,
Yao X, Gao J and Zhang Q: Daurisoline alleviated experimental
colitis in vivo and in vitro: Involvement of NF-κB and
Wnt/β-catenin pathway. Int Immunopharmacol.
108(108714)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ma B and Hottiger MO: Crosstalk between
Wnt/β-catenin and NF-κB signaling pathway during inflammation.
Front Immunol. 7(378)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Caioni G, Viscido A, d'Angelo M, Panella
G, Castelli V, Merola C, Frieri G, Latella G, Cimini A and
Benedetti E: Inflammatory bowel disease: New insights into the
interplay between environmental factors and PPARγ. Int J Mol Sci.
22(985)2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang N, Kong R, Han W, Bao W, Shi Y, Ye L
and Lu J: Honokiol alleviates ulcerative colitis by targeting
PPAR-γ-TLR4-NF-κB signaling and suppressing gasdermin-D-mediated
pyroptosis in vivo and in vitro. Int Immunopharmacol.
111(109058)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Fang J, Wang H, Xue Z, Cheng Y and Zhang
X: PPARγ: The central mucus barrier coordinator in ulcerative
colitis. Inflamm Bowel Dis. 27:732–741. 2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Venkataraman B, Ojha S, Belur PD, Bhongade
B, Raj V, Collin PD, Adrian TE and Subramanya SB: Phytochemical
drug candidates for the modulation of peroxisome
proliferator-activated receptor γ in inflammatory bowel diseases.
Phytother Res. 34:1530–1549. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Selim MA, Mosaad SM and El-Sayed NM:
Lycopene protects against Bisphenol A induced toxicity on the
submandibular salivary glands via the upregulation of PPAR-γ and
modulation of Wnt/β-catenin signaling. Int Immunopharmacol.
112(109293)2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Jeon KI, Phipps RP, Sime PJ and Huxlin KR:
Antifibrotic actions of peroxisome proliferator-activated receptor
γ ligands in corneal fibroblasts are mediated by
β-catenin-regulated pathways. Am J Pathol. 187:1660–1669.
2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Oda M, Hatano Y and Sato T: Intestinal
epithelial organoids: Regeneration and maintenance of the
intestinal epithelium. Curr Opin Genet Dev.
76(101977)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Dunleavy KA, Raffals LE and Camilleri M:
Intestinal barrier dysfunction in inflammatory bowel disease:
Underpinning pathogenesis and therapeutics. Dig Dis Sci.
68:4306–4320. 2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Rath T, Atreya R and Neurath MF: A
spotlight on intestinal permeability and inflammatory bowel
diseases. Expert Rev Gastroenterol Hepatol. 17:893–902.
2023.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Li G, Zhang B, Hao J, Chu X, Wiestler M,
Cornberg M, Xu CJ, Liu X and Li Y: Identification of novel
population-specific cell subsets in chinese ulcerative colitis
patients using single-cell RNA sequencing. Cell Mol Gastroenterol
Hepatol. 12:99–117. 2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liang SJ, Li XG and Wang XQ: Notch
signaling in mammalian intestinal stem cells: Determining cell fate
and maintaining homeostasis. Curr Stem Cell Res Ther. 14:583–590.
2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zipper L, Jassmann D, Burgmer S, Görlich B
and Reiff T: Ecdysone steroid hormone remote controls intestinal
stem cell fate decisions via the PPARγ-homolog Eip75B in
Drosophila. Elife. 9(e55795)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Kong L, Pokatayev V, Lefkovith A, Carter
GT, Creasey EA, Krishna C, Subramanian S, Kochar B, Ashenberg O,
Lau H, et al: The landscape of immune dysregulation in Crohn's
disease revealed through single-cell transcriptomic profiling in
the ileum and colon. Immunity. 56:444–458.e5. 2023.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Smillie CS, Biton M, Ordovas-Montanes J,
Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M,
Waldman J, et al: Intra- and inter-cellular rewiring of the human
colon during ulcerative colitis. Cell. 178:714–730.e22.
2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhou S and Shu Y: Transcriptional
regulation of solute carrier (SLC). Drug transporters. Drug Metab
Dispos. 50:1238–1250. 2022.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chen L, Jiao T, Liu W, Luo Y, Wang J, Guo
X, Tong X, Lin Z, Sun C, Wang K, et al: Hepatic cytochrome P450 8B1
and cholic acid potentiate intestinal epithelial injury in colitis
by suppressing intestinal stem cell renewal. Cell Stem Cell.
29:1366–1381.e9. 2022.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Kapur N, Alam MA, Hassan SA, Patel PH,
Wempe LA, Bhogoju S, Goretsky T, Kim JH, Herzog J, Ge Y, et al:
Enhanced mucosal mitochondrial function corrects dysbiosis and
OXPHOS metabolism in IBD. bioRxiv [Preprint]: 2024.03.14.584471,
2024.
|
|
59
|
Yin X, Farin HF, van Es JH, Clevers H,
Langer R and Karp JM: Niche-independent high-purity cultures of
Lgr5+ intestinal stem cells and their progeny. Nat Methods.
11:106–112. 2014.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Hageman JH, Heinz MC, Kretzschmar K, van
der Vaart J, Clevers H and Snippert HJG: Intestinal regeneration:
Regulation by the microenvironment. Dev Cell. 54:435–446.
2020.PubMed/NCBI View Article : Google Scholar
|