Introduction
Chronic myeloid leukemia (CML) is a hematopoietic
stem cell disorder characterized by marked increase in
granulocytes, bone marrow hyperplasia, and splenomegaly. The
current annual incidence of CML has been reported to be 1 to 2
cases per 100,000 people, and accounts for 15% of adult leukemias
diagnosed worldwide (1). The
molecular hallmark of CML is the Philadelphia (Ph) chromosome, a
reciprocal translocation t (9:22) (q34; q11) between the
proto-oncogene ABL on chromosome 9 and the BCR gene on chromosome
22. This abnormality has been clinically detected in 95% of CML
patients, and is considered the primary risk factor for CML
development (2). The Ph
translocation creates a fusion BCR-ABL oncogene, which
encodes a functional protein with constitutive tyrosine kinase
activity that aberrantly modulates tumorigenic pathways.
Imatinib is the first chemotherapeutic agent
described as a specific BCR-ABL tyrosine kinase inhibitor. Its
proven efficacy for the treatment of patients with CML has led to
its preferred use as first-line treatment, supplanting the
traditional single agents busulphan, hydroxyurea and interferon-α
(3). The molecular mechanism of
imatinib has been described: imatinib potently binds to the
inactive form of BCR-ABL, thereby blocking the ATP-binding site and
its enzymatic activity (4).
Imatinib treatment of CML patients has produced estimated
cumulative best rates of complete hematologic response (CHR) and
complete cytogenetic response (CCyR) of 98 and 87%, respectively
(5). Despite the significant
improvements in the outcome for patients with CML following
treatment with imatinib, the exact mechanism of its anti-leukemic
effect remains to be completely elucidated (6).
In a previous study, we discovered that imatinib
treatment of K562 leukemia cells produced a significant change in
the ratio of Bcl-x splice variants. The Bcl-x gene undergoes
alternative splicing at exon 2 to produce two isoforms with
distinctive anti- and pro-apoptotic properties. The
Bcl-xS isoform promotes apoptosis, while the
Bcl-xL inhibits apoptosis. Imatinib treatment of K562
cells led to a transcriptional shift towards the Bcl-xS
isoform, which in turn promoted apoptosis in the treated cells.
Accordingly, this finding suggested that imatinib could regulate
the alternative splicing of other apoptotic genes in K562
cells.
Alternative splicing is a common mechanism used by
the human system to generate diversity in the transcriptome and
proteome. Through alternative splicing, multiple different
transcripts can be generated from a single mRNA precursor. Recent
studies have suggested that up to 94% of all human genes undergo
alternative splicing, and aberrant pre-mRNA splicing has been
implicated in the pathogenesis of neoplasia (7,8).
Likewise, studies have demonstrated that most of the currently used
chemotherapeutic agents not only induce an apoptotic response, but
also affect the alternative splicing of some genes. Shkreta et
al (9) determined that 20 of
the mainstream anticancer drugs could influence the production of
Bcl-x splice isoforms. In addition, these drugs could alter a
subset of alternative splicing events in some cell lines,
suggesting that the influence of anticancer drugs on alternative
splicing is a very common phenomenon.
In recent years, powerful techniques for genome-wide
identification and analysis of alternative splicing isoforms have
been developed. These large-scale high-throughput analytical
methods have been successfully applied for the identification of
differential splicing events in various cancer tissues (10). Exon arrays, which contain both
known and predicted exons, are rapidly gaining popularity and
becoming a standard for both gene- and exon-level expression
analyses (11). The Affymetrix
GeneChip® Human Exon 1.0 ST array (Human Exon 1.0 ST
array) is the latest product in the family of exon arrays. It
contains approximately 5.4 million probes grouped into 1.4 million
probe sets, allowing the interrogation of over 1 million exon
clusters, which are exon annotations from various sources that
overlap by genomic location. For each gene, the average number of
probes on the exon array is 35, and these probes are usually
distributed along the entire transcript sequence. The commercial
availability of specific software for exon-level expression
analysis has provided a powerful tool for the study of alternative
splicing for every known and predicted exon. At the same time, exon
arrays provide robust gene-level expression analysis (Affymetrix
Technical notes, Identifying and Validating Alternative Splicing
Events) facilitating identification of relatively subtle
differences in the isoform expression ratios.
In our current study, we employed the Human Exon 1.0
ST Array to investigate the comprehensive profile of apoptotic
genes in leukemia K562 cells that are differentially spliced in
response to imatinib treatment.
Materials and methods
Cell culture
Human myeloid leukemia cells K562 were grown in
Roswell Park Memorial Institute 1640 (RPMI-1640; Hyclone, USA)
media supplemented with 10% fetal bovine serum (FBS; Hyclone) in a
5% CO2 humidified atmosphere at 37°C. Experimental
cultures were initiated at a density of 2×105 cells/ml.
Imatinib (Novartis Co., China) was added from a 1 mM stock solution
in DMSO to achieve various final concentrations in culture. The
control non-treated cells were generated by adding an equal volume
of DMSO alone. Cells were incubated for up to 24 h before cell
proliferation was assessed by WST-1 reagent based assay kit
(Millipore, USA) and apoptosis was assessed by flow cytometry. For
details of the two methods, refer to our previous study (12).
RNA extraction and array
hybridization
Total-RNA was extracted from 107 K562
cells by using the TRIzol reagent (Tiangen, China) following the
manufacturer’s instructions. The extracted RNA was further purified
by DNase I treatment using an RNeasy mini kit and an RNase-Free
DNase Set (Qiagen, China), according to the manufacturer’s
protocols. The concentration and purity of total-RNA were
determined by spectrophotometric measurements at an optical density
(OD) of 260/280 nm and electrophoresis on 1% agarose gel. An
OD260/OD280 ratio of 1.8–2.0 was considered
to indicate pure RNA. RNA samples were immediately frozen and
stored at −80°C until required for use in further microarray
studies.
Two 1-μg RNA samples from untreated control K562
cells and imatinib-treated K562 cells (1 μM of imatinib for 24 h)
were labeled according to the GeneChip whole transcript (WT) Sense
Target Labeling Assay as provided by the manufacturer (http://www.affymetrix.com). Labeled RNA was hybridized
to the Human Exon 1.0 ST Arrays for 16 h, following the Affymetrix
protocol. Afterwards, arrays were scanned using the Affymetrix GCS
3000 7G Scanner and digital images were processed by the
GeneChip® Operating Software v1.4 to produce raw CEL
intensity files for analysis of differential expression.
Data analysis
Data from the exon arrays were processed using the
easyExon software (http://microarray.ym.edu.tw/easyexon). This tool
provides a user-friendly, platform-independent and efficient
processing method to explore Affymetrix exon array data. Two
parallel analyses (gene-level and exon-level) were carried out by
this software. To improve the processing efficiency, we downloaded
the related human gene database and meta probeset files from the
software homepage (http://microarray.ym.edu.tw:8080/easyexon/index.jsp?mode=support)
and installed each as a local copy.
After data loading of the CEL files, gene/exon
signal summarizations were calculated automatically by the
Affymetrix Power Tools (APT) package, which can execute the binary
file ‘apt-probeset-summarize’ to generate both exon-level and
gene-level summary files. Two of the most commonly used signal
estimation algorithms, RMA and PLIER, were implemented to combine
information from probes belonging to the same transcript, or exon,
to generate a summarized expression signal value of the single
represented gene or exon (13).
Background noise was detected using the ‘Detection above Background
(DABG)’ algorithm. Normalization was performed by the ‘quantile
normalization’ algorithm for both the exon- and gene-level.
Reverse transcription-PCR (RT-PCR) and
sequencing validation
RT-PCR and direct sequencing were used to confirm
the differentially expressed alternatively spliced genes detected
by the Human Exon 1.0 ST Arrays. Reverse transcription was
performed with 1 μg of total-RNA using a reverse transcription kit
(Takara Bio, Inc., Japan), following the manufacturer’s
instructions. For subsequent PCR analysis, 1 μl of cDNA was used.
The primers and reaction conditions for each transcript are shown
in Table I. The amplified
products were electrophoresed through 1.5–2.5% agarose gels,
according to amplified fragment size requirements for resolution.
Images of the gels were analyzed by the Quantity One 4.6.2 software
(Bio-Rad, USA). Ratios between splice variants expressed in
untreated and imatinib-treated cells were calculated. Distinct gel
bands were purified with a gel extraction kit (Tiangen), sequenced
using the same primers as for PCR and the BigDye Terminator v3.1
Cycle Sequencing kit (Applied Biosystems, USA), and analyzed on an
ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
 | Table IPrimers and amplification conditions
used in this study. |
Table I
Primers and amplification conditions
used in this study.
| Gene name | Sequence
(5′→3′) | Reaction
condition | Splicing
isoform | Product size
(bp) |
|---|
| Bcl-x | F:
ATGGCAGCAGTAAAGCAAGCG | 32 cycles at 94°C
for 30 sec, 55°C for 30 sec, and 72°C for 1 min |
Bcl-xL | 456 |
| R:
TCATTTCCGACTGAAGAGTGA |
Bcl-xS | 267 |
| RNF34 | F:
TGAGTTTCCTGGTAGAGCC | 35 cycles at 94°C
for 30 sec, 55°C for 30 sec, and 72°C for 1 min | Variant L | 173 |
| R:
TTCCCATGACTTCATTCAGC | Variant S | 107 |
| SNCA | F:
GTTGGAGGAGCAGTGGTG | 35 cycles at 94°C
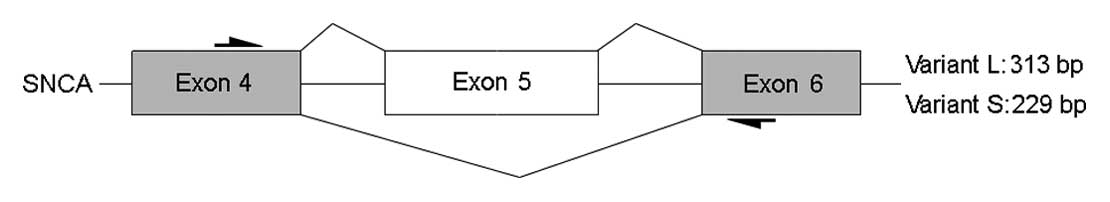
for 30 sec, 64.5°C for 45 sec, and 72°C for 1 min | Variant L | 313 |
| R:
ATGACTGGGCACATTGGA | Variant S | 229 |
| β-actin | F:
CGGGAAATCGTGCGTGAC | 35 cycles at 94°C
for 30 sec, 55°C for 30 sec, and 72°C for 1 min | - | 443 |
| R:
TGGAAGGTGGACAGCGAGG | | |
Results
Exon-level expression differences induced
by imatinib treatment
The pro-apoptotic activity of imatinib on K562 cells
was first confirmed by a WST-1 assay and a double fluorescence
assay to analyze cell proliferation and apoptosis, respectively.
Our results revealed that imatinib exposure rapidly induced
proliferation inhibition and apoptosis of K562 cells in a
time-dependent manner [data not shown; identical to that of our
previous study (12)].
The expression of alternatively spliced genes in the
K562 cells in response to treatment with imatinib were then
assessed via exon array and easyExon analysis. Before performing
statistical filtration, the limitation number of probesets in a
transcript cluster was set from 4 to 40. For any transcript cluster
with probesets greater than the setting number, only the setting
number of probesets was included in the latter analysis. Three
statistical methods for exon-level filtration in the easyExon
software were available: Affymetrix MIDAS (Microarray Detection of
Alternative Splicing), Partek AS ANOVA (Alternative Splice Analysis
of Variance), and PAC (Pattern-Based Correlation) for exon
analysis. We used the default Affymetrix MIDAS. For selecting
alternatively spliced genes, the criteria for probeset filtration
were set as follows, fold-change (FC) ≥2 or ≤0.5, with a
statistical significance of P<0.05.
A total of 331 genes represented by one or several
probesets met the above criteria. Most of these probesets presented
change in expression of less than 3-fold. The genes affiliated with
these probesets were investigated by the easyExon transcript
cluster filtration tool to determine which were related to cell
apoptosis. Briefly, the easyExon ‘GO Biological Process’ parameter
was set for ‘apoptosis’ related to transcript-affiliated gene name
or accession number. The program identified 175 apoptosis-related
genes from the original set of 331. Each of these genes contained
at least one probeset meeting the above criteria, for a total of
430 probesets. Tables II and
III list the 20 most
upregulated and most downregulated probesets.
 | Table IIAlternative splice probesets that
were significantly upregulated in K562 cells in response to
imatinib treatment (20 most differentially expressed)a. |
Table II
Alternative splice probesets that
were significantly upregulated in K562 cells in response to
imatinib treatment (20 most differentially expressed)a.
| Gene probe ID | Exon probe ID | Splice index | Gene title | Gene symbol | Accession no. |
|---|
| 4007617 | 4007634 | 15.12 | Pim-2 oncogene |
Pimatinib2 | NM_006875 |
| 2638676 | 2638692 | 9.74 | ELL associated
factor 2 | EAF2 | NM_018456 |
| 3939545 | 3939549 | 9.47 | Macrophage
migration inhibitory factor (glycosylation-inhibiting factor) | MIF | NM_002415 |
| 2574752 | 2574785 | 8.85 | Excision repair
cross-complementing rodent repair deficiency, complementation group
3 (xeroderma pigmentosum group B complementing) | ERCC3 | NM_000122 |
| 3818468 | 3818480 | 7.39 | Tumor necrosis
factor (ligand) superfamily, member 9 | TNFSF9 | NM_003811 |
| 2939034 | 2939056 | 6.77 | Serpin peptidase
inhibitor, clade B (ovalbumin), member 6 |
SERPINB6 | NM_004568 |
| 3127703 | 3127731 | 6.05 | Tumor necrosis
factor receptor superfamily, member 10b |
TNFRSF10B | NM_003842 |
| 3736290 | 3736292 | 6.00 | Baculoviral IAP
repeat-containing 5 (survivin) | BIRC5 | NM_001168 |
| 2524743 | 2524769 | 5.58 | FAST kinase domains
2 | FASTKD2 | NM_014929 |
| 3816509 | 3816515 | 5.30 | Growth arrest and
DNA-damage-inducible, β | GADD45B | NM_015675 |
| 3454662 | 3454675 | 5.05 | Family with
sequence similarity 130, member A1 |
FAM130A1 | NM_030809 |
| 3868183 | 3868242 | 4.80 | Interleukin 4
induced 1 | IL4I1 | NM_152899 |
| 3790704 | 3790707 | 4.62 |
Phorbol-12-myristate-13-acetate-induced
protein 1 | PMAIP1 | NM_021127 |
| 3956433 | 3956483 | 4.58 | CHK2 checkpoint
homolog (S. pombe) | CHEK2 | NM_001005735 |
| 2940920 | 2940959 | 4.50 | Eukaryotic
translation elongation factor 1 epsilon 1 | EEF1E1 | NM_004280 |
| 3886704 | 3886711 | 4.50 | Serine/threonine
kinase 4 | STK4 | NM_006282 |
| 3426257 | 3426261 | 4.47 | Suppressor of
cytokine signaling 2 | SOCS2 | NM_003877 |
| 2638676 | 2638690 | 4.36 | ELL associated
factor 2 | EAF2 | NM_018456 |
| 2940920 | 2940962 | 4.29 | Eukaryotic
translation elongation factor 1 epsilon 1 | EEF1E1 | NM_004280 |
| 2589214 | 2589215 | 4.27 | Protein kinase,
interferon-inducible double stranded RNA dependent activator | PRKRA | NM_003690 |
 | Table IIIAlternative splice probesets that
were significantly down-regulated in K562 cells in response to
imatinib treatment (20 most differentially expressed). |
Table III
Alternative splice probesets that
were significantly down-regulated in K562 cells in response to
imatinib treatment (20 most differentially expressed).
| Gene probe ID | Exon probe ID | Splice index | Gene title | Gene symbol | Accession no. |
|---|
| 3742783 | 3742800 | 0.21 | NLR family, pyrin
domain containing 1 | NLRP1 | NM_033004 |
| 3481410 | 3481444 | 0.21 | Tumor necrosis
factor receptor superfamily, member 19 |
TNFRSF19 | NM_018647 |
| 3426257 | 3426279 | 0.24 | Suppressor of
cytokine signaling 2 | SOCS2 | NM_003877 |
| 3873160 | 3873177 | 0.26 | Tribbles homolog 3
(Drosophila) | TRIB3 | NM_021158 |
| 3865301 | 3865328 | 0.27 | Excision repair
cross-complementing rodent repair deficiency, complementation group
2 (xeroderma pigmentosum D) | ERCC2 | NM_000400 |
| 3742783 | 3742808 | 0.27 | NLR family, pyrin
domain containing 1 | NLRP1 | NM_033004 |
| 3873160 | 3873179 | 0.29 | Tribbles homolog 3
(Drosophila) | TRIB3 | NM_021158 |
| 3426257 | 3426280 | 0.3 | Suppressor of
cytokine signaling 2 | SOCS2 | NM_003877 |
| 3481410 | 3481459 | 0.3 | Tumor necrosis
factor receptor superfamily, member 19 |
TNFRSF19 | NM_018647 |
| 3873160 | 3873178 | 0.3 | Tribbles homolog 3
(Drosophila) | TRIB3 | NM_021158 |
| 3838795 | 3838806 | 0.31 | Bcl-2-like 12
(proline rich) | BCL2L12 | NM_138639 |
| 3699508 | 3699557 | 0.31 | Craniofacial
development protein 1 | CFDP1 | NM_006324 |
| 3924144 | 3924180 | 0.31 | Collagen, type
XVIII, α 1 | COL18A1 | NM_030582 |
| 3873160 | 3873176 | 0.31 | Tribbles homolog 3
(Drosophila) | TRIB3 | NM_021158 |
| 3742783 | 3742792 | 0.32 | NLR family, pyrin
domain containing 1 | NLRP1 | NM_033004 |
| 3465409 | 3465413 | 0.33 | B-cell
translocation gene 1, anti-proliferative | BTG1 | NM_001731 |
| 3441849 | 3441861 | 0.33 | Tumor necrosis
factor receptor superfamily, member 1A |
TNFRSF1A | NM_001065 |
| 3441849 | 3441863 | 0.33 | Tumor necrosis
factor receptor superfamily, member 1A |
TNFRSF1A | NM_001065 |
| 3194896 | 3194952 | 0.33 | TNF
receptor-associated factor 2 | TRAF2 | NM_021138 |
| 2487412 | 2487447 | 0.34 | Annexin A4 | ANXA4 | NM_001153 |
Detection of alternative splicing events
in response to imatinib treatment
EasyExon provides a module to visualize and
interpret selected subsets of exons and transcripts that meet
specified criteria. The normalized, log-transformed and variance
stabilized probeset intensities of a transcript are plotted as
signal mean ± standard error in an intensity plot. Probesets marked
with asterisks by the program represent statistically significant
alternative splicing events which pass the defined MIDAS/Partek AS
ANOVA threshold. Probesets colored in bold by the program indicate
cases in which the intensity FC between groups was greater than the
threshold (by at least 2-fold). The degree of alternative splicing
can also be visualized by the splicing index (SI) plot in the same
panel. Different probesets for the same exon group together and are
visually indicated by the program using a triangle or trapezoid.
Adjacent exons are distinguished by different colors. From the
graphic presentation of analysis results, we were able to identify
whether a specific isoform was up or downregulated in the
imatinib-treated cells (Fig. 1B)
(13).
After identifying probesets that were differentially
spliced in response to imatinib treatment, we randomly chose three
internal exons (RNF34, DAPK1 and SNCA) for experimental validation.
In addition, we included Bcl-x as the positive control, which was
shown to be differentially spliced in K562 cells treated with
imatinib in our previous study (12).
We introduce here a workflow for detecting
alternative splicing events between untreated K562 cells and
imatinib-treated K562 cells by using the easyExon program. In Bcl-x
(ID 3902489), the probeset (ID 3902522, in bold) targeting the 3′
region of exon 2 was found to be downregulated in imatinib-treated
K562 cells. The expression fold-change of this probeset was larger
than 2 (labeled in black) and the difference was statistically
significant (marked by an asterisk by the program). This indicated
that there were two Bcl-x isoforms in K562 cells, one with both the
5′ and 3′ region of exon 2 and another with only the 5′ region of
exon 2, and the latter one was upregulated in imatinib-treated K562
cells (Fig. 1B). In RNF34 (ID
3434823), the second probeset (ID 3434832, in bold) targeting exon
2 was found to be downregulated in imatinib-treated K562 cells. In
SNCA (ID 2777714), the probeset (ID 2777722) targeting exon 5 was
found to be upregulated in imatinib-treated K562 cells. Both RNF34
and SNCA had two spliceosomes in K562 cells and the alternative
splicing was affected by imatinib (Figs. 2B and 3B). Similarly, we were able to identify
other transcripts that undergo alternative splicing by using this
process.
RT-PCR and sequencing validation
The expression of Bcl-x, RNF34 and SNCA mRNAs in
K562 cells before and after treatment with imatinib were evaluated
by RT-PCR using primers targeting the exons that flank the exon of
interest. Amplification results were visualized by gel
electrophoresis. When two alternatively spliced isoforms were
present, two different bands would be produced by the same primer
pair between the two groups (Figs. 1A
and C, 2A and C, 3A and C). The relative quantity of the
transcripts was calculated by the Quantity One software. The
results indicated a difference in the expression ratio of the
isoforms in response to imatinib treatment for probesets 3902522
(Bcl-x), 3434832 (RNF34) and 2777722 (SNCA). The relative quantity
analyses for Bcl-x, RNF34 and SNCA were consistent with the trend
of SI values calculated from the exon array data (Figs. 1C, 2C and 3C). To further validate the differences
between the spliceosomes, we performed sequencing validation of the
distinct bands resolved by gel electrophoresis. The sequencing
results for RNF34 and SNCA confirmed the alternative splicing
events that were predicted by the exon array data (Figs. 2D and 3D).
For DAPK1, the sequencing validation did not support
the presence of alternatively spliced isoforms even though two gel
bands that appeared to be in the correct position were detected
(data not shown).
Discussion
Alternative splicing of mRNA precursors is a nearly
ubiquitous and extremely flexible point of gene control in humans
(14). Abnormal alternative
splicing has been shown to be associated with many human cancers
(15). In addition, previous
studies have demonstrated that some anticancer drugs can affect
alternative splicing of genes in cancer cells (9,16).
These results may provide a novel rationale for the effectiveness
of anticancer drugs.
Apoptosis is a major pathway involved in the complex
homeostatic balance between cellular proliferation and cell death.
Previous studies have also demonstrated that transcripts from a
significant number of genes involved in apoptosis are alternatively
spliced, often resulting in isoforms with opposing roles in
promoting or preventing cell death (17–19). Examples of this type are found in
every category of apoptotic regulators, from transmembrane
receptors (Fas, Fas ligand and LARD) and adaptor molecules (Bcl-x,
Bak, Apaf1, survivin, Mcl-1 and TRAF2) to caspases (caspase-1, -2,
-6, -7, -8 and-9) and executors (FLIP and ICAD) (20). Although alternative splicing is an
important mechanism regulating apoptosis, the effect of
chemotherapeutic agents on the alternative splicing of apoptotic
genes has rarely been examined (9).
In previous studies, we have demonstrated that one
of the most commonly used chemotherapeutic agents, imatinib, which
is considered the most effective and a relatively safe drug for the
treatment of the chronic phase of CML, could regulate alternative
splicing of the apoptotic gene Bcl-x in K562 cells through the
activation of PP1 (12). In the
present study, we employed the Human Exon 1.0 ST Array, together
with the easyExon software, to assess the comprehensive profile of
alternatively spliced apoptotic genes in imatinib-treated K562
cells. A total of 175 (175/661, 26.48%) differentially spliced
apoptosis-related transcripts were identified, several of which
were successfully validated by RT-PCR and sequencing. Our results
revealed that imatinib treatment led to aberrant alternative
splicing events in K562 cells.
Table IV
summarizes the previous studies in the literature that have
demonstrated the association of some of these alternatively spliced
isoforms with apoptotic events. These genes belong to different
cell death protein families, including adaptor proteins, Bcl-2
family, and caspases, in which the splicing mechanism has been
shown to modulate both intrinsic (mitochondrial) and extrinsic
(death receptor) apoptosis pathways. These two signaling routes to
cell death are linked via the Bcl-2 family member Bid, whose
alternative splicing was also detected by the exon array in our
study. Thus, it is likely that imatinib has wide-ranging effects on
apoptosis of K562 cells. In addition, our results suggest the
existence of a novel mechanism by which imatinib can induce cell
apoptosis, which is the regulation of alternative splicing of a
series of apoptotic genes.
 | Table IVApoptotic genes previously shown to
be regulated by alternative splicing. |
Table IV
Apoptotic genes previously shown to
be regulated by alternative splicing.
| Gene symbol
(ref.) | Gene title | Accession no. | Gene probe ID | Exon probe ID | Splice index |
|---|
| Adaptor proteins
and regulators |
| TRAF2
(22) | TNF
receptor-associated factor 2 | NM_021138 | 3194896 | 3194919 | 0.50 |
| 3194950 | 0.50 |
| 3194944 | 0.48 |
| 3194952 | 0.33 |
| Apaf-1
(23) | Apoptotic peptidase
activating factor 1 | NM_181868 | 3427876 | 3427877 | 3.68 |
| 3427936 | 3.00 |
| 3427937 | 2.46 |
| 3427881 | 2.26 |
| 3427889 | 2.08 |
| Bcl-2 family |
| Bcl-x
(24) | Bcl-2-like 1 | NM_001191 | 3902489 | 3902523 | 0.4 |
| Bak
(25) |
Bcl-2-antagonist/killer 1 | NM_001188 | 2950753 | 2950768 | 2.14 |
| Bax
(26) | Bcl2-associated X
protein | NM_004324 | 3838067 | 3838084 | 0.48 |
| Bid
(18) | BH3 interacting
domain death agonist | NM_197966 | 3951927 | 3951950 | 3.91 |
| 3951929 | 3.64 |
| 3951928 | 2.63 |
| 3951930 | 2.15 |
| Bmf
(17) | Bcl-2 modifying
factor | NM_001003940 | 3619229 | 3619242 | 3.59 |
| Caspases and
caspase-like proteins |
| Caspase-3
(19) | Caspase 3,
apoptosis-related cysteine peptidase | NM_004346 | 2796484 | 2796499 | 2.00 |
| 2796505 | 2.00 |
| Caspase-10
(27) | Caspase 10,
apoptosis-related cysteine peptidase | NM_032977 | 2522693 | 2522719 | 2.33 |
| 2522707 | 2.07 |
| 2522715 | 2.00 |
Human Exon 1.0 ST Arrays, which are comprised of
probesets that cover the entire length of all known transcripts,
facilitate the study of gene expression at the exon level (probeset
expression) and predict the likelihood of encountering alternative
splicing for a given gene. This platform has also enabled detection
of specific changes at the gene expression level (transcript
cluster expression). Exon array technology has several key
advantages over conventional analytical methods, including
high-throughput, updated content, probesets designed to span the
entire length of a given transcript, and expression analysis at the
exon level. In the present study, using RT-PCR and sequencing, two
of the three randomly-chosen exons (67%) from the 430 probesets
were able to be validated for cell differences in abundance of the
respective transcript isoforms. Though the exon array technology is
reliable and sensitive enough to detect differential expression at
the exon level, there are some important limitations that exist and
should be considered when analyzing the exon array results. The
sensitivity and specificity of the Affymetrix exon arrays for
detecting exon splicing across the whole genome has not yet been
defined, at least based on the current published data. Thus,
confirmation of the detected exon splicing events requires PCR with
primers that flank the exons of interest or RNA deep sequencing to
demonstrate consistent differential splicing (21).
In conclusion, using exon arrays, we have discovered
that imatinib can produce a wide-ranging effect on the alternative
splicing of apoptotic factors in K562 leukemic cells. This
information may help to improve the mechanism of imatinib therapy
in patients with CML. Even though several of the imatinib-induced
alternative splicing events were successfully validated by RT-PCR
and sequencing in our study, a more comprehensive validation will
be necessary to provide a more accurate estimation of the current
findings.
Acknowledgements
This study was supported by a grant from the
National Science Foundation of China (no. 30700338).
References
|
1
|
S FaderlM TalpazZ EstrovS O’BrienR
KurzrockHM KantarjianThe biology of chronic myeloid leukemiaN Engl
J Med341164172199910.1056/NEJM199907153410306
|
|
2
|
CL SawyersChronic myeloid leukemiaN Engl J
Med34013301340199910.1056/NEJM19990429340170610219069
|
|
3
|
WY AuPB CaguioaC ChuahChronic myeloid
leukemia in AsiaInt J
Hematol891423200910.1007/s12185-008-0230-019101781
|
|
4
|
T SchindlerW BornmannP PellicenaWT MillerB
ClarksonJ KuriyanStructural mechanism for STI-571 inhibition of
abelson tyrosine
kinaseScience28919381942200010.1126/science.289.5486.193810988075
|
|
5
|
JM GoldmanHow I treat chronic myeloid
leukemia in the imatinib
eraBlood11028282837200710.1182/blood-2007-04-03894317626839
|
|
6
|
A PuissantP ColosettiG RobertJP CassutoS
RaynaudP AubergerCathepsin B release after imatinib-mediated
lysosomal membrane permeabilization triggers BCR-ABL cleavage and
elimination of chronic myelogenous leukemia
cellsLeukemia24115124201010.1038/leu.2009.233
|
|
7
|
ET WangR SandbergS LuoAlternative isoform
regulation in human tissue
transcriptomesNature456470476200810.1038/nature0750918978772
|
|
8
|
JP VenablesAberrant and alternative
splicing in cancerCancer
Res6476477654200410.1158/0008-5472.CAN-04-191015520162
|
|
9
|
L ShkretaU FroehlichER PaquetJ ToutantSA
ElelaB ChabotAnticancer drugs affect the alternative splicing of
Bcl-x and other human apoptotic genesMol Cancer
Ther713981409200810.1158/1535-7163.MCT-08-019218566212
|
|
10
|
JA CalarcoAL SaltzmanJY IpBJ
BlencoweTechnologies for the global discovery and analysis of
alternative splicingAdv Exp Med
Biol6236484200710.1007/978-0-387-77374-2_518380341
|
|
11
|
H ChenY GuoM HuW DuanG ChangC
LiDifferential expression and alternative splicing of genes in
lumbar spinal cord of an amyotrophic lateral sclerosis mouse
modelBrain
Res13405269201010.1016/j.brainres.2010.03.07520362558
|
|
12
|
Y XiaoH XiongJ LiImatinib regulates the
alternative pre-mRNA splicing of Bcl-x in K562 cellsAsian Biomed(In
press)
|
|
13
|
TY ChangYY LiCH JeneasyExon - a Java-based
GUI tool for processing and visualization of Affymetrix exon array
dataBMC Bioinformatics9432200810.1186/1471-2105-9-43218851762
|
|
14
|
CJ DavidJL ManleyAlternative pre-mRNA
splicing regulation in cancer: pathways and programs unhingedGenes
Dev2423432364201010.1101/gad.197301021041405
|
|
15
|
Z KalninaP ZayakinK SilinaA
LineAlterations of pre-mRNA splicing in cancerGenes Chromosomes
Cancer42342357200510.1002/gcc.2015615648050
|
|
16
|
JG ChangDM YangWH ChangSmall molecule
amiloride modulates oncogenic RNA alternative splicing to
devitalize human cancer cellsPLoS
One6e18643201110.1371/journal.pone.001864321694768
|
|
17
|
AA MoralesA OlssonF CelsingA OsterborgM
JondalLM OsorioExpression and transcriptional regulation of
functionally distinct Bmf isoforms in B-chronic lymphocytic
leukemia cellsLeukemia184147200410.1038/sj.leu.240318314574334
|
|
18
|
SA RenshawCE DempseyFA BarnesThree novel
Bid proteins generated by alternative splicing of the human Bid
geneJ Biol Chem27928462855200410.1074/jbc.M30976920014583606
|
|
19
|
Y HuangNH ShinY SunKK WangMolecular
cloning and characterization of a novel caspase-3 variant that
attenuates apoptosis induced by proteasome inhibitionBiochem
Biophys Res Commun283762769200110.1006/bbrc.2001.487111350049
|
|
20
|
C SchwerkK Schulze-OsthoffRegulation of
apoptosis by alternative pre-mRNA splicingMol
Cell19113200510.1016/j.molcel.2005.05.02615989960
|
|
21
|
Y TianIH LiaoX ZhanExon expression and
alternatively spliced genes in Tourette SyndromeAm J Med Genet B
Neuropsychiatr Genet156B7278201110.1002/ajmg.b.3114021184586
|
|
22
|
R BrinkHF LodishTumor necrosis factor
receptor (TNFR)-associated factor 2A (TRAF2A), a TRAF2 splice
variant with an extended RING finger domain that inhibits
TNFR2-mediated NF-kappaB activationJ Biol
Chem27341294134199810.1074/jbc.273.7.41299461607
|
|
23
|
MA BenedictY HuN InoharaG NunezExpression
and functional analysis of Apaf-1 isoforms. Extra Wd-40 repeat is
required for cytochrome c binding and regulated activation of
procaspase-9J Biol
Chem27584618468200010.1074/jbc.275.12.846110722681
|
|
24
|
LH BoiseM Gonzalez-GarciaCE Postemabcl-x,
a bcl-2-related gene that functions as a dominant regulator of
apoptotic cell
deathCell74597608199310.1016/0092-8674(93)90508-N8358789
|
|
25
|
YF SunLY YuM SaarmaT TimmuskU
ArumaeNeuron-specific Bcl-2 homology 3 domain-only splice variant
of Bak is anti-apoptotic in neurons, but pro-apoptotic in
non-neuronal cellsJ Biol
Chem2761624016247200110.1074/jbc.M01041920011278671
|
|
26
|
ZN OltvaiCL MillimanSJ KorsmeyerBcl-2
heterodimerizes in vivo with a conserved homolog, Bax, that
accelerates programmed cell
deathCell74609619199310.1016/0092-8674(93)90509-O8358790
|
|
27
|
PW NgAG PorterRU JanickeMolecular cloning
and characterization of two novel pro-apoptotic isoforms of
caspase-10J Biol
Chem2741030110308199910.1074/jbc.274.15.1030110187817
|