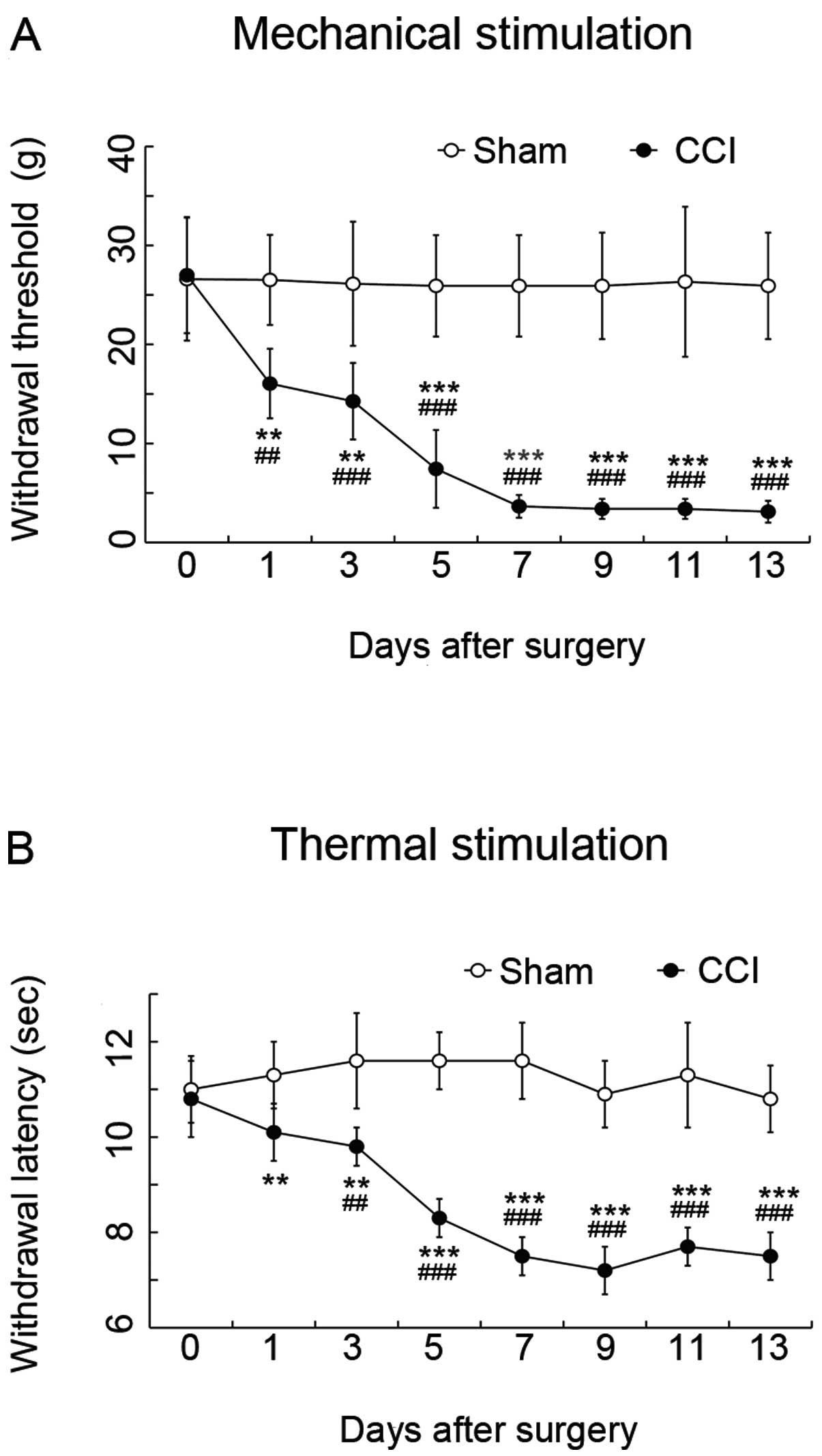
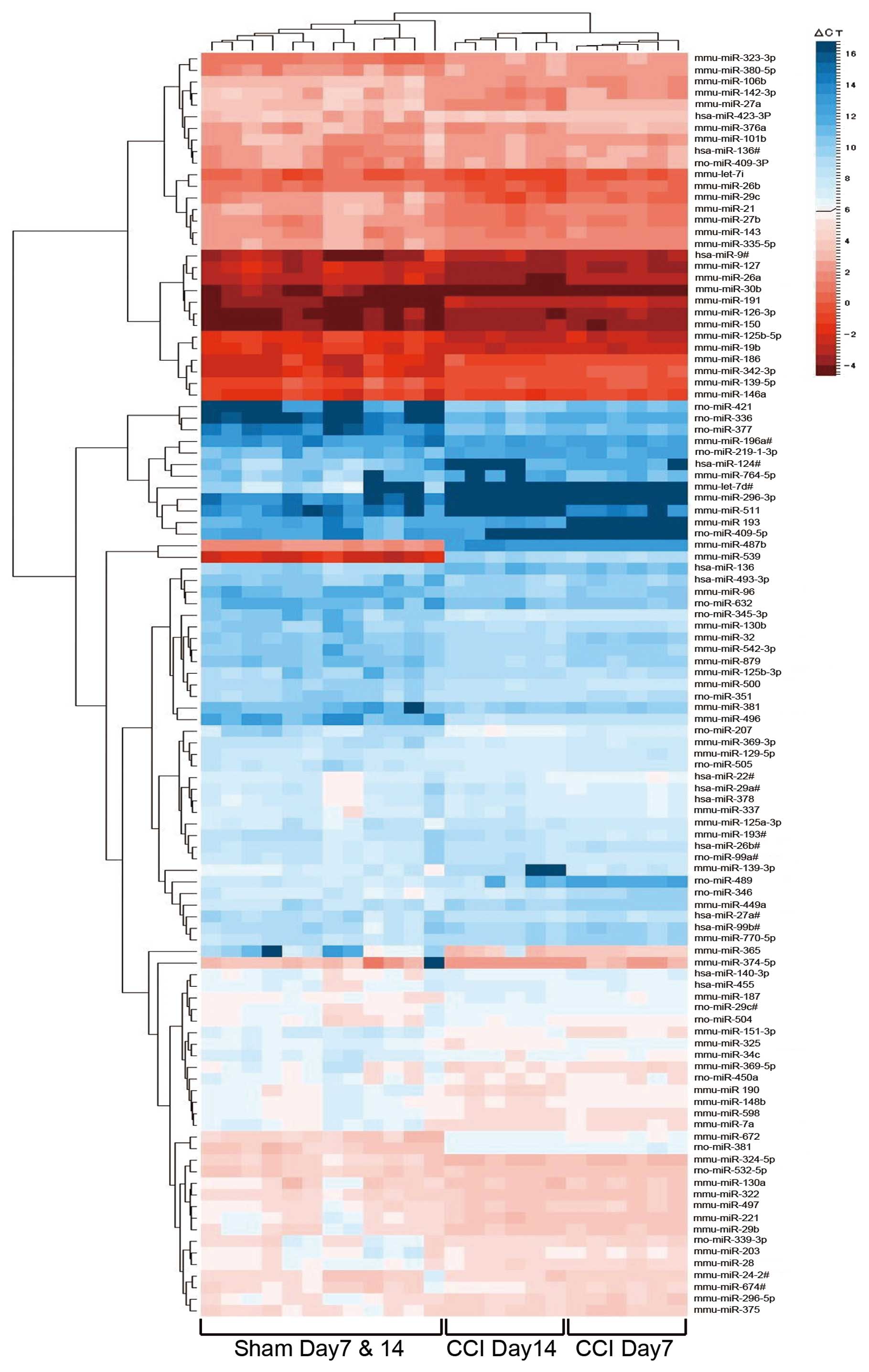
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grishok A, Pasquinelli AE, Conte D, Li N,
Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G and Mello CC: Genes
and mechanisms related to RNA interference regulate expression of
the small temporal RNAs that control C. elegans developmental
timing. Cell. 106:23–34. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hutvagner G and Zamore PD: A microRNA in a
multiple-turnover RNAi enzyme complex. Science. 297:2056–2060.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mehler MF and Mattick JS: Non-coding RNAs
in the nervous system. J Physiol. 575:333–341. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kosik KS: The neuronal microRNA system.
Nat Rev Neurosci. 7:911–920. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kusuda R, Cadetti F, Ravanelli MI, Sousa
TA, Zanon S, De Lucca FL and Lucas G: Differential expression of
microRNAs in mouse pain models. Mol Pain. 7:172011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aldrich BT, Frakes EP, Kasuya J, Hammond
DL and Kitamoto T: Changes in expression of sensory organ-specific
microRNAs in rat dorsal root ganglia in association with mechanical
hypersensitivity induced by spinal nerve ligation. Neuroscience.
164:711–723. 2009. View Article : Google Scholar
|
|
9
|
Imai S, Saeki M, Yanase M, Horiuchi H, Abe
M and Narita M, Kuzumaki N, Suzuki T and Narita M: Change in
microRNAs associated with neuronal adaptive responses in the
nucleus accumbens under neuropathic pain. J Neurosci.
31:15294–15299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
von Schack D, Agostino MJ, Murray BS, Li
Y, Reddy PS, Chen J, Choe SE, Strassle BW, Li C, Bates B, Zhang L,
Hu H, Kotnis S, Bingham B, Liu W, Whiteside GT, Samad TA, Kennedy
JD and Ajit SK: Dynamic changes in the microRNA expression profile
reveal multiple regulatory mechanisms in the spinal nerve ligation
model of neuropathic pain. PLoS One. 6:e176702011.PubMed/NCBI
|
|
11
|
Covino BG, Dubner R and Gybels J: Ethical
standards for investigations of experimental pain in animals. Pain.
9:141–143. 1980. View Article : Google Scholar
|
|
12
|
Jarvis MF and Boyce-Rustay JM: Neuropathic
pain: models and mechanisms. Curr Pharm Des. 15:1711–1716. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bennett GJ and Xie YK: A peripheral
mononeuropathy in rat that produces disorders of pain sensation
like those seen in man. Pain. 720:111–119. 1988.
|
|
14
|
Sato C, Sakai A, Ikeda Y, Suzuki H and
Sakamoto A: The prolonged analgesic effect of epidural ropivacine
in a rat model of neuropathic pain. Anesth Analg. 106:313–320.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okabe T, Sato C and Sakamoto A: Changes in
neuropeptide Y gene expression in the spinal cord of chronic
constrictive injury model rats after electroconvulsive stimulation.
Biomed Res. 31:287–292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shibata M, Wakisaka S, Inoue T and Yoshiya
I, Shimizu T and Yoshiya I: The effect of electroconvulsive
treatment on thermal hyperalgesia and mechanical allodynia in a rat
model of peripheral neuropathy. Anesth Analg. 86:584–587.
1998.PubMed/NCBI
|
|
17
|
Kodani M, Yang G, Conklin LM, Travis TC,
Whitney CG, Anderson LJ, Schrag SJ, Taylor TH Jr, Beall BW, Breiman
RF, Feikin DR, Njenga MK, Mayer LW, Oberste MS, Tondella ML,
Winchell JM, Lindstrom SL, Erdman DD and Fields BS: Application of
TaqMan low-density arrays for simultaneous detection of multiple
respiratory pathogens. J Clin Microbiol. 49:2175–2182. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stanton MC, Chen SC, Jackson JV,
Rojas-Triana A, Kinsley D, Cui L, Fine JS, Greenfeder S, Bober LA
and Jenh CH: Inflammatory signals shift from adipose to liver
during high fat feeding and influence the development of
steatohepatitis in mice. J Inflamm (Lond). 8:82011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang B, Howel P, Bruheim S, Ju J, Owen LB,
Oystein F and Xi Y: Systematic evaluation of three microRNA
profiling platforms: microarray, beads array, and quantitative
real-time PCR array. PLoS One. 6:e171672011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu B, Zhou S, Qian T, Wang Y, Ding F and
Gu X: Altered microRNA expression following sciatic nerve resection
in dorsal root ganglia of rats. Acta Biochim Biophys Sin
(Shanghai). 43:909–915. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu B, Zhou S, Wang Y, Qian T, Ding G, Ding
F and Gu X: miR-221/222 promote Schwann cell proliferation and
migration by targeting LASS2 following sciatic nerve injury. J Cell
Sci. 125:2675–2683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bai G, Ambalavanar R, Wei D and Dessem D:
Downregulation of selective microRNAs in trigeminal ganglion
neurons following inflammatory muscle pain. Mol Pain. 3:152007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao J, Lee MC, Momin A, Cendan CM,
Shepherd ST, Baker MD, Asante C, Bee L, Bethry A, Perkins JR,
Nassar MA, Abrahamsen B, Dickenson A, Cobb BS, Merkenschlager M and
Wood JN: Small RNAs control sodium channel expression, nociceptor
excitability, and pain thresholds. J Neurosci. 30:10860–10871.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Smirnova L, Gräfe A, Seiler A, Schumacher
S, Nitsch R and Wulczyn FG: Regulation of miRNA expression during
neural cell specification. Eur J Neurosci. 21:1469–1477. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hohjoh H and Fukushima T: Expression
profile analysis of microRNA (miRNA) in mouse central nervous
system using a new miRNA detection system that examines
hybridization signals at every step of washing. Gene. 391:39–44.
2007. View Article : Google Scholar
|
|
26
|
Bak M, Silahtaroglu A, Møller M,
Christensen M, Rath MF, Skryabin B, Tommerup N and Kauppinen S:
MicroRNA expression in the adult mouse central nervous system. RNA.
14:432–444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang X, Gal J, Zhuang X, Wang W, Zhu H and
Tang G: A simple array platform for microRNA analysis and its
application in mouse tissues. RNA. 13:1803–1822. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Strickland ER, Hook MA, Balaraman S, Huie
JR, Grau JW and Miranda RC: MicroRNA dysregulation following spinal
cord contusion: implications for neural plasticity and repair.
Neuroscience. 186:146–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu NK, Wang XF, Lu QB and Xu XM: Altered
microRNA expression following traumatic spinal cord injury. Exp
Neurol. 219:424–429. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Madathil SK, Nelson PT, Saatman KE and
Wilfred BR: MicroRNAs in CNS injury: potential roles and
therapeutic implications. Bioessays. 33:21–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu G, Detloff MR, Miller KN, Santi L and
Houlé JD: Exercise modulates microRNAs that affect the PTEN/mTOR
pathway in rats after spinal cord injury. Exp Neurol. 233:447–456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Purohit V, Rapaka RS, Rutter J and
Shurtleff D: Do opioids activate latent HIV-1 by down-regulating
anti-HIV microRNAs? J Neuroimmune Pharmacol. 7:519–523. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leone V, D'Angelo D, Ferraro A, Pallante
P, Rubio I, Santoro M, Croce CM and Fusco A: A TSH-CREB1-microRNA
loop is required for thyroid cell growth. Mol Endocrinol.
25:1819–1830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao C, Huang C, Weng T, Xiao X, Ma H and
Liu L: Computational prediction of MicroRNAs targeting GABA
receptors and experimental verification of miR-181, miR-216 and
miR-203 targets in GABA-A receptor. BMC Res Notes. 5:912012.
View Article : Google Scholar : PubMed/NCBI
|