Introduction
Neuropathic pain is defined as pain arising as a
direct consequence of a lesion or disease affecting either the
peripheral or central nervous system (1,2).
Although pain has been investigated in depth for decades,
neuropathic pain is still frequently under-treated (3), due to poor understanding of its
pathophysiological and molecular mechanisms. Several regions of the
nociceptive pathway, including the anterior cingulate cortex (ACC),
hippocampus, spinal dorsal horn (SDH) and dorsal root ganglion
(DRG), are involved in the development and maintenance of
neuropathic pain (4–9). Several recent studies have shown
that peripheral and central sensitization are associated with
global changes in gene expression in different regions of the pain
transmission pathway, and that these changes may be part of the
mechanisms behind neuropathic pain (10–13). In order to elucidate the molecular
mechanisms underlying neuropathic pain, it is essential to
determine how gene expression patterns are altered by nerve
injuries and how these alterations lead to the development and
maintenance of chronic pain.
miRNAs are a large class of short non-coding RNAs
(~22 nt long), many of which are expressed either predominantly or
exclusively in the nervous system (14–21). Furthermore, changes (either
increases or decreases) in miRNA expression have been found in many
disease states (22–24). Bilateral sciatic nerve chronic
constriction injury (bCCI) is commonly used as a model for studying
human neuropathic pain, in which chromic gut sutures are used to
ligate each sciatic nerve. This model is characterized by
long-lasting cold allodynia and mechanical hypersensitivity
(25,26); however, no information is
available on how miRNA expression patterns in the nociceptive
system are altered.
The aim of the present study was to examine the
differential expression patterns of miRNAs in the DRG, SDH,
hippocampus and ACC using a rat model of neuropathic pain induced
by bCCI. Utilizing a microarray platform, we identified miRNAs with
a 2-fold change in expression in the DRG and SDH due to bCCI. We
then confirmed the results in the DRG, SDH, hippocampus and ACC
using quantitative reverse transcriptase-polymerase chain reaction
(qRT-PCR). Our results demonstrate that miRNAs are differentially
expressed at the neuroanatomical level under neuropathic pain
conditions in vivo.
Materials and methods
Animals
Specific pathogen-free (SPF) adult female
Sprague-Dawley rats (Shanghai Laboratory, Animal Research Center,
Shanghai, China) weighing 150–180 g were randomly divided into 3
groups (naïve, sham-operated and bCCI) and housed 2–3 per cage in a
climate-controlled environment on a 12 h light/dark cycle with food
and water ad libitum. Behavioral experiments were performed
between 9:00 a.m. and 4:00 p.m. Animal handling and experimental
procedures were performed in accordance with the policies and
recommendations of the Guide for the Care and Use of Laboratory
Animals, and were approved by the Ethics Committee for Animal
Experimentation of the Peking Union Medical College Hospital,
Beijing, China. The minimum number of rats was used and every
effort was made to reduce their discomfort and stress.
Neuropathic pain model
bCCI was induced under aseptic conditions in rats
anesthetized using a mixture of ketamine and xylazine (60 and 8
mg/kg, respectively). The sciatic nerve on each side was exposed
via a mid-thigh incision followed by the separation of the heads of
the biceps femoris muscle. Each sciatic nerve was identified above
the trifurcation and freed from the surrounding loose connective
tissue before 4 snug ligatures of 4-0 chromic gut suture were
placed around them. The sham-operated animals had their sciatic
nerves exposed but not ligated, and the rats in the naïve group
were not operated upon.
Mechanical withdrawal test
The thresholds for paw withdrawal in response to
mechanical stimuli were assessed using Von Frey filaments at 1, 2
and 3 days pre-operation and 1, 3, 7 and 14 days post-operation
according to previously described procedures (27). At the end of the behavioral
testing, rats were euthanized and the bilateral L4–6 DRG, L2–4 SDH,
hippocampus and ACC were chronologically harvested and rapidly
frozen at −180°C.
Acetone test
Cold allodynia was evaluated on 3 continuous days
pre-operation and on days 1, 3, 7 and 14 post-operation. A drop
(0.1 ml) of room temperature acetone was gently applied to each
hindpaw through polyethylene (PE; (10 standard specification)
plastic tubing connected to a 1-ml syringe. A rapid withdrawal of
the hindpaw in response to the spread of the acetone over the
plantar surface was considered a sign of cold allodynia. The test
was repeated 5 times for each hindpaw (alternating hindpaws) for a
total of 10 trials/day with an interval of ~2 min between each
test. The results were graded as a percentage of applications that
evoked a response of hindpaw withdrawal. An increase in the
percentage of applications that elicited a withdrawal response
compared to the control was interpreted as the development of
increased cold sensitivity.
RNA isolation
Total RNA was isolated using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) and a miRNeasy
mini kit (Qiagen, Hilden, Germany) in accordance with the
manufacturer’s instructions. This procedure efficiently recovered
all RNA species, including miRNAs. RNA quality and quantity were
measured using a NanoDrop spectrophotometer (ND-1000; NanoDrop
Technologies, Wilmington, DE, USA), and RNA integrity was evaluated
by gel electrophoresis.
Microarray analysis
Following RNA isolation, the miRCURY™ Hy3™/Hy5™
Power labeling kit (Exiqon, Vedbaek, Denmark) was used according to
the manufacturer’s specifications for miRNA labeling. Once the
labeling procedure was completed, the Hy3-labeled samples were
hybridized on a miRCURY LNA Array (v.16.0) (Exiqon) according to
the manufacturer’s instructions. Scanned images of the hybridized
arrays were then imported into GenePix Pro 6.0 software (Axon) for
grid alignment and data extraction. Replicated miRNAs were
averaged, and miRNAs with intensities ≥50 in all samples were
chosen for calculating the normalization factor. Expression results
were normalized using a median normalization. After normalization,
significantly differentially expressed miRNAs (as determined by
ANOVA followed by a Student-Newman-Keuls multiple comparison test)
were identified through volcano plot filtering.
qRT-PCR analysis of miRNAs
Two-fold increases or decreases in gene expression
observed in the microarrays were validated by qRT-PCR analysis.
Complementary DNA (cDNA) was generated from 20 ng of total RNA
using a universal cDNA synthesis kit (Exiqon). The cDNA template
was then amplified using mature miRNA-specific LNA™-enhanced
forward and reverse primers (rno-miR-341#130384, rno-miR203#204285,
rno-miR-181a-1*#204110 and rno-miR-541#130392;
rno-RNU5G#203908, Exiqon). A SYBR®-Green Master mix kit
(Exiqon) was used for detection. Real-time PCR was performed on an
ABI PRISM® 7500 Sequence Detection System (Applied
Biosystems, Foster City, CA, USA) and data were analyzed using ABI
PRISM 7500 Sequence Detection System Software, version 2.0.1
(Applied Biosystems). Levels of mature miRNAs (miR-341, miR-203,
miR-181a-1* and miR-541*) in the DRG, SDH,
hippocampus and ACC were calculated in relation to the levels of U5
RNA (used as an internal control) using the 2−ΔΔCt
method as previously described (28). Samples from the naïve group were
used as a calibrator. Each set of PCR reactions included a
no-template control and an RNA spike-in (a synthetic control
template). We analyzed 5 biological replicates and 3 technical
replicates. The ratios of miRNA amounts were compared among the
samples using a one-way analysis of variance (ANOVA) followed by a
Student-Newman-Keuls multiple comparison test.
Statistical analysis
The data for the mechanical threshold are expressed
as the means ± SEM. Kruskal-Wallis ANOVA was used to analyze the
differences among treatment groups. Acetone test data were analyzed
using a Pearson’s χ2 test. The changes in miRNA
expression among the groups were analyzed by ANOVA followed by a
Student-Newman-Keuls multiple comparison test. All statistical
tests were performed using SPSS version 17.0 software. Data were
analyzed and are expressed as the means ± SEM, as stated in the
figures.
Results
Mechanical hypersensitivity test
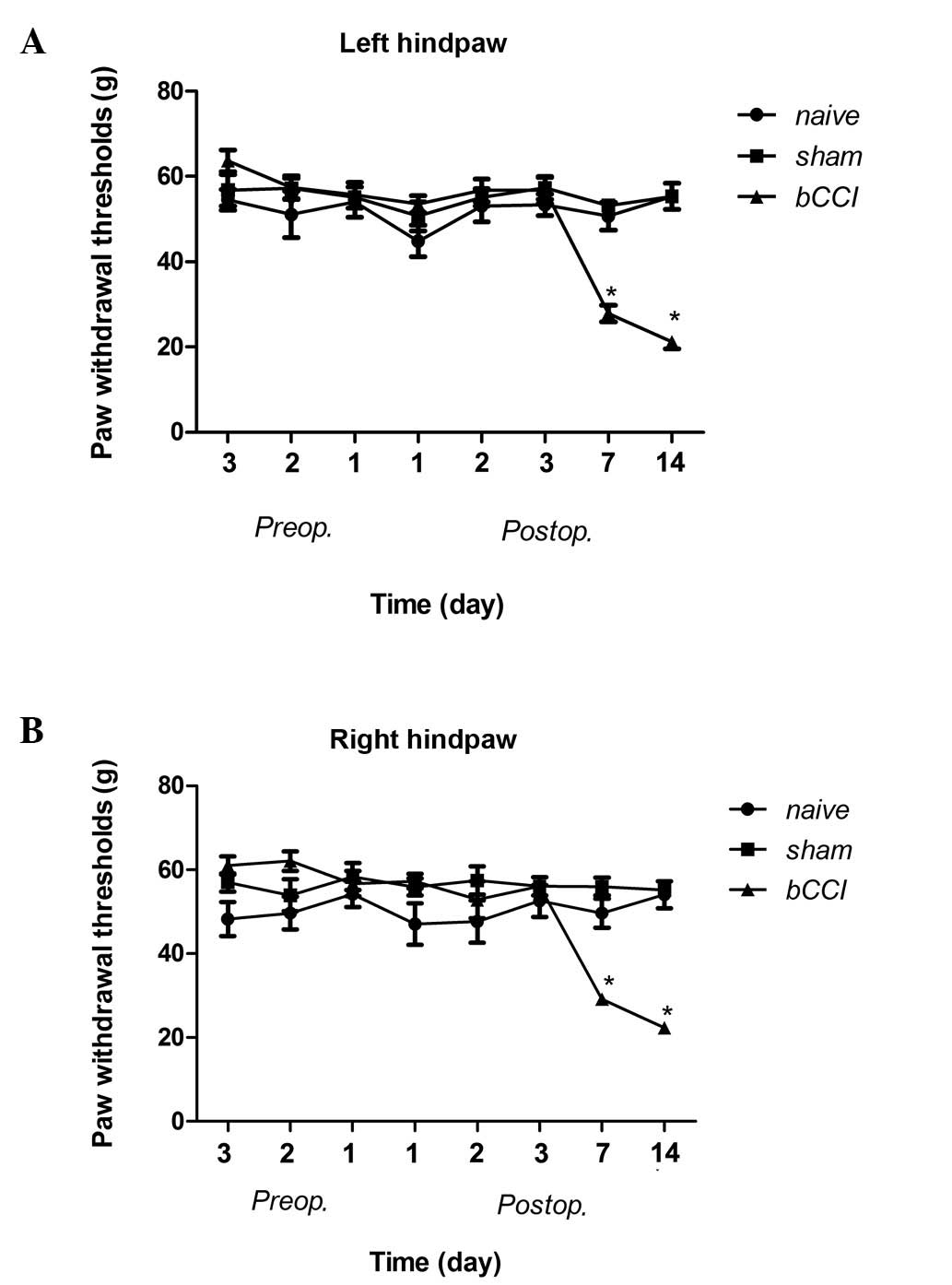
The mechanical sensitivity threshold of the rats in
the bCCI group on post-operative days 7 and 14 was significantly
lower than that in the naïve and sham-operated group rats (Fig. 1; Kruskal-Wallis ANOVA, P≤0.001).
The responses were not significantly different among the groups on
pre-operative days or postoperative days 1 or 3.
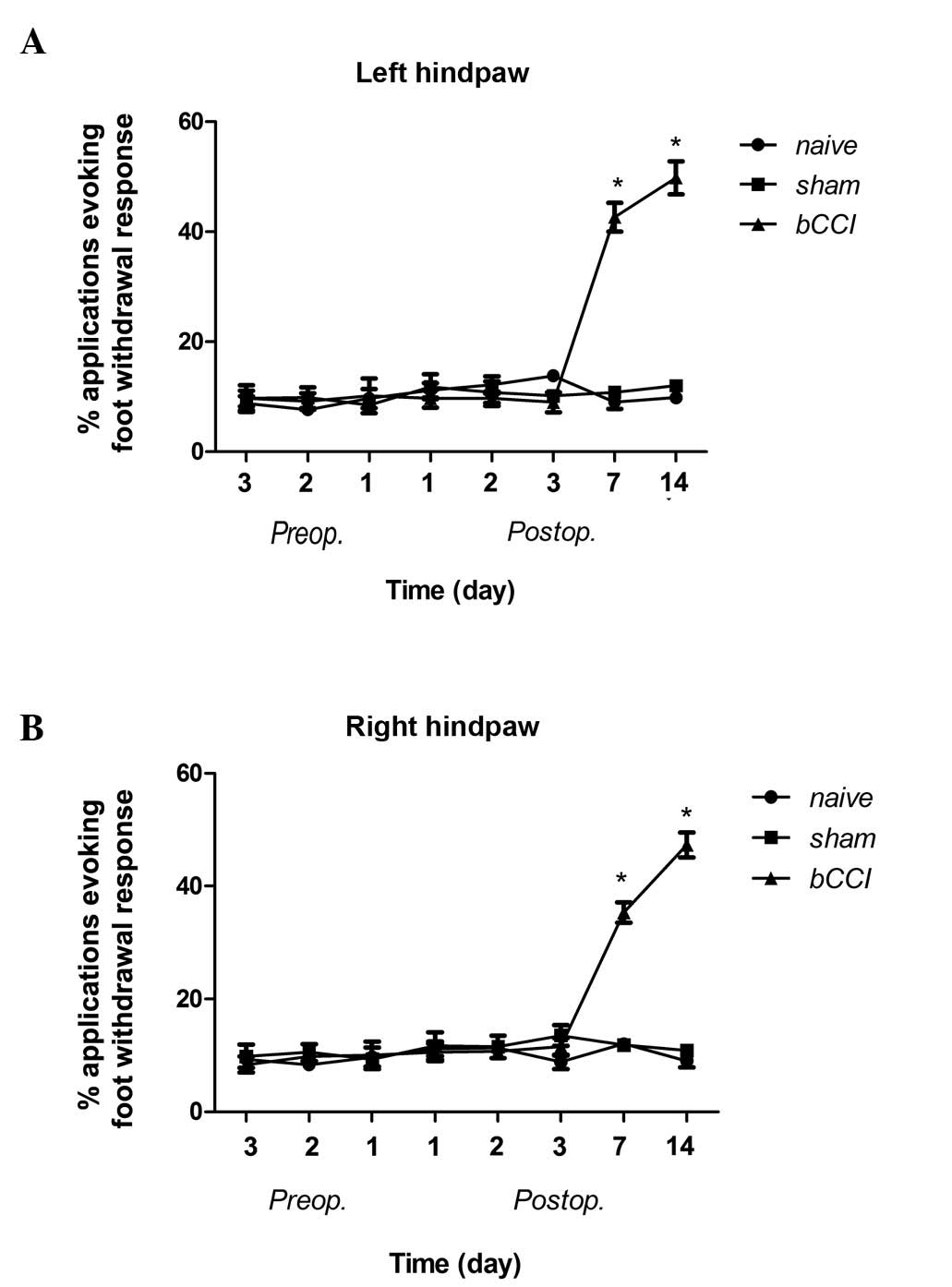
Acetone test
The bCCI group rats displayed cold allodynia on days
7 and 14 post-ligation, whereas the sham-operated and naïve group
rats showed no change as regards cold allodynia (Fig. 2; Pearson’s χ2 test
P<0.001).
Microarray analysis
Using miRCURY LNA expression arrays we found that
the expression of miR-341 was substantially upregulated in the DRG
of the bCCI group rats compared to the sham-operated and naïve
group rats (Table I; one-way
ANOVA, fold change >2, P<0.05). By contrast, the expression
of miR-203, miR-181a-1* and miR-541* in the
SDH of the bCCI group rats was significantly downregulated compared
to the sham-operated and naïve group rats (Table II; one-way ANOVA, fold change
>2, P<0.05).
 | Table I.miRNA expression profiling data in
the dorsal root ganglion of the rats in the naïve, sham-operated
and bCCI groups through one-way ANOVA followed by a
Student-Newman-Keuls multiple comparison test. |
Table I.
miRNA expression profiling data in
the dorsal root ganglion of the rats in the naïve, sham-operated
and bCCI groups through one-way ANOVA followed by a
Student-Newman-Keuls multiple comparison test.
| Name | Average signal in
each group | P-value one-way
ANOVA |
|---|
|
|---|
| Naïve | Sham-operated | bCCI |
|---|
|
rno-miR-3559-5p | 0.0022455 | 0.006415 | 0.008763 | 0.024940839 |
| rno-miR-146b | 0.9553484 | 0.8539158 | 1.8513 | 0.046685766 |
| rno-miR-101a | 1.2790858 | 1.9181574 | 2.5265632 | 0.027562292 |
| rno-miR-187 | 0.034373 | 0.0091985 | 0.014633 | 0.008399584 |
| rno-miR-341 | 0.1797986 | 0.170857 | 0.519283 | 0.001411632 |
| rno-miR-33 | 0.6025086 | 0.9127382 | 1.0853938 | 0.03558754 |
| rno-miR-30c | 2.6648084 | 3.891379 | 4.7367974 | 0.010422544 |
| rno-miR-345-5p | 0.1649046 | 0.2183336 | 0.2121974 | 0.024897078 |
| rno-miR-182 | 0.4810526 | 0.5580232 | 1.0227522 | 0.018045938 |
|
rno-miR-3583-5p | 0.281121 | 0.0023605 | 0.044797 | 0.03715006 |
|
rno-miR-410* | 0.0249235 | 0.005856667 | 0.001947 | 0.008571975 |
|
rno-let-7i* | 0.168998 | 0.2825604 | 0.2546902 | 0.021318724 |
|
rno-miR-3597-3p | 0.0094055 | 0.002546 | 0.0018845 | 0.005867282 |
|
rno-miR-106b* | 0.0120186 | 0.02215325 | 0.0227986 | 0.035722364 |
|
rno-miR-411* | 0.1184006 | 0.1171084 | 0.2318458 | 0.016256798 |
|
rno-miR-129-1* | 0.4003384 | 0.2062314 | 0.2269198 | 0.010098178 |
| rno-miR-503 | 0.287767 | 0.2432734 | 0.3782772 | 0.008755002 |
| rno-miR-140 | 0.2425574 | 0.259532 | 0.5147134 | 0.001715675 |
| rno-miR-124 | 5.1789616 | 3.183408 | 6.2611188 | 0.034531422 |
|
rno-miR-140* | 0.3500766 | 0.3628198 | 0.6690048 | 0.04393541 |
| rno-miR-195 | 0.6586048 | 0.8834174 | 1.1171146 | 0.02132432 |
|
rno-miR-146a* | 0.12624275 | 0.007429667 | 0.02289 | 0.016607437 |
| rno-miR-300-3p | 0.0615604 | 0.070172 | 0.1147072 | 0.025517702 |
|
rno-miR-883* | 0.4047586 | 0.2715158 | 0.2310512 | 0.044802193 |
| rno-let-7a | 1.940091 | 1.30881 | 0.5369012 | 0.043364115 |
| rno-miR-100 | 0.7489904 | 0.628444 | 0.3956312 | 0.016141195 |
| rno-miR-20a | 0.3330146 | 0.2114048 | 0.2921848 | 0.00734766 |
| rno-miR-185 | 0.2947272 | 0.1182896 | 0.3695192 | 0.04991285 |
| rno-miR-146a | 0.3531998 | 0.2088992 | 0.482795 | 0.04055271 |
| rno-miR-760-5p | 0.003096 | 0.014082 | 0.012536333 | 0.008632696 |
| rno-miR-28 | 0.112624 | 0.0823878 | 0.0488122 | 0.007563461 |
| rno-miR-154 | 0.0859528 | 0.0647452 | 0.129833 | 0.047474157 |
|
rno-miR-337* | 0.06191125 | 0.0365376 | 0.0997064 | 0.005767338 |
 | Table II.miRNA expression profiling data in
the SDH of rats in the naïve, sham-operated and bCCI groups through
one-way ANOVA followed by a Student-Newman-Keuls multiple
comparison test. |
Table II.
miRNA expression profiling data in
the SDH of rats in the naïve, sham-operated and bCCI groups through
one-way ANOVA followed by a Student-Newman-Keuls multiple
comparison test.
| Name | Average signal in
each group | P-value one-way
ANOVA |
|---|
|
|---|
| Naïve | Sham-operated | bCCI |
|---|
| rno-miR-872 | 0.088160721 | 0.052155876 | 0.027305913 | 0.013864466 |
| rno-miR-196b | 0.121384594 | 0.050332544 | 0.033891158 | 0.004570896 |
|
rno-miR-425* | 0.023008365 | 0.01885867 | 0.009661174 | 0.044888683 |
| rno-miR-598-3p | 0.20572949 | 0.119244638 | 0.047441445 | 0.041713364 |
| rno-miR-194 | 0.072227716 | 0.038316478 | 0.021800709 | 0.03436289 |
| rno-miR-203 | 0.024348097 | 0.034445658 | 0.002642154 | 0.025992041 |
| rno-miR-221 | 0.139988347 | 0.070956641 | 0.063008119 | 0.046209376 |
| rno-miR-130b | 0.008131009 | 0.015276097 | 0.00664774 | 0.027954211 |
|
rno-miR-181a-1* | 0.027279688 | 0.043180838 | 0.00897442 | 0.009224762 |
| rno-miR-34b | 0.219309204 | 0.098465466 | 0.094511268 | 0.021039538 |
|
rno-miR-434* | 0.165626184 | 0.088637773 | 0.064209862 | 0.02409819 |
| rno-miR-485 | 0.134299181 | 0.042754007 | 0.052200311 | 0.039111715 |
|
rno-miR-3584-3p | 0.005339837 | 0.028103025 | 0.014496631 | 0.026070911 |
| rno-miR-466c | 0.026500144 | 0.060297098 | 0.010574433 | 4.78E-04 |
| rno-miR-200c | 0.008538573 | 0.019556966 | 0.092025084 | 0.020312184 |
|
rno-miR-146a* | 0.04261538 | 0.015903939 | 0.02441009 | 0.022401344 |
|
rno-miR-448* | 0.031652286 | 0.049919515 | 0.014945385 | 0.006336924 |
| rno-miR-324-3p | 0.067771106 | 0.033641862 | 0.033705427 | 0.022043822 |
| rno-miR-223 | 0.052215506 | 0.019387663 | 0.028017393 | 0.012133583 |
|
rno-miR-218a-2* | 0.011025996 | 0.035931925 | 0.003778643 | 0.04053875 |
|
rno-miR-541* | 0.008237646 | 0.014807682 | 0.001920307 | 0.040937595 |
| rno-miR-488 | 0.046445858 | 0.020892955 | 0.02520577 | 0.045676824 |
|
rno-miR-10b* | 0.041413909 | 0.021220252 | 0.026583696 | 0.047183 |
| rno-miR-146a | 0.105815309 | 0.060321471 | 0.133724172 | 0.03959339 |
|
rno-miR-376c* | 0.089361893 | 0.056806745 | 0.04675663 | 0.04180455 |
qRT-PCR validation of the differentially
expressed miRNAs in the pain transmission pathway
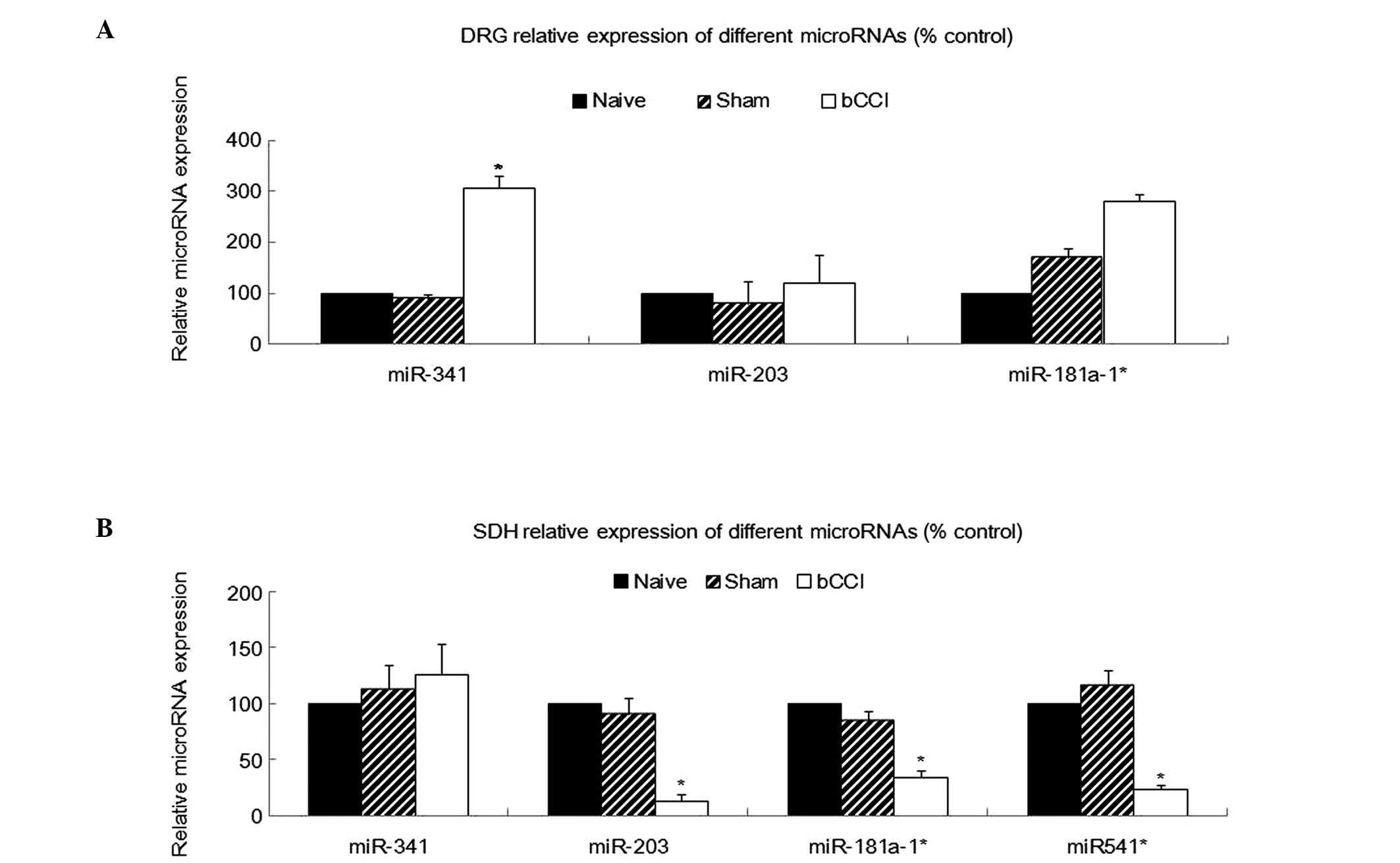
Using RT-PCR, we analyzed miR-341, miR-203,
miR-181a-1* and miR-541* expression in total
RNA isolated from the DRG, SDH, hippocampus and ACC from the naïve,
sham-operated and bCCI group rats (n=5 for each category). A
comparison of the relative miRNA expression levels among the
different treatment groups revealed a significant upregulation of
miR-341 in the DRG of the bCCI group rats, in agreement with the
microarray data. The remaining miRNAs did not significantly differ
in expression among the groups (Fig.
3A; one-way ANOVA, P<0.05).
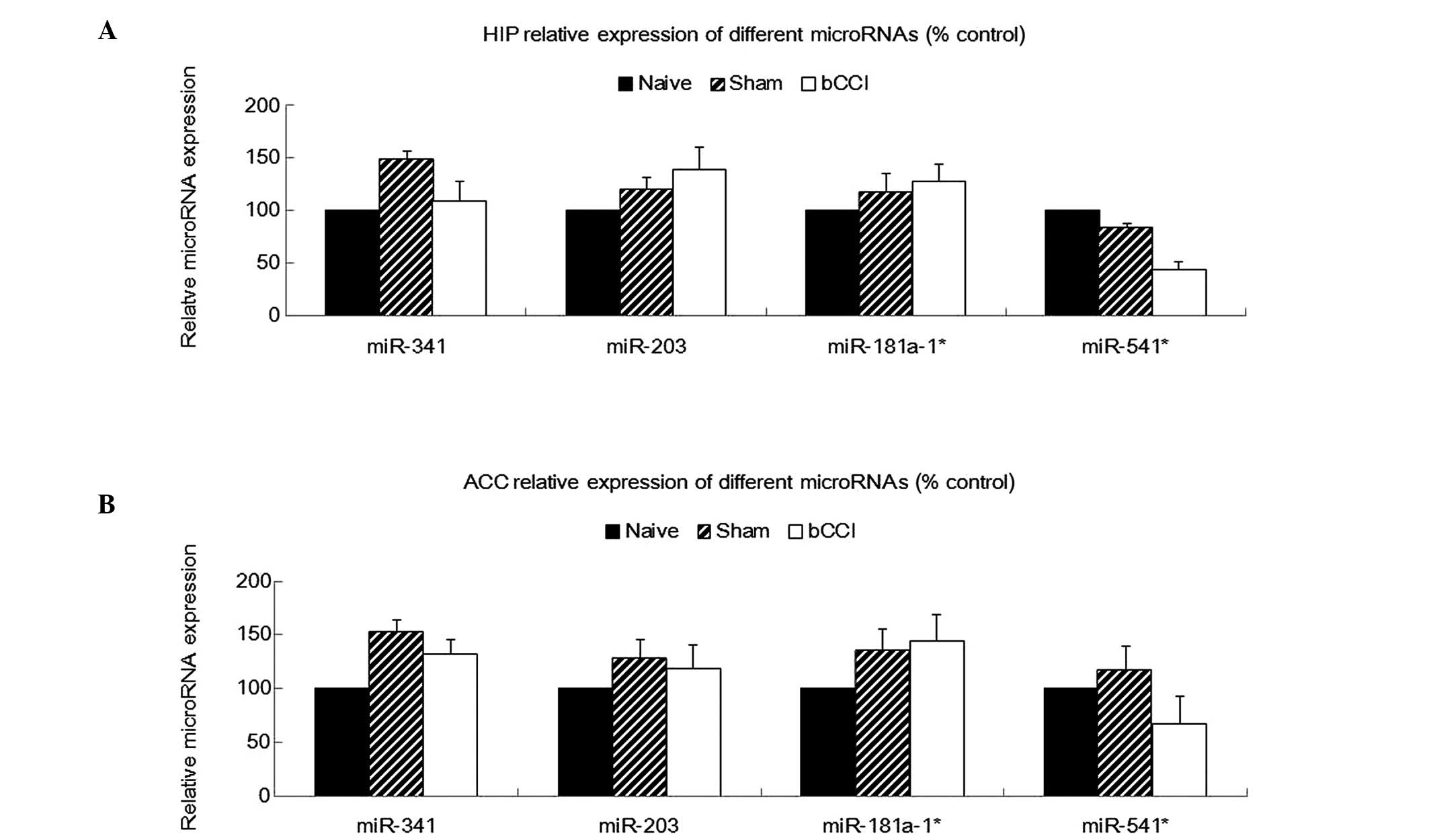
Real-time PCR analysis also indicated that miR-203,
miR-181a-1* and miR-541* were significantly
downregulated among in the bCCI group rats (Fig. 3B; one-way ANOVA, P<0.05, n=5),
consistent with the microarray analysis. However, none of the above
4 miRNAs showed any significant differences in expression among the
naïve, sham-operated and bCCI group rats in the hippocampus or ACC
(Fig. 4; one-way ANOVA, P≥0.05,
n=5).
Discussion
In this study, we analyzed miRNA expression in
different areas of the nociceptive pathway in rats with bCCI
compared to naïve and sham-operated rats. We found that the DRG and
SDH had a specific and restricted expression of miRNAs that may be
associated with the neuropathic pain model. These changes appear to
be the result of the specific regulation of miRNAs in individual
tissues or organs along the pain transmission pathway, rather than
a global change in miRNA or small RNA levels, as the miRNA changes
observed after the induction of bCCI were unique to their specific
region. Previous studies have determined that miRNA expression in
the nociceptive system is not only temporally and spatially
specific, but also stimulus-dependent (14,22). However, to our knowledge, little
is known about the differential expression of miRNAs in the
nociceptive pathway during bCCI, and our is the first study to
investigate this.
Neuropathic pain is currently under-recognized and
under-treated. An increasing body of evidence shows that miRNAs
play fundamental roles in neurogenesis, neuron survival, dendritic
outgrowth and spine formation (29–31). Aberrant miRNA expression has also
been linked to a variety of diseases, including several nervous
system diseases (32–34). For example, miR-219 modulates
N-methyl-D-aspartate (NMDA) receptor-mediated
neurobehavioral dysfunction, which is implicated in schizophrenia
and autism (35), and the
expression of the sensory organ-specific miR-183 family has been
shown to be altered following spinal nerve ligation (14). Moreover, small RNAs have been
shown to play critical roles in altering pain thresholds through
controlling sodium-channel expression during inflammatory pain
(36). Therefore, miRNAs may be
novel therapeutic targets for treating these diseases, although we
still do not fully understand the molecular mechanisms by which
miRNAs regulate gene expression nor do we know the complete
repertoire of mRNAs that each miRNA targets.
In this study, using a novel microarray-based
approach, we found for the first time that miRNA expression was
altered in the DRG and SDH of rats with bCCI. This 6th-generation
miRNA array contains more than 1891 capture probes, covering all
human, mouse and rat miRNAs annotated in miRBase 16.0 (the older
version of the miRNA database), as well as all viral miRNAs related
to these species. In addition, this array contains capture probes
for 66 new miRPlus™ human miRNAs. These are proprietary miRNAs not
found in miRBase, and therefore may lead to the discovery of
changes in new miRNAs in different disease models.
DRG neurons are primary sensory organs that
selectively respond to noxious or potentially tissue-damaging
stimuli. They can be sensitized, which is one of the critical
mechanisms behind neuropathic pain (37,38). Previous studies have observed that
changes in the expression of voltage-gated sodium channel 1.8 and
tissue inhibitors of metalloproteinase (TIMPs) occurred in the DRG
of rats in a neuropathic pain model (39,40). As shown in our study, miR-341 was
upregulated exclusively in the DRG of rats with bCCI, and not in
the SDH, hippocampus and ACC. By contrast, miR-541*
expression was not detected (even after 40 PCR cycles), indicating
that this miRNA either is not expressed, or has a very low
expression, in the DRG. Additionally, neither miR-203 nor
miR-181a-1* expression was significantly altered in the
DRG of the rats with bCCI. Prior to our study, Koturbash et
al investigated the differential expression of miRNAs
associated with X-ray irradiation (41). They found that changes in miRNA
expression were tissue-specific in the hippocampus, frontal cortex
and cerebellum of female mice. Furthermore, Rao et al
observed that tissue-specific RNA interference knocked down Wilms’
tumor 1 (WT1) expression in a tissue-specific manner, affecting
germ cell survival and spermatogenesis in vivo (42). These results indicate that many
miRNAs exhibit tissue- or organ-specific expression patterns and
functions in several disease conditions. Our results from profiling
the nervous system of rats with bCCI are consistent with this model
for miRNA behavior.
miR-203, miR-181a-1* and
miR-541* exhibited differential expression only in the
SDH of rats with bCCI, but not the DRG, hippocampus or ACC. This
suggests that these miRNAs are involved in regulating neuropathic
pain, and that their effects are limited to that specific
region.
miR-203 has been shown to be downregulated in tumors
(43); however, the pathological
interactions between neuropathic pain and tumorigenesis require
further investigation. Changes in miR-341, miR-541* and
miR181a-1* expression have previously only shown a limited
association with abnormal states. Therefore, further studies are
required to determine the significance of the observed differences
in the bCCI model. These findings have implications for neuropathic
pain management by miRNA replacement therapy, which can replenish
the miRNAs lost or reduced in neuropathic pain by the addition of
miRNA mimics. This may also minimize toxicity while retaining
potency against the intended miRNA targets.
The 4 miRNAs whose expression was either increased
or reduced in the DRG or SDH exhibited no statistically different
changes in the hippocampus or ACC. These results suggest that these
areas may have their own unique miRNA expression patterns that can
influence the development and maintenance of the neuropathic pain
condition. However, this conclusion needs to be confirmed by
further studies. The realization that the inappropriate production
of individual miRNAs in a specific region of the pain transmission
pathway contributes to neuropathic pain has reinvigorated antisense
oligonucleotide (ASO) drug development (44,45).
In conclusion, in our study, using a rat model of
bCCI, we found that miR-341 may play a role in the pathogenesis of
neuropathic pain in the DRG, while miR-203, miR-181a-1*
and miR-541* may play critical roles in the SDH. Since
miRNAs have strong therapeutic potentials and have relatively easy
accessibility for systemic or regional delivery in gene therapy,
the miRNAs identified in this study may be considered as potential
candidates for novel treatment strategies. We expect that our
results will help elucidate the molecular mechanisms involved in
neuropathic pain and may provide preliminary experimental evidence
for the use of miRNAs in the gene therapy treatment of neuropathic
pain.
Acknowledgements
This study was supported by a grant
from the National Natural Science Foundation of China (C31070930).
We thank Dr Jian Guan of the Pathology Department of the Peking
Union Medical College Hospital for her assistance with the animal
surgeries.
Abbreviations:
|
bCCI
|
bilateral sciatic nerve chronic
constriction injury;
|
|
SDH
|
spinal dorsal horn;
|
|
DRG
|
dorsal root ganglion;
|
|
ACC
|
anterior cingulate cortex
|
References
|
1.
|
Treede RD, Jensen TS, Campbell JN, et al:
Neuropathic pain: redefinition and a grading system for clinical
and research purposes. Neurology. 70:1630–1635. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Haanpää M, Attal N, Backonja M, et al:
NeuPSIG guidelines on neuropathic pain assessment. Pain. 152:14–27.
2011.
|
|
3.
|
Green L and McGhie J: Assessment of acute
and chronic pain. Anaesth Intensive Care Med. 12:9–11. 2011.
View Article : Google Scholar
|
|
4.
|
Bie B, Brown DL and Naguib M: Increased
synaptic GluR1 subunits in the anterior cingulate cortex of rats
with peripheral inflammation. Eur J Pharmacol. 653:26–31. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Boroujerdi A, Zeng J, Sharp K, Kim D,
Steward O and Luo ZD: Calcium channel alpha-2-delta-1 protein
upregulation in dorsal spinal cord mediates spinal cord
injury-induced neuropathic pain states. Pain. 152:649–655. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Chou CW, Wong GT, Lim G, et al: Peripheral
nerve injury alters the expression of NF-kappaB in the rat’s
hippocampus. Brain Res. 1378:66–71. 2011.PubMed/NCBI
|
|
7.
|
Emery EC, Young GT, Berrocoso EM, Chen L
and McNaughton PA: HCN2 ion channels play a central role in
inflammatory and neuropathic pain. Science. 333:1462–1466. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Jaggi AS and Singh N: Role of different
brain areas in peripheral nerve injury-induced neuropathic pain.
Brain Res. 1381:187–201. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Vogt BA: Pain and emotion interactions in
subregions of the cingulate gyrus. Nat Rev Neurosci. 6:533–544.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Hodgdon KE, Hingtgen CM and Nicol GD:
Dorsal root ganglia isolated from Nf1+/− mice exhibit
increased levels of mRNA expression of voltage-dependent sodium
channels. Neuroscience. 206:237–244. 2012.PubMed/NCBI
|
|
11.
|
Kim DS, Figueroa KW, Li KW, Boroujerdi A,
Yolo T and Luo ZD: Profiling of dynamically changed gene expression
in dorsal root ganglia post peripheral nerve injury and a critical
role of injury-induced glial fibrillary acetic protein in
maintenance of pain behaviors. Pain. 143:114–122. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Cady RJ, Glenn JR, Smith KM and Durham PL:
Calcitonin gene-related peptide promotes cellular changes in
trigeminal neurons and glia implicated in peripheral and central
sensitization. Mol Pain. 7:942011. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Uchida H, Ma L and Ueda H: Epigenetic gene
silencing underlies C-fiber dysfunctions in neuropathic pain. J
Neurosci. 30:4806–4814. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Aldrich BT, Frakes EP, Kasuya J, Hammond
DL and Kitamoto T: Changes in expression of sensory organ-specific
microRNAs in rat dorsal root ganglia in association with mechanical
hyper-sensitivity induced by spinal nerve ligation. Neuroscience.
164:711–723. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Bak M, Silahtaroglu A, Moller M, et al:
MicroRNA expression in the adult mouse central nervous system. RNA.
14:432–444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Bastian I, Tam Tam S, Zhou X-F, et al:
Differential expression of microRNA-1 in dorsal root ganglion
neurons. Histochem Cell Biol. 135:37–45. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
von Schack D, Agostino MJ, Murray BS, et
al: Dynamic changes in the microRNA expression profile reveal
multiple regulatory mechanisms in the spinal nerve ligation model
of neuropathic pain. PLoS One. 6:e176702011.PubMed/NCBI
|
|
18.
|
Chiang HR, Schoenfeld LW, Ruby JG, et al:
Mammalian microRNAs: experimental evaluation of novel and
previously annotated genes. Genes Dev. 24:992–1009. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Hua YJ, Tang ZY, Tu K, et al:
Identification and target prediction of miRNAs specifically
expressed in rat neural tissue. BMC Genomics. 10:2142009.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Kosik KS: The neuronal microRNA system.
Nat Rev Neurosci. 7:911–920. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Wheeler G, Ntounia-Fousara S, Granda B,
Rathjen T and Dalmay T: Identification of new central nervous
system specific mouse microRNAs. FEBS Lett. 580:2195–2200. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Kusuda R, Cadetti F, Ravanelli MI, et al:
Differential expression of microRNAs in mouse pain models. Mol
Pain. 7:172011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Harraz MM, Dawson TM and Dawson VL:
MicroRNAs in Parkinson’s disease. J Chem Neuroanat. 42:127–130.
2011.
|
|
24.
|
Zhang HY, Zheng SJ, Zhao JH, et al:
MicroRNAs 144, 145, and 214 are down-regulated in primary neurons
responding to sciatic nerve transection. Brain Res. 1383:62–70.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Datta S, Chatterjee K, Kline RH and Wiley
RG: Behavioral and anatomical characterization of the bilateral
sciatic nerve chronic constriction (bCCI) injury: correlation of
anatomic changes and responses to cold stimuli. Mol Pain. 6:72010.
View Article : Google Scholar
|
|
26.
|
Vierck CJ, Acosta-Rua AJ and Johnson RD:
Bilateral chronic constriction of the sciatic nerve: a model of
long-term cold hyper-algesia. J Pain. 6:507–517. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Chaplan SR, Bach FW, Pogrel JW, Chung JM
and Yaksh TL: Quantitative assessment of tactile allodynia in the
rat paw. J Neurosci Methods. 53:55–63. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative CT method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Krichevsky AM, King KS, Donahue CP,
Khrapko K and Kosik KS: A microRNA array reveals extensive
regulation of microRNAs during brain development. RNA. 9:1274–1281.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Schratt GM, Tuebing F, Nigh EA, et al: A
brain-specific microRNA regulates dendritic spine development.
Nature. 439:283–289. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Mercader JM, Gonzalez JR, Lozano JJ, et
al: Aberrant brain microRNA target and miRISC gene expression in
the anx/anx anorexia mouse model. Gene. 497:181–190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Wei L, Wang M, Qu X, et al: Differential
expression of microRNAs during allograft rejection. Am J
Transplant. 12:1113–1123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Liu PT, Wheelwright M, Teles R, et al:
MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway
in leprosy. Nat Med. 18:267–273. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Kocerha J, Faghihi MA, Lopez-Toledano MA,
et al: MicroRNA-219 modulates NMDA receptor-mediated
neurobehavioral dysfunction. Proc Natl Acad Sci USA. 106:3507–3512.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Zhao J, Lee MC, Momin A, et al: Small RNAs
control sodium channel expression, nociceptor excitability, and
pain thresholds. J Neurosci. 30:10860–10871. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Kuner R: Central mechanisms of
pathological pain. Nature Med. 16:1258–1266. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Gold MS and Gebhart GF: Nociceptor
sensitization in pain pathogenesis. Nature Med. 16:1248–1257. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Ruangsri S, Lin A, Mulpuri Y, Lee K,
Spigelman I and Nishimura I: Relationship of axonal voltage-gated
sodium channel 1.8 (NaV1.8) mRNA accumulation to sciatic nerve
injury-induced painful neuropathy in rats. J Biol Chem.
286:39836–39847. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Huang B, Zhao X, Zheng LB, Zhang L, Ni B
and Wang YW: Different expression of tissue inhibitor of
metalloproteinase family members in rat dorsal root ganglia and
their changes after peripheral nerve injury. Neuroscience.
193:421–428. 2011. View Article : Google Scholar
|
|
41.
|
Koturbash I, Zemp F, Kolb B and Kovalchuk
O: Sex-specific radiation-induced microRNAome responses in the
hippocampus, cerebellum and frontal cortex in a mouse model. Mutat
Res. 722:114–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Rao MK, Pham J, Imam JS, et al:
Tissue-specific RNAi reveals that WT1 expression in nurse cells
controls germ cell survival and spermatogenesis. Genes Dev.
20:147–152. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Bo J, Yang G, Huo K, et al: microRNA-203
suppresses bladder cancer development by repressing bcl-w
expression. FEBS J. 278:786–792. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Elmén J, Lindow M, Schütz S, et al:
LNA-mediated microRNA silencing in non-human primates. Nature.
452:896–899. 2008.PubMed/NCBI
|
|
45.
|
Broderick JA and Zamore PD: MicroRNA
therapeutics. Gene Ther. 18:1104–1110. 2011. View Article : Google Scholar : PubMed/NCBI
|