Introduction
Lung cancer is the most common malignancy worldwide,
with approximately 1.3 million new cases and 300,000 deaths each
year, as estimated by the World Health Organization (1). As non-small cell lung cancer (NSCLC)
accounts for 80–85% of all lung cancer cases, understanding the
pathogenic mechanism of NSCLC is critical (2).
Survivin, a member of the inhibitor of apoptosis
protein (IAP) family, is a key regulator of mitosis and programmed
cell death. Although minimally expressed in normal adult tissues,
survivin is highly expressed in most human tumors, such as melanoma
and cancer of the lung, esophagus, stomach, intestine, pancreas and
breast (3). Of note, survivin is
associated with tumor progression, angiogenesis, poor patient
prognosis, resistance to radiation and drug treatments, and
increased rate of cancer relapse (3–5).
Several pharmacological and environmental stimuli, such as UVB
exposure, chemotherapeutic agents, hypoxia and vascular injury, can
increase survivin expression (6);
survivin has also become a therapeutic target and a potentially
important prognostic marker for numerous types of tumor.
Hypoxia is a unique microenvironment in solid
tumors, including lung cancer. Since vasculature in tumors is
dysfunctional, rapid growth of tumor cells results in insufficient
oxygen supply (7). Hypoxia is
associated with increased malignancy, resistance to therapy and
distant metastasis (8–10). Hypoxia-inducible factor-1 (HIF-1),
a master transcription factor of oxygen-regulated genes, mediates a
wide range of cellular and physiological adaptive responses to
changes in oxygen tension (11).
HIF-1 is composed of two subunits, HIF-1α and HIF-1β (12). HIF-1α is the main active subunit,
which can induce a vast array of gene products that control energy
metabolism, neovascularization, survival and cell migration and is
a strong promoter of tumor growth (13). In our previous study, we found
that HIF-1α and survivin were widely expressed in both A549 cells
and fresh NSCLC tissue samples, and increased significantly in
hypoxia compared with normoxia (14). This finding is consistent with
other studies that show survivin expression is induced by hypoxia
(15). We speculate that HIF-1α
could become an important target for lung cancer therapy. Herein,
we showed HIF-1α expression knocked down by miRNA to inhibit
proliferation and promote apoptosis under hypoxia, to increase cell
migration in both hypoxia and normoxia, to reduce survivin
expression and to trigger apoptosis in vitro and in
vivo. We also confirmed that HIF-1α mediates survivin
overexpression by direct binding to the survivin promoter
region.
Materials and methods
Cell lines and culture conditions
Human lung adenocarcinoma cell lines were obtained
from the Cell Culture Center, Chinese Academy of Medical Sciences
(Shanghai, China). A549 cells were maintained in Ham’s F12 medium
supplemented with 10% fetal bovine serum. Cells were incubated
under normoxic (20% O2) or hypoxic (cobalt chloride, a
hypoxia-mimicking agent, the maximum expression of HIF-1α was with
150 μmol/l CoCl2) conditions.
HIF-1α miRNA construct and cotransfection
with survivin expression vectors
For the miRNA construct, one target sequence
(5′-GCAGGTCATAGTTTTGGCCACTG-3′) was selected corresponding to the
open reading frame of the human HIF-1α gene (NM-001530). The
construct containing a scrambled sequence
(5′-CGTGGAGACGTTTTGGCCACTGA-3′) (Scrambled) was also included as a
negative control; it has no significant homology with human gene
sequences. They were synthesized by Invitrogen and inserted into
pcDNA6.2-GW/EmGFP eukaryotic expression vectors to construct miRNA
or negative control vectors, which were termed HIF-1α-miRNA and
Scrambled, respectively. For gene transfection, 2×105
cells per well were set into 6-well plates and grown overnight
until they were 50–80% confluent. Plasmids HIF-1α-miRNA and
Scrambled were transfected into A549 cells by Lipofectamine 2000
reagent (Invitrogen) as per the manufacturer’s instructions. Cells
were subcultured at a 1:5 dilution in 300 mg/ml G418-containing
medium. Positive stable transfections were selected and expanded
for further study. The pCLEN plasmid encoding full-length survivin
was a kind gift from Dr Feng Qian (Department of Pharmacology,
University of Illinois, Chicago, IL, USA). Cells were transfected
twice with 2 μg of expression vector or empty pCRII-TOPO control
(Invitrogen) 6 and 24 h after HIF-1α-miRNA transfection (described
above) using the FuGENE 6 Transfection Reagent (Roche Diagnostics)
as per the manufacturer’s recommendations. Cells were harvested 24
h after transfection for western blotting.
Cotransfection of survivin
promoter-luciferase reporter vectors and HIF-1α expression
vectors
Constructs were removed from pGL3-basic by
restriction endonuclease MluI/HindIII, following
procedures described in our previous study (14). Reporter vectors were constructed
by T4 DNA ligase, known as pGL3-SVP-230-luc. The plasmid encoding
HIF-1α, known as pcDNA3-HIF-1α, was a kind gift from Dr Feng Qian.
Cells were plated at 5×105 cells per well in 6-well
dishes and allowed to settle overnight. The following morning,
cells were cotransfected with constructs (pLuc-surP-230 and
pcDNA3-HIF-1α or pcDNA3) using Lipofectamine 2000 according to the
manufacturer’s protocols; 30 h after transfection, cells were
harvested and lysed with 1X lysis buffer (Promega); 20 μl of cell
extract was then assayed for luciferase activity using the
Dual-Luciferase assay kit (Promega) according to the manufacturer’s
instructions. Relative levels of reporter gene expression were
expressed as ratios of firefly luciferase activity to Renilla
luciferase (LU/RL).
Chromatin immunoprecipitation (ChIP)
To demonstrate direct binding of HIF-1α protein to
the survivin promoter region in A549 cells under both
normoxic and hypoxic conditions, ChIP was performed using the
ChIP-IT Express kit (Active Motif) according to the manufacturer’s
protocols. Briefly, A549 cells were transfected with pcDNA3-HIF-1α
or pcDNA3 prior to fixation with 1% formaldehyde for 10 min. Cells
were then washed, lysed, and sonicated to reduce DNA lengths to a
range of 300–600 bp. The HIF-1α/DNA complexes were incubated with
mouse antibody against HIF-1α (Santa Cruz Biotechnology, Santa
Cruz, CA, USA), or normal mouse IgG (Santa Cruz Biotechnology) for
18 h at 4°C. The immune complexes were precipitated, eluted,
reverse-crosslinked and treated with proteinase K. The resulting
DNA samples were amplified using primers for the putative HIF-1α
site in the human survivin promoter region confirmed by our
previous study (15) (F,
5′-GCGTTCTTTGAAAGCAGT-3′ and R, 5′-ATCTGGCGGTTAATGGCG-3′).
Reverse transcription-PCR
Total RNA was isolated using TRIzol reagent
(Invitrogen) according to the manufacturer’s instructions.
Concentration of total RNA was detected by UV spectrophotometry.
RT-PCR was performed by the two-step method. Synthesis of cDNA was
performed using the cDNA Synthesis kit (Thermo, Shanghai, China).
The PCR reaction conditions were: 95°C for 5 min, 94°C for 30 sec,
56°C for 30 sec, 72°C for 30 sec for 35 cycles; the total volume
was 20 μl. For quantitative analysis of HIF-1α and
survivin mRNA, expression of the housekeeping gene
GAPDH was used as an internal standard. The primers used in
this study were: F, 5′-AGCCAGACGATCATGCAGCTACTA-3′ and R,
5′-TGTGGTAATCCACTTTCATCCATTG-3′ for HIF-1α (167 bp); F,
5′-AGGTCATCTCGGCTGTTCCTG-3′ and R, 5′-TCATCCTCACTGCGGCTGTC-3′, for
survivin (147 bp); and F, 5′-GGTCTCCTCTGACTTCAACA-3′ and R,
5′-AGCCAAATTCGTTGTCATAC-3′ for GAPDH (375 bp). Primers were
synthesized by Shanghai Sangon Biological Engineering Technology
& Services Co., Ltd. PCR fragments were separated and
visualized in 20 g/l agarose gels stained with ethidium bromide.
Semi-quantitative analysis was performed with Gis gel analysis
software (Shanghai, China). All experiments were performed in
triplicate. Ratios of photo-density of RT-PCR products of target
genes and GAPDH were used to identify expression intensity
of target genes.
Western blot analysis
Tumor tissues were ground and sonicated with
supersonic lytic buffer that contained 50 mmol/l
NaH2PO4, 100 mmol/l Tris-HCl, 250 mmol/l
NaCl, 100 mg/l PMSF, 1 mg/l aprotinin, pH 8.0, and then centrifuged
at 12,000 × g for 40 min. A Bio-Rad standard curve was used to
determine protein concentration in each lysate. Loading buffer was
added to each lysate, which was then boiled for 5 min and
electrophoresed by SDS-PAGE. The proteins were mixed with 2X
loading buffer to the same volume prior to electrophoresis. After
transferring onto nitrocellulose, proteins were incubated with
antibodies (anti-HIF-1α, anti-survivin and β-actin, purchased from
Santa Cruz Biotechnology), and then with peroxidase-conjugated
secondary antibody (Santa Cruz Biotechnology). Detection was
performed with an enhanced chemiluminescence agent. Analysis was
performed with Bandscan analysis software (Sterling, VA, USA). All
experiments were carried out in triplicate. Ratios of HIF-1α,
survivin and β-actin proteins were used to identify expression
intensity.
Cell viability CCK-8 assay
After G418 selection for 4–5 weeks, HIF-1α-miRNA,
Scrambled and untreated cells were exposed to CoCl2 at
150 μmol/l in 96-well plates for 24, 48 and 72 h. Cell viability
was detected by Cell Counting Kit-8 (CCK-8). Following treatment,
10 μl of CCK-8 solution was added to each well; the 96-well plate
was continuously incubated at 37°C for 1 h, then OD values for each
well were read on a microplate reader (Multiskan, Thermo, USA) at
450 nm to determine cell viability. The assay was repeated 3 times.
Cell viability was calculated as follows: % cell
viability=[(ODexperiment -
ODblank)/(ODcontrol - ODblank)]
×100%.
FACS assay
Transfected cells and control cells in the log
growth phase were harvested by trypsinization at 48 h under
normoxic and hypoxic conditions for flow cytometry. Apoptotic cells
in early and late stages were detected using an Annexin V-FITC
Apoptosis Detection kit from BioVision (Mountain View, CA, USA). In
brief, 5.0×105 cells were transfected with oligos at
various concentrations in the presence of Lipofectin (7 mg/ml) for
48 h. Media and cells were then collected. Cells harvested by
centrifugation were washed with serum-free media and re-suspended
in Annexin V Binding Buffer (500 ml); Annexin V-FITC (5 ml) and
then propidium iodide (PI; 5 ml) were added. Samples were incubated
in the dark for 5 min at room temperature (25.8°C) and then
analyzed using a Becton Dickinson FACSCalibur (Ex=488 nm; Em=530
nm). Cells positive for Annexin V-FITC alone (early apoptosis) and
for Annexin V-FITC and PI (late apoptosis) were counted. Each assay
was repeated 3 times.
Transwell invasion assay
Transwell filters (Costar, USA) were coated with
Matrigel (3.9 mg/ml, 60–80 ml) on the upper surface of the
polycarbonic membrane (diameter: 6.5 mm; pore size: 8 mm). After
incubating at 37°C for 30 min, Matrigel became solidified and
served as the extracellular matrix for tumor cell invasion
analysis. Harvested cells (1×105) in 100 ml of
serum-free Ham’s F-12 were added into the upper compartment of the
chamber. A total of 200 ml conditioned medium derived from A549
cells was used as a source of chemoattractant and placed in the
bottom compartment of the chamber. After 24 h of incubation at 37°C
with 5% CO2, the medium was removed from the upper
chamber. Non-invading cells on the upper side of the chamber were
scraped off with a cotton swab. Cells that had migrated from
Matrigel into pores of the inserted filter were fixed with 100%
methanol, stained with hematoxylin, mounted and dried at 80°C for
30 min. The number of cells invading through the Matrigel was
counted in 3 randomly selected visual fields each from the central
and peripheral portions of the filter, using an inverted microscope
at ×200 magnification. Each assay was repeated 3 times.
Subcutaneous tumor model
Male immune-deficient nude mice (4 weeks old)
(BALB/c-nu) were purchased from Shanghai Slac Laboratory Animal
Co., Ltd., bred at the facility of laboratory animals, Bengbu
Medical College, and housed in micro-isolator individually
ventilated cages with water and food. All experimental procedures
were carried out according to the regulations and internal
biosafety and bioethics guidelines of Bengbu Medical College and
the Bengbu Municipal Science and Technology Commission. Mice were
divided into 3 groups of 8 mice each. Each mouse was simultaneously
injected subcutaneously with 1×107 of A549 cells
transfected with HIF-lα miRNA, Scrambled miRNA (control) or A549
cells untreated. Mice were monitored daily and all formed
subcutaneous tumors. Tumor dimensions of 3 groups were measured
every day with a sliding caliper using the formula: volume = length
× width2 ×0.52. When tumor volume reached ~50
mm3, tumor dimensions were measured every three days. At
58 days after injection, tumors were surgically removed and
weighed. Animals were monitored by general observation and
determination of body weight until they were euthanized.
TUNEL assay
Tumor tissues were fixed with 10% formalin for 4 h
and then embedded in paraffin. Slices were deparaffinized in water
and placed in 3% H2O2 for 10 min at room
temperature. The TUNEL assay was carried out according to the
manufacturer’s instructions (Beyotime Institute of Biotechnology,
Beijing, China). Positive results showed brown nuclear
staining.
Statistical analyses
All assays were repeated 3 times to ensure
reproducibility. For comparisons of the 3 assays and between groups
ANOVA and Student’s t-test were used, respectively. All tests were
performed using SPSS 11.5. Results are displayed as the means ± SD.
P<0.05 was considered to indicate a statistically significant
difference.
Results
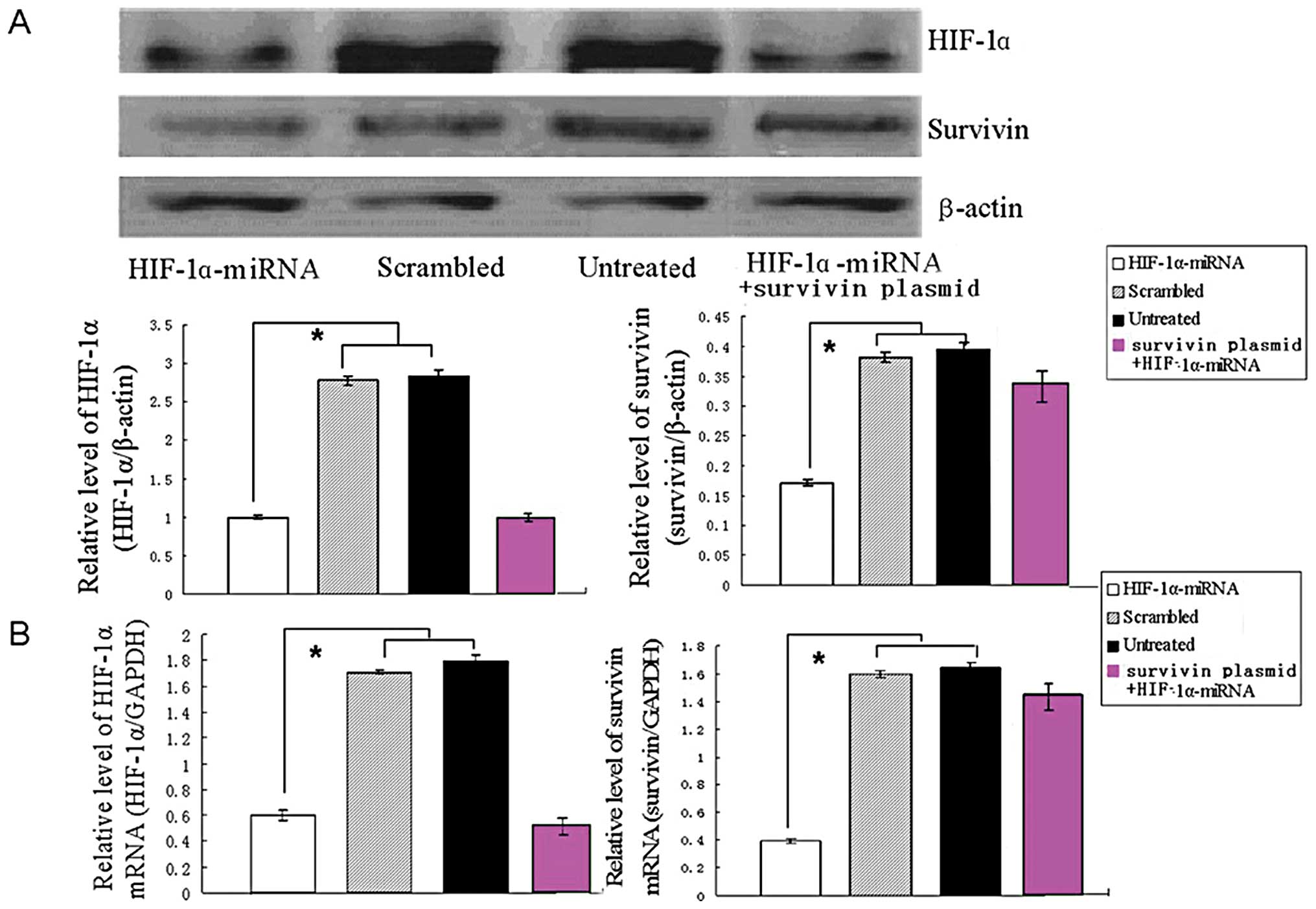
Effect of HIF-lα-miRNA on survivin
expression in A549 cells
To compare the effects of miRNA targeting HIF-lα on
survivin expression, two constructs were prepared and transfected
into human A549 cells. After selection for 4 weeks, G418-resistant
cells were obtained. Western blotting showed that transfection of
the control vector had little effect on HIF-lα expression. However,
expression of HIF-lα mRNA was markedly downregulated by 67%
in cells transfected with HIF-lα-miRNA, and the survivin expression
was also downregulated to 75% (Fig.
1B). Western blot analysis showed similar downregulation of
HIF-lα and survivin protein expression (Fig. 1A). Furthermore, transfection of
survivin expression vectors in HIF-lα knockdown cells rescued
survivin expression (Fig. 1).
Such an effect was not observed in control cells transfected with
empty pCRII-TOPO (data not shown). These results suggest that
HIF-lα-miRNA can potently and specifically inhibit endogenous
survivin expression in A549 cells.
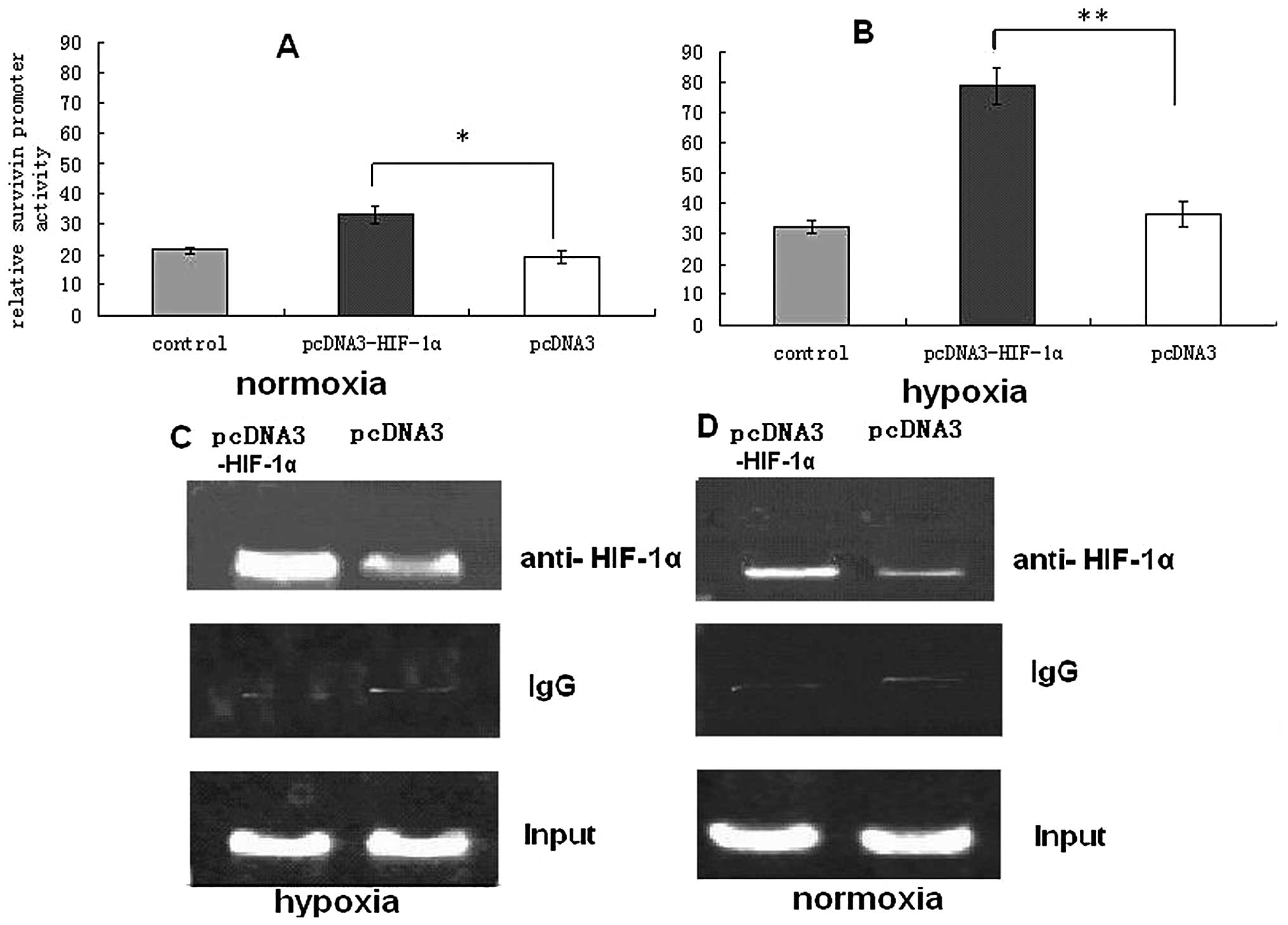
Effect of HIF-lα expression on survivin
promoter activity in A549 cells
To determine if survivin overexpression is mediated
by HIF-1α transcriptional activity, we first performed
cotransfection experiments under normoxic and hypoxic conditions.
Transfection with expression vector pcDNA3-HIF-1α vs. a control led
to a 3–4-fold induction of survivin promoter activity
following cotransfection under hypoxic conditions, which suggests
transcriptional regulation of survivin by HIF-1α under hypoxic
conditions (Fig. 2B). Although
A549 cells transfected with pcDNA3-HIF-1α significantly increased
survivin promoter activity under normoxic and hypoxic
conditions, compared with untreated cells and those transfected
with pcDNA3, survivin promoter activity under hypoxic conditions
was significantly higher than under normoxic conditions (Fig. 2B). Our previous study detected a
putative binding site for HIF-1α, located at −16 to
−19 bp in the proximal promoter region of the human
survivin gene (14). To
show HIF-1α binds to the survivin promoter in living cells,
we performed a ChIP assay in A549 cells under normoxia and hypoxia.
In the chromatin fraction pulled down by an anti-HIF-1α antibody,
we detected higher expression of survivin promoter PCR
fragments in pcDNA3-HIF-1α-transfected cells than in
pcDNA3-transfected cells under both normoxia and hypoxia (Fig. 2C and D). However, survivin
promoter PCR fragments were not found in samples pulled down by a
control IgG antibody. This further indicated that HIF-1α affects
survivin transcription by direct binding to an HIF-1α site
in the survivin core promoter.
Effect of HIF-lα-miRNA on A549 cell
proliferation
To verify if specific blockade of HIF-lα inhibits
cell proliferation under normoxia and hypoxia, we assayed cell
viability in each group of cells transfected with HIF-lα miRNA,
Scrambled miRNA (control) or untreated (PBS), for 24, 48 and 72 h.
Specific blockade of HIF-lα by miRNA inhibited proliferation at 24
h in A549 cells under hypoxia; following treatment with 150 μmol/l
CoCl2 for 48 and 72 h, the difference remained (Fig. 3B). Conversely, effects of
HIF-1α-miRNA on proliferation did not statistically differ between
Scrambled and untreated groups under normoxia (Fig. 3A). These results indicate HIF-lα
only promotes lung cancer proliferation under hypoxia, whereas
survivin expression vector transfection in HIF-lα knockdown cells
partly revived proliferation under hypoxia (Fig. 3).
Effect of HIF-lα-miRNA on A549 cell
apoptosis
Apoptosis was analyzed by flow cytometry under both
hypoxic and normoxic conditions. The apoptosis ratio of
miRNA-transfected cells in hypoxic conditions was 22.34±3.27%,
which was significantly higher than in untreated and Scrambled
cells (Fig. 4). However, under
normoxia, apoptosis rates in the HIF-1α-miRNA group did not
statistically differ from those of Scrambled and untreated cells
(data not shown). Furthermore, survivin expression vector
transfection in HIF-lα knockdown cells rescued the apoptotic
phenotype under hypoxic but not under normoxic conditions (Fig. 4).
Effect of HIF-lα-miRNA on A549 cell
invasion
To evaluate the anti-invasive effect of miRNA on
A549 cells under normoxic and hypoxic conditions, we used a
Transwell assay. Representative micrographs of Transwell filters
are shown in Fig. 5. A549 cells
transfected with miRNA constructs were significantly less invasive
under normoxic and hypoxic conditions, compared with untreated and
Scrambled cells (Fig. 5).
Meanwhile, the number of invasive cells under hypoxic conditions
was significantly higher than under normoxic conditions (Fig. 5). The result suggests that human
HIF-lα expression knocked down by miRNA can significantly reduce
A549 cell invasion under normoxic and hypoxic conditions, but
re-expression of survivin by transfection of expression vectors in
the HIF-lα knockdown cells does not promote invasive activity
regardless of hypoxic or normoxic conditions (Fig. 5).
Antitumor effect of HIF-lα-miRNA on an
A549 cell xenograft model
To further study the antitumor effect of
HIF-lα-miRNA on A549 cells in vivo, we used an A549
xenograft model and lipofectamine-mediated gene therapy as
indicated in Materials and methods. Tumors were established
subcutaneously in the axillary cavities of 24 mice by inoculating
cultured cells in the 3 groups. The tumor formation rate in nude
mice was 100%. Following inoculation, nodules could be felt
subcutaneously in the control and Scrambled groups at 4–5 days, but
not until 6–7 days in the HIF-1α-miRNA group. The standard (~50
mm3) occurred after 10 days; volume of each tumor was
measured by sliding calipers every 3 days. HIF-lα gene
silencing resulted in statistically significant reduction of tumor
volumes compared with the untreated and the Scrambled groups
(P<0.01; Fig. 6A). After mice
were observed for 58 days, tumor samples were excised and weighed.
Tumor weight in the HIF-1α-miRNA group was 1.14±0.08 g,
significantly lower than that in the untreated (1.71±0.18 g) and
the Scrambled group (1.75±0.26 g) (Fig. 6B, P<0.01), but differences
between the untreated and Scrambled groups were not significant
(P>0.05).
RT-PCR was used to detect expression of
HIF-1α mRNA and survivin mRNA in tumor tissues
(Fig. 7A). Where expression of
HIF-1α was knocked down by miRNA, survivin expression was
significantly lower than in the Scrambled and untreated groups.
Western blot results were consistent with the PCR results (Fig. 7B). TUNEL staining showed that
apoptosis was prominently increased in the HIF-1α-miRNA group
compared with the untreated and Scrambled groups (Fig. 8; P<0.01), Thus, these data show
that silencing HIF-1α expression by miRNA significantly inhibits
expression of HIF-1α mRNA and protein, and suppresses growth of
human pulmonary adenocarcinoma in tumor-bearing nude mice.
Decreased survivin expression is responsible for these results.
Nude mice in the HIF-1α miRNA group did not differ in body weight
gain, feed uptake or locomotive activity from the other groups. No
deaths occurred in any groups.
Discussion
The data presented in this study clearly indicate
that HIF-1α mediates survivin expression in vitro and in
vivo. First, assays revealed that inhibition of HIF-1α by miRNA
in A549 cells led to decreased survivin expression under normoxia
and hypoxia. We next showed that HIF-1α activated the
survivin promoter by direct interaction with binding sites
in the promoter region. In addition, HIF-1α-miRNA induced cell
apoptosis and inhibited cell proliferation in A549 cells under
hypoxic, but not normoxic, conditions. Cell migration was
substantially suppressed by HIF-1α silencing both under normoxia
and hypoxia. Transfection of survivin expression vectors in HIF-lα
knockdown cells partly rescued the apoptotic phenotype and cell
proliferation under hypoxic conditions. By contrast, expression
vectors had only slight effect on cell migration. Finally, we
confirmed that silencing HIF-1α expression downregulates survivin
expression in lung cancer xenografts.
Previous studies have shown that survivin
promoter activity is significantly increased in tumor cells
(16,17). This suggests that survivin
expression is transcriptionally regulated. Our recent data
suggested that Sp1 strongly affects upregulation of survivin in
lung cancer cells at the transcriptional level (18). However, how survivin
transcription is regulated by other, possibly cis-acting elements
is unclear. Notably, a putative HIF-1α binding site lies within the
survivin core promoter (19), as
confirmed by our previous results, which found site-directed
mutagenesis of the HIF-1α binding site reduced survivin
transcriptional activity by 36.60% (14). The mechanism by which HIF-1α
activates survivin expression is unclear. Survivin levels are also
strongly upregulated in A549 cells by hypoxia compared with
normoxia, as described in our previous study (15,20). This could be explained by the
involvement of HIF-1α, (a member of the basic helix-loop-helix-PAS
protein family) which is induced predominantly by hypoxia and
subsequently translocates into the nucleus where it dimerizes with
HIF-1β, consequently regulating a series of gene expression events
critical for cellular function under hypoxic conditions (21).
Our study confirmed that HIF-1α and survivin are
co-overexpressed in the lung cancer cell line A549. This finding is
consistent with studies that show positive rate of HIF-lα is 58.33%
and positive rate of survivin is 81.60% in lung cancer tissue, and
their expressions correlate with one another (14), indicating that HIF-1α regulates
survivin expression. Thus, we tested the impact of HIF-1α on
survivin expression in lung cancer cells. As anticipated, our data
showed that the silencing of HIF-1α by miRNA inhibited
survivin expression in A549 cells under hypoxic conditions, which
is in accordance with another study showing HIF-1α siRNA to
block EGF-induced survivin upregulation and to increase apoptosis
induced by docetaxel in breast cancer cell lines under normoxia
(19). However, our earlier study
did not show this effect by transient transfection of HIF-1α
siRNA in A549 under normoxic conditions. We suspect that different
tumor cells and different stimuli may result in HIF-1α showing
different effects on survivin gene expression under
normoxia.
To further investigate the mechanism by which HIF-1α
regulates survivin expression, we performed cotransfection
experiments under normoxic and hypoxic conditions to test the
effect of HIF-1α on survivin promoter activity. The
survivin promoter was markedly activated in A549 cells
transfected with pcDNA3-HIF-1α under hypoxic conditions, but was
only slightly activated under normoxic conditions, suggesting
HIF-1α upregulates survivin expression at the transcription level
under hypoxic conditions. Our previous study detected a putative
binding site for HIF-1α, located at −16 to
−19 bp in the proximal promoter region of the human
survivin gene (14). In
light of this, we used a ChIP assay to determine if HIF-1α can
directly bind to the above survivin promoter region binding
sites, indicating that HIF-1α exerts its effect on the
survivin promoter by direct interaction, consistent with our
previous electrophoretic mobility shift assay (EMSA), which
indicated that nuclear extracts of A549 could bind to the
r-32P-labeled 18-bp probe (nucleotides −26 to
−9 of the survivin core promoter) which includes
binding sites for HIF-1α (22).
The mechanism for HIF-1α-mediated transcriptional activation of the
survivin gene is currently under investigation.
Our laboratory recently demonstrated that HIF-1α
cooperated with Notch-1 signaling to increase survivin expression
through its direct association with N1ICD, thus accelerating
survivin transcription (20).
Understanding the molecular mechanism is crucial and urgent for the
development of new and improved therapeutic strategies for
NSCLC.
Previous findings suggest that survivin is critical
to both the initiation of cell proliferation and the inhibition of
apoptosis in lung cancer cells. We tested the downstream effects of
HIF-1α miRNA on cell growth and apoptosis. Our study showed
miRNA-mediated downregulation of HIF-1α expression in A549 cells
resulted in significant decline in cell proliferation and increased
spontaneous apoptosis under hypoxic conditions. However, these
changes did not occur with HIF-1α miRNA under normoxic conditions.
This finding is consistent with studies showing HIF-1α to exert
anti-apoptotic effects in human umbilical vascular endothelial
cells (23), cardiomyocytes
(24) and breast cells (19).
Although other studies support our results, Luo
et al had conflicting observations suggesting that
HIF-1α siRNA inhibited A549 cell apoptosis by involving the
glycolysis pathway (25).
Compared with siRNAs used by Luo et al, we consider that
miRNA used in our experiment silences target genes in
vector-infected cells more effectively (26,27). Moreover, in this experiment we
designed two other miRNA sequences to confirm that our results were
not caused by an off-target effect (data not shown). Also, compared
with their study which only utilized transient transfection with
HIF-1α siRNA plasmids, we adopted both transient
transfection and stable transfection methods to ensure silencing
effects in the previous and present experiments. Significantly,
whereas Luo et al did not further investigate the role of
HIF-1α in apoptosis in vivo, we confirmed that silencing
HIF-1α gene expression using miRNA can increase apoptosis in
nude mice, which has not previously been reported.
To further investigate the effect of survivin on
apoptosis induction and cell proliferation inhibition by
HIF-1α-miRNA in A549 cells under hypoxic conditions, we transfected
survivin expression vectors into HIF-lα-knockdown cells.
Re-expression of survivin in the HIF-lα knockdown cells partly
revived cell proliferation and rescued the apoptotic phenotype
under hypoxic conditions (Fig.
3), indicating that upregulation of survivin is a cause of the
protective effects exerted by HIF-1α in A549. Our results also
suggested that gene silencing does not affect cell proliferation
and apoptosis under normoxia. These may be due to the lower HIF-1α
expression under normoxia (which is inadequate to activate
survivin), or the dynamic balance between apoptosis and
anti-apoptosis signaling pathways regulated by HIF-1α. Our
hypotheses are supported by our previous study that expression of
HIF-1α and survivin is increased significantly in hypoxia compared
with normoxia (15).
Of note, HIF-1α miRNA inhibits A549 cell migration
under both normoxia and hypoxia. Furthermore, more cells migrated
under hypoxia than under normoxia (data not shown). This result is
consistent with that of Shyu et al(28), which suggests that HIF-1α
overexpression promotes migration of lung cancer cells. However,
survivin re-expression in HIF-lα knockdown cells does not promote
invasive activity in either hypoxic or normoxic conditions,
suggesting that survivin is not related to the effect of HIF-lα on
migration. The mechanism by which HIF-1α induces migration warrants
further study; establishing a clear link between HIF-1α and
survivin in A549 cells could provide new information on the
mechanisms by which HIF-1α promotes tumor growth.
We thus evaluated, for the first time, whether in
vitro effects can be obtained in vivo in nude mice
bearing A549 cells. We used cells transfected with
Scrambled-sequence plasmid and eukaryotic expression plasmid to
construct transplanted tumors, indicating that survivin
downregulation, tumor inhibition and apoptosis induced by HIF-1α
miRNA are in accordance with in vitro data. Furthermore,
animals in this study presented no mortality from the treatments.
Notably, HIF-1α miRNA used in our experiments inhibited tumor
growth more effectively than survivin RNA interference, as shown in
a previous study (29).
We speculate that HIF-1α also mediates tumor
progression by a survivin-independent mechanism; this is supported
by evidence that HIF-1α regulates expression of approximately 40
genes, such as angiogenic factors, glucose transporters, glycolytic
enzymes, survival and invasion factors, which may be critical for
tumor progression (30). The
benefits of elucidating the HIF-1α pathway in tumorigenesis may
lead to development of novel approaches for the prevention of tumor
progression and for lung cancer therapies. Long-term effects of
HIF-1α miRNA therapy are currently unknown and require further
investigation. Our findings in vivo, therefore, both
corroborate a possible mechanism for upregulated survivin
expression in A549, and provide a basis to target the HIF-1α
pathway as a lung cancer therapy.
In conclusion, these results show that silencing
HIF-1α gene expression using miRNA can increase apoptosis
and suppress growth of A549 cells by inhibiting expression of
survivin in vitro and in vivo. This suggests that
HIF-1α is an important transcription factor involved in the
regulation of survivin expression.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (no. 30772532).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Vermeulen K, Van Bockstaele DR and
Berneman ZN: Apoptosis: mechanisms and relevance in cancer. Ann
Hematol. 84:627–639. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li F and Ling X: Survivin study: an update
of ‘what is the next wave’? J Cell Physiol. 208:476–486. 2006.
|
|
4
|
Altieri DC: The molecular basis and
potential role of survivin in cancer diagnosis and therapy. Trends
Mol Med. 7:542–547. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rodel F, Hoffmann J, Distel L, et al:
Survivin as a radioresistance factor, and prognostic and
therapeutic target for radiotherapy in rectal cancer. Cancer Res.
65:4881–4887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Altieri DC: Validating survivin as a
cancer therapeutic target. Nat Rev Cancer. 3:46–54. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Knowles HJ and Harris AL: Hypoxia and
oxidative stress in breast cancer. Hypoxia and tumourigenesis.
Breast Cancer Res. 3:318–322. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harrison L and Blackwell K: Hypoxia and
anemia: factors in decreased sensitivity to radiation therapy and
chemotherapy. Oncologist. 9:31–40. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Denko NC: Hypoxia, HIF1 and glucose
metabolism in the solid tumour. Nat Rev Cancer. 8:705–713. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ogawa K, Chiba I, Morioka T, et al:
Clinical significance of HIF-1α expression in patients with
esophageal cancer treated with concurrent chemoradiotherapy.
Anticancer Res. 31:2351–2359. 2011.
|
|
11
|
Hochachka PW, Buck LT, Doll CJ and Land
SC: Unifying theory of hypoxia tolerance: molecular/metabolic
defense and rescue mechanisms for surviving oxygen lack. Proc Natl
Acad Sci USA. 93:9493–9498. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Semenza GL: Regulation of mammalian
O2homeostasis by hypoxia-inducible factor 1. Annu Rev
Cell Dev Biol. 15:551–578. 1999.
|
|
13
|
Jiang CQ, Fan LF, Liu ZS, et al:
Expression levels and significance of hypoxia inducible factor-1
alpha and vascular endothelial growth factor in human colorectal
adenocarcinoma. Chin Med J. 117:1541–1546. 2004.
|
|
14
|
Chen YQ, Zhao CL and Li W: Effect of
hypoxia-inducible factor-1alpha on transcription of survivin in
non-small cell lung cancer. J Exp Clin Cancer Res. 28:292009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang L, Cao Z, Li F, et al: Tumor-specific
gene expression using the survivin promoter is further increased by
hypoxia. Gene Ther. 11:1215–1223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Estève PO, Chin HG and Pradhan S:
Molecular mechanisms of transactivation and doxorubicin-mediated
repression of survivin gene in cancer cells. J Biol Chem.
282:2615–2625. 2007.PubMed/NCBI
|
|
17
|
Kawamura K, Yu L, Tomizawa M, et al:
Transcriptional regulatory regions of the survivin gene
activate an exogenous suicide gene in human tumors and enhance the
sensitivity to a prodrug. Anticancer Res. 27:89–93. 2007.PubMed/NCBI
|
|
18
|
Chen Y, Wang X, Li W, et al: Sp1
upregulates survivin expression in adenocarcinoma of lung cell line
A549. Anat Rec. 294:774–780. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peng XH, Karna P, Cao Z, Jiang BH, Zhou M
and Yang L: Cross-talk between epidermal growth factor receptor and
hypoxia-inducible factor-1α signal pathways increases resistance to
apoptosis by up-regulating survivin gene expression. J Biol Chem.
281:25903–25914. 2006.
|
|
20
|
Chen Y, Li D, Liu H, et al: Notch-1
signaling facilitates survivin expression in human non-small cell
lung cancer cells. Cancer Biol Ther. 11:14–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Semenza GL, Agani F, Feldser D, et al:
Hypoxia, HIF-1, and the pathophysiology of common human diseases.
Adv Exp Med Biol. 475:123–130. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li W, Chen YQ, Sun Y, Zhao CL and Wang XJ:
Regulation of survivin expression by hypoxia-inducible factor-1α in
non-small cell lung cancer. Chin Oncol. 21:567–574. 2011.
|
|
23
|
Yu EZ, Li YY, Liu XH, Kagan E and McCarron
RM: Antiapoptotic action of hypoxia-inducible factor-1α in human
endothelial cells. Lab Invest. 84:553–561. 2004.
|
|
24
|
Malhotra R and Brosius FC: Glucose uptake
and glycolysis reduce hypoxia-induced apoptosis in cultured
neonatal rat cardiac myocytes. J Biol Chem. 274:12567–12575. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo F, Liu X, Yan N, et al:
Hypoxia-inducible transcription factor-1α promotes hypoxia-induced
A549 apoptosis via a mechanism that involves the glycolysis
pathway. BMC Cancer. 6:262006.
|
|
26
|
Nakahara K and Carthew RW: Expanding roles
for miRNAs and siRNAs in cell regulation. Curr Opin Cell Biol.
16:127–133. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shyu KG, Hsu FL, Wang MJ, Wang BW and Lin
S: Hypoxia-inducible factor 1α regulates lung adenocarcinoma cell
invasion. Exp Cell Res. 313:1181–1191. 2007.
|
|
29
|
Liu GF, Zhao QG, Si L, Cao YG, Li GY and
Wang LX: Effects of survivin interference RNA on non-small cell
lung carcinoma. Clin Invest Med. 32:E2252009.PubMed/NCBI
|
|
30
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar
|