Introduction
Lung cancer is the leading cause of cancer-related
death worldwide, causing an estimated 1.4 million deaths in 2010
(1). The discovery of mutations
in the epidermal growth factor receptor (EGFR) kinase, and
fusions involving anaplastic lymphoma kinase (ALK), has
improved the treatment of patients with lung adenocarcinoma, the
most common type of lung cancer (2–4).
The discovery of gene fusions involving ROS1 and RET
may also facilitate the treatment of adenocarcinomas (5–7);
however, targeted agents developed for lung adenocarcinoma are
largely ineffective against the second most common type of lung
cancer, lung squamous cell carcinoma (SqCC).
The comprehensive genomic characterization study of
SqCC has identified several somatic mutations of nuclear factor
(erythroid derived 2)-like 2 (NFE2L2) (8). NRF2 (gene name NFE2L2) is a
master transcriptional activator of genes encoding numerous
cytoprotective enzymes that are induced in response to
environmental and endogenously derived oxidative/electrophilic
agents (9–11). In normal cells, NRF2 is a cap ‘n’
collar basic leucine zipper transcription factor.
NFE2L2-deficient mice are highly susceptible to chemically
induced carcinogenesis of multiple organs (12,13). A previous study showed that
RNAi-mediated silencing of NRF2 gene expression in non-small cell
lung cancer inhibited tumor growth (14). NRF2 gene promoter polymorphism has
been identified and may be correlated with carcinogenesis (15). More recently, an NRF2 gene
mutation was identified in lung cancer cell lines and carcinoma
(16). Somatic mutations
occurring in the coding region of the NRF2 gene were more common
among patients with a history of smoking or suffering from SqCC and
were correlated with poor prognosis (16,17).
The standard for experimental detection of mutations
is direct sequencing of DNA samples from tissues. For known
mutations, real-time polymerase chain reaction followed by melting
curve analysis, using hybridization probes, is highly sensitive,
rapid, and an efficient alternative approach to mutation detection
(18–20). To determine the NRF2 gene status
in Japanese lung carcinoma patients for screening and diagnostic
purpose, we investigated the NRF2 gene mutation status, an
N-terminal domain, using real-time reverse transcription-PCR assay
using LightCycler (20). With
this method, 32 samples were genotyped within 1 h without the need
of any post-PCR sample manipulation. The findings were compared to
the clinico-pathologic features of the lung cancers.
Patients and methods
Patients
The study group included 262 lung cancer patients
who had undergone surgery at the Department of Surgery, Nagoya City
University Hospital between 2005 and 2012. The lung tumors were
classified according to the General Rule for Clinical and
Pathological Record of Lung Cancer (7th edition) in Japan. All
tumor samples were immediately frozen and stored at −80°C until
assayed.
The clinical and pathological characteristics of the
262 lung cancer patients were as follows: 176 cases at stage I, 47
at stage II, and 39 at stage III-IV. The mean patient age was 68.2
years (range, 22–86). Among the 262 lung cancer patients, 164
(61.8%) were males and 156 (59.5%) were nonsmokers. Sixty were
diagnosed as having SqCC.
PCR analysis of the NRF2 gene
Total RNA was extracted from lung cancer tissues
using the Isogen kit (Nippon Gene, Tokyo, Japan) according to the
manufacturer’s instructions. The RNA concentration was determined
by a spectrophotometer and adjusted to a concentration of 200
ng/ml. Approximately 10 cases were excluded for each assay since
tumor cells were too few to sufficiently extract tumor RNA. RNA (1
μg) was reverse transcribed by SuperScript II enzyme (Gibco-BRL,
Gaithersburg, MD, USA) with 0.5 μg oligo(dT)12–16
(Amersham Pharmacia Biotech Inc., Piscataway, NJ, USA). The
reaction mixture was incubated at 42°C for 50 min and then at 72°C
for 15 min. We then used 1 μl of each DNA for PCR analyses.
Initially, 281 (between 1997–2006) samples were sequenced as
reported in our previous study (16). These sets of RNA were used as a
positive and negative control for genotyping.
The primer sequences for the NRF2 gene in the DLG
motif were as follows: the forward primer, 5′-GGACATGGATTTGA
TTGACATAC-3′ and the reverse primer, 5′-CTCCTTTTGG AGTTGTTCTTGT-3′
(151 bp). For DLG motif genotyping, sensor (LC Red
640-TCTCGACTTACTCCAAGATCTAT) and anchor
(CAGCTCATACTCTTTCCGTCGCTGACTGA AGTCAAATAC-Fluorescein) probes were
used. The cycling conditions were as follows: initial denaturation
at 95°C for 10 min, followed by 45 cycles at 95°C for 1 sec, 59°C
for 10 sec, and 72°C for 7 sec. The primer sequences for the NRF2
gene in the ETGE motif were as follows: the forward primer,
5′-CCAAAAGGAGCAAGAGAAAGC-3′ and the reverse primer,
5′-GCAGTCATCAAAGTACAAAGCAT-3′ (172 bp). For ETGE motif genotyping,
sensor (LC Red AAATTCACC TGTCTCTTCATCTAG) and anchor (GATGTGCTGGGCT
GGCTGAATTGGGA-Fluorescein) probes were used. The cycling conditions
were as follows: initial denaturation at 95°C for 10 min, followed
by 45 cycles at 95°C for 1 sec, 62°C for 10 sec, and 72°C for 7
sec. The positive products were purified using the Qiagen PCR
purification kit (Qiagen, Valencia, CA, USA). The positive samples
were sequenced by ABI PRISM 3100 analyzer (Applied Biosystems Japan
Ltd., Tokyo, Japan) and analyzed by BLAST and chromatograms by
manual review.
Statistical analysis
Statistical analyses were performed using the
Mann-Whitney U-test for unpaired samples and Wilcoxon’s signed rank
test for paired samples. Linear relationships between variables
were determined by means of simple linear regression. Correlation
coefficients were determined by rank correlation using Spearman’s
test and χ2 test. The overall survival of lung cancer
patients was examined by the Kaplan-Meier methods, and differences
were examined by the log-rank test. All analysies were conducted
using the StatView software package (Abacus Concepts Inc.,
Berkeley, CA, USA), and a P-value <0.05 was considered to
indicate a statistically significant result.
Results
Genotyping of the NRF2 gene mutation at
the DLG and ETGE motif in lung cancer
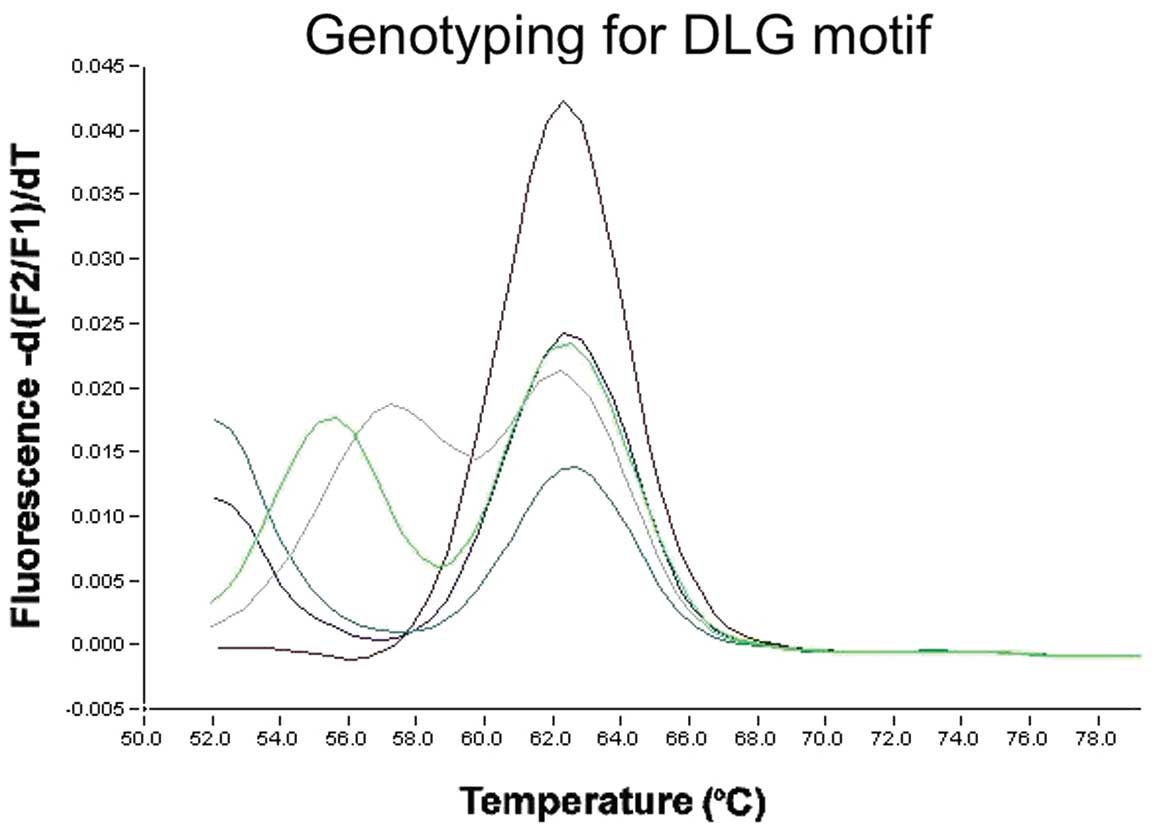
Using the primer sets for the DLG motif, the anchor
probe was matched for wild-type. For the DLG motif in exon 2, the
homozygous wild-type PCR product showed a single peak at 63°C,
whereas the heterozygous products (mutant) showed an additional
peak at a lower temperature (Fig.
1). Of the 262 lung cancer cases, 4 had an NRF2 gene mutation.
All were male nonsmokers with SqCC. For the ETGE motif genotyping,
the anchor probe was matched for wild-type. The homozygous
wild-type PCR product showed a single peak at 63°C, whereas the
heterozygous products (mutant) showed an additional peak at a lower
temperature (Fig. 2). Two
patients had mutations. In total, 6 of the 262 patients had a
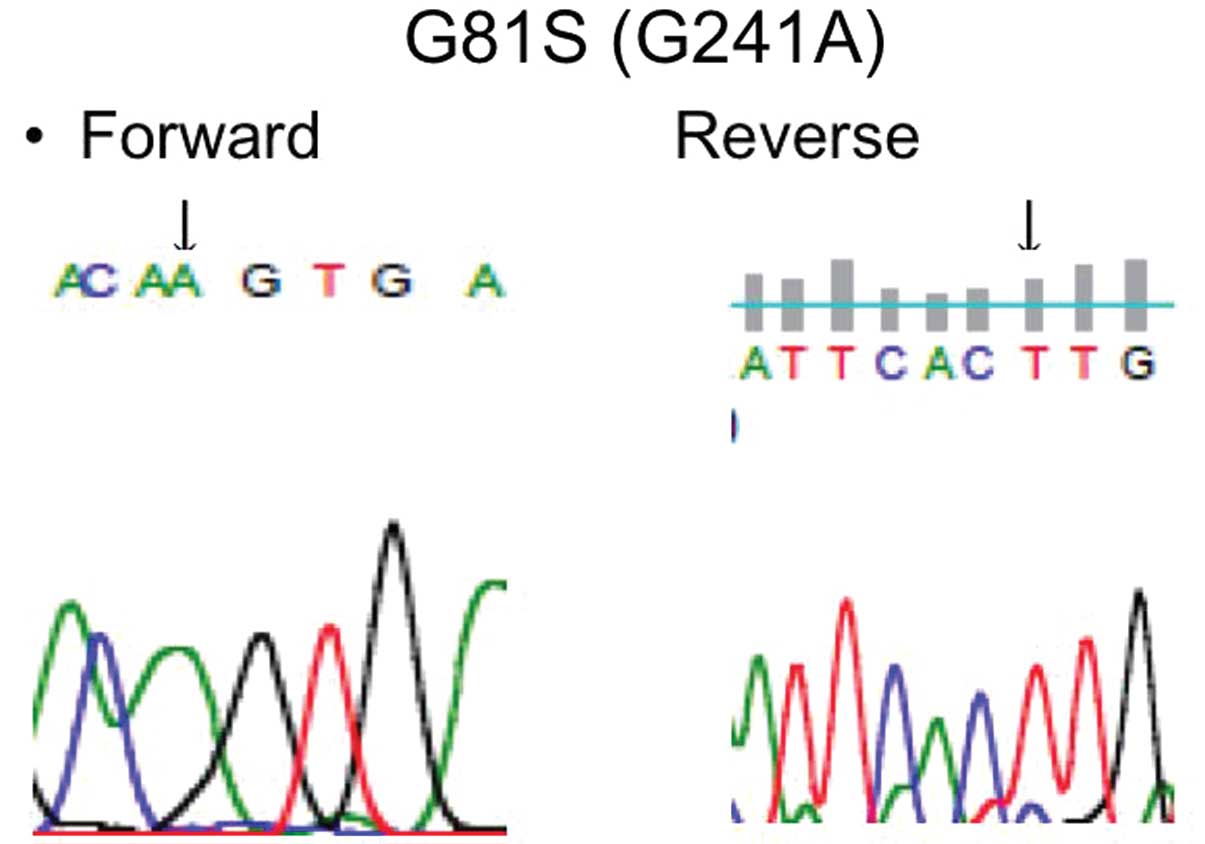
mutation (Table I). Five were
known mutations (D29H, G31A, R34G and E79K) and 1 was a novel
mutation (G81S and G241A) (Fig.
3). The NRF2 gene mutations were clustered in exon 2 and coded
for amino acid changes in either the DLG or ETGE motifs of the
regulatory Neh2 domain (21). All
were male smokers with SqCC. NRF2 gene mutations were paricularly
noted in smokers (P=0.04) and cases of squamous histology
(P=0.0001). In addition, the NRF2 mutation displayed a tendency
towards having a higher frequency in males (P=0.0868). In this
analysis, 88 patients had an EGFR mutation, 3 had an
erbB2 mutation (22), 2
had ALK translocations and 2 had RET translocations
(23). The NRF2 gene mutation was
completely exclusive of these mutations.
 | Table IClinicopathological data of the 262
lung cancer patients. |
Table I
Clinicopathological data of the 262
lung cancer patients.
| NRF2 gene status |
|---|
|
|
|---|
| Characteristics | Wild-type
patients | Mutant patients | P-value |
|---|
| Total no. of
patients | 256 | 6 | |
| Mean age, 68.2±9.4
years |
| Age, n (%), in
years |
| >65 | 80 (31.3) | 2 (33.3) | 0.9999 |
| ≥65 | 176 (68.8) | 4 (66.7) | |
| Gender, n (%) |
| Male | 158 (61.7) | 6 (100) | 0.0868 |
| Female | 98 (38.3) | 0 (0) | |
| Stage, n (%) |
| I | 172 (67.2) | 4 (66.7) | 0.9999 |
| II-IV | 84 (22.8) | 2 (33.3) | |
| Lymph node
metastasis, n (%) |
| N0 | 193 (75.4) | 4 (66.7) | 0.6399 |
| N+ | 63 (24.6) | 2 (33.3) | |
| Smoking, n (%) |
| Never smoker | 156 (60.9) | 0 (0) | 0.04 |
| Smoker | 100 (39.1) | 6 (100) | |
| EGFR mutation, n
(%) |
| Wild-type | 168 (65.6) | 6 (100) | 0.1834 |
| Mutation | 88 (34.4) | 0 (0) | |
| Pathological
subtypes, n (%) |
| SqCC | 54 (21.1) | 6 (100) | 0.0001 |
| Non-SqCC | 202 (78.9) | 0 (0) | |
The overall survival of the 543 lung cancer patients
from Nagoya City University Hospital, with follow-up through
October 31, 2012, was studied in reference to the NRF2 gene
mutation status. Patients with an NRF2 gene mutation in the coding
region (n=22, 11 succumbed to disease; mean survival, 54.94 months)
had a significantly worse prognosis than the patient with the
wild-type NRF2 gene (n=521, 98 succumbed to disease; mean survival,
80.67 months) (log-rank test, P<0.0001, Breslow-Gehan-Wilcoxon
test; P=0.0001) (Fig. 4). The
multivariate analyses revealed that pathological stage
(P<0.0001; hazard ratio, 3.888; 2.585–5.849) and NRF2 gene
mutation (P=0.0028; hazard ratio, 2.600; 1.389–4.867) were
significant prognostic factors.
Discussion
Our findings revealed that the NRF2 gene mutation
status was correlated with squamous histology and smoking status.
This was in agreement with previous studies (17,24). Previous studies have documented
that genetic alterations in lung cancer are frequent in smokers,
such as mutations of the TP53 and Kras genes
(25). The cause of these somatic
mutations in cancer cells is shaped by multiple factors, such as
exposure to mutagens, selective pressures in the tissue
microenvironment and genomic stability (26), while tobacco smoking results in
deposits of many hundreds of chemicals in the airways and lung.
Continuous chronic exposure of tissues of the
respiratory tract to cigarette smoke generally activates cellular
defense systems and the deposits trigger a pleiotropic adaptive
response, aimed at restoring tissue homeostasis. A previous study
suggests that a hallmark of this defense system is the activation
of the transcription factor NRF2, consequent to its established
role as master regulator of the cellular antioxidant response
(27). NRF2 regulates the
expression of several genes encoding antioxidant and detoxification
proteins (9). In addition,
constitutive expression in a tumorigenic situation could provide a
survival advantage to invasive and metastatic cancer cells. A
previous study found that the RNAi-mediated silencing of NRF2 gene
expression in lung cancer inhibited tumor growth (14). NRF2 gene promoter polymorphism was
identified and was suggested to correlate with carcinogenesis
(15). The adaptation to the
microenvironment and the development of chemoresistance in cancer
cells are also known to occur in tumors under hypoxia (28,29). These influence a worse prognosis
in NRF2 mutant patients. The expression of multidrug
resistance-associated proteins (MRPs), drug efflux proteins, was
also found to be significantly reduced in NRF2 gene-silenced A549
cells (30). A recent report
showed that MRP3 gene expression was correlated with NRF2 gene
mutations in lung SqCC (31).
Oxidative stress-regulated lentiviral gene therapy may overcome the
resistance of lung cancer to treatment (32).
Real-time PCR assay allows for easy identification
of new mutations and provides the best means for pre-therapeutic
genotyping in a clinical setting at present. Therefore, we
developed two different PCRs to detect NRF2 gene mutations. The
rapid PCR method and elimination of additional steps to analyze PCR
products saves time. Handling is facilitated and potentially toxic
reagents, such as ethidium bromide stain, are avoided. Using the
LightCycler reverse transcription-PCR assay described here,
determination of the NRF2 gene mutation status may be of clinical
importance in predicting the prognosis or determining additional
therapy for lung cancer patients. Using this method, 32 samples
were genotyped within 1 h without the need of any post-PCR sample
manipulation.
Acknowledgements
The authors would like to thank Mrs. Yuka Toda for
her excellent technical assistances. This study was supported by
Grants-in-Aid for Scientific Research, Japan Society for the
Promotion of Science (JSPS) (nos. 24692097, 23659674 and
21591820).
References
|
1
|
World Health Organization. Cancer. Fact
Sheet. 297:http://www.who.int/mediacentre/factsheets/fs297/en/.
Accessed February, 2012
|
|
2
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soda H, Choi YL, Enomoto M, et al:
Identification of the transforming EML4-ALK fusion gene in
non-small-cell lung cancer. Nature. 448:561–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lipson D, Capelletti M, Yelensky R, et al:
Identification of new ALK and RET gene fusions from colorectal and
lung cancer biopsies. Nat Med. 18:382–384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kohno T, Ishikawa H, Totoki Y, et al:
KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 18:375–377.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takeuchi K, Soda M, Togashi Y, et al: RET,
ROS and ALK fusions in lung cancer. Nat Med. 18:378–381. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cancer Genome Atlas Research Network.
Comprehensive genomic characterization of squamous cell cancers.
Nature. 489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Itoh K, Chiba T, Takahashi S, et al: An
Nrf2/small Maf heterodimer mediates the induction of phase II
detoxifying enzyme genes through antioxidant response elements.
Biochem Biophys Res Commun. 236:313–322. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rushmore TH and Kong AN: Pharmacogenomics,
regulation and signaling pathways of phase I and II detoxifying
enzymes. Curr Drug Metab. 3:481–490. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ngyyen T, Yang CS and Pickett CB: The
pathways and molecular mechanisms regulating Nrf2 activation in
response to chemical stress. Free Radic Biol Med. 37:433–441. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fahey JW and Kensler TW: Role of dietary
supplements/nutraceuticals in chemoprevention through induction of
cytoprotective enzymes. Chem Res Toxicol. 20:572–576. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kensler TW, Wakabayashi N and Biswal S:
Cell survival responses to environmental stresses via the
Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 47:89–116.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh A, Boldin-Adamsky S, Thimmulappa RK,
et al: RNAi-mediated silencing of nuclear factor
erythroid-2-related factor 2 gene expression in non-small cell lung
cancer inhibits tumor growth and increases efficacy of
chemotherapy. Cancer Res. 68:7975–7984. 2008. View Article : Google Scholar
|
|
15
|
Arisawa T, Tahara T, Shibata T, et al:
Nrf2 gene promoter polymorphism and gastric carcinogenesis.
Hepatogastroenterology. 55:750–754. 2008.PubMed/NCBI
|
|
16
|
Shibata T, Ohta T, Tong KI, et al: Cancer
related mutations in NRF2 impair its recognition by Keap1-Cul3 E3
ligase and promote malignancy. Proc Natl Acad Sci USA.
105:13568–13573. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasaki H, Hikosaka Y, Okuda K, et al:
NFE2L2 gene mutation in male Japanese squamous cell carcinoma of
the lung. J Thorac Oncol. 5:786–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wittwer CT, Herrmann MG, Moss AA and
Rasmussen RP: Continuous fluorescence monitoring of rapid cycle DNA
amplification. Biotechniques. 22:130–138. 1997.PubMed/NCBI
|
|
19
|
Pals G, Pindolia K and Worsham MJ: A rapid
sensitive approach to mutation detection using real-time polymerase
chain reaction and melting curve analyses, using BRCA1 as an
example. Mol Diagn. 4:241–246. 1999. View Article : Google Scholar
|
|
20
|
Sasaki H, Endo K, Konishi A, et al: EGFR
mutation status in Japanese lung cancer patients: genotyping
analysis using LightCycler. Clin Cancer Res. 11:2924–2929. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Itoh K, Wakabayashi N, Katoh Y, et al:
Keap1 represses nuclear activation of antioxidant responsive
elements by Nrf2 through binding to the amino-terminal Neh2 domain.
Genes Dev. 13:76–86. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sasaki H, Shimizu S, Endo K, et al: EGFR
and erbB2 mutation status in Japanese lung cancer patients. Int J
Cancer. 118:180–184. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yokota K, Sasaki H, Okuda K, et al:
KIF5B/RET fusion gene in surgically-treated Japanese adenocarcinoma
of the lung. Oncol Rep. 28:1187–1192. 2012.PubMed/NCBI
|
|
24
|
Hu Y, Ju Y, Lin D, et al: Mutation of the
Nrf2 gene in non-small cell lung cancer. Mol Biol Rep.
39:4743–4747. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahrendt SA, Decker PA, Alawi EA, et al:
Cigarette smoking is strongly associated with mutation of the Kras
gene in patients with primary adenocarcinoma of the lung. Cancer.
92:1525–1530. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stratton MR, Campbell PJ and Futreal PA:
The cancer genome. Nature. 458:719–724. 2009. View Article : Google Scholar
|
|
27
|
Muller T and Hengstermann A: Nrf2: friend
and foe in preventing cigarette smoking-dependent lung disease.
Chem Res Toxicol. 25:1805–1824. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar
|
|
29
|
Zhou J, Scmid T, Schnitzer S and Brune B:
Tumor hypoxia and cancer progression. Cancer Lett. 237:10–21. 2006.
View Article : Google Scholar
|
|
30
|
Homma S, Ishii Y, Morishima Y, et al: Nrf2
enhances cell proliferation and resistance to anticancer drugs in
human lung cancer. Clin Cancer Res. 15:3423–3432. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sasaki H, Shitara M, Yokota K, et al: MRP3
gene expression correlates with NRF2 mutations in lung squamous
cell carcinomas. Mol Med Rep. 6:705–708. 2012.PubMed/NCBI
|
|
32
|
Leinonen HM, Ruotsalainen AK, Määttä AM,
et al: Oxidative stress-regulated lentiviral TK/GCV gene therapy
for lung cancer treatment. Cancer Res. 72:6227–6235. 2012.
View Article : Google Scholar : PubMed/NCBI
|