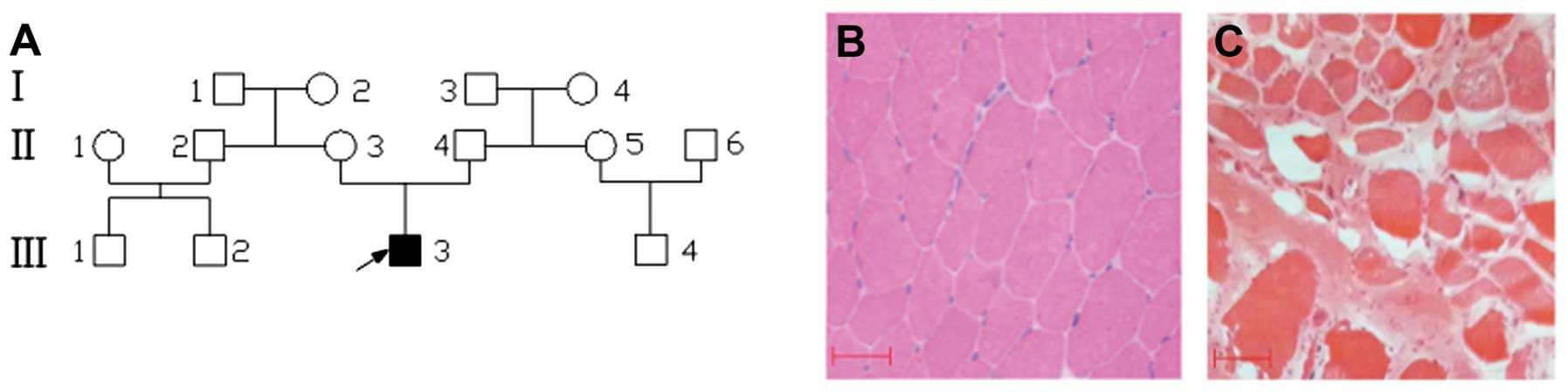
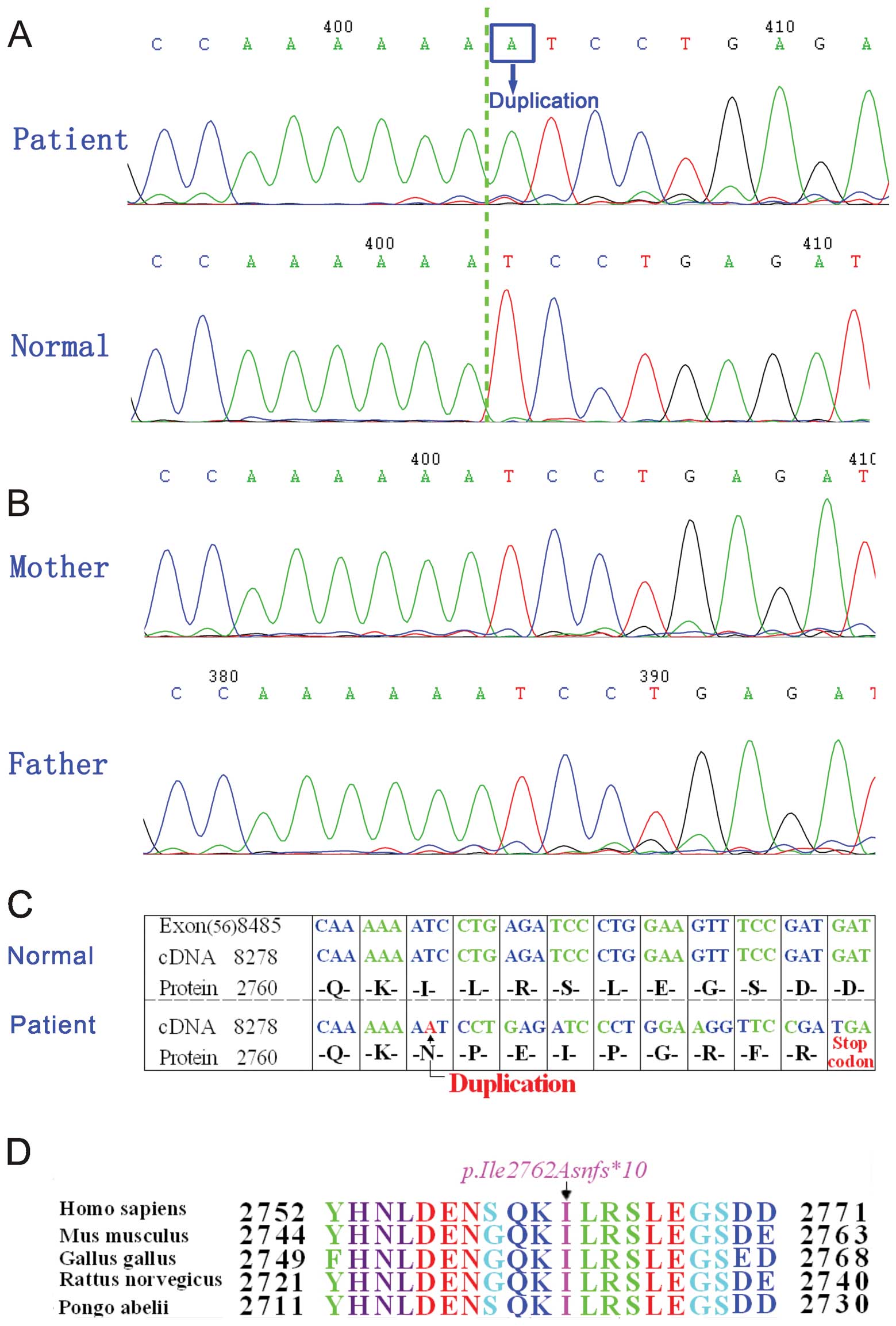
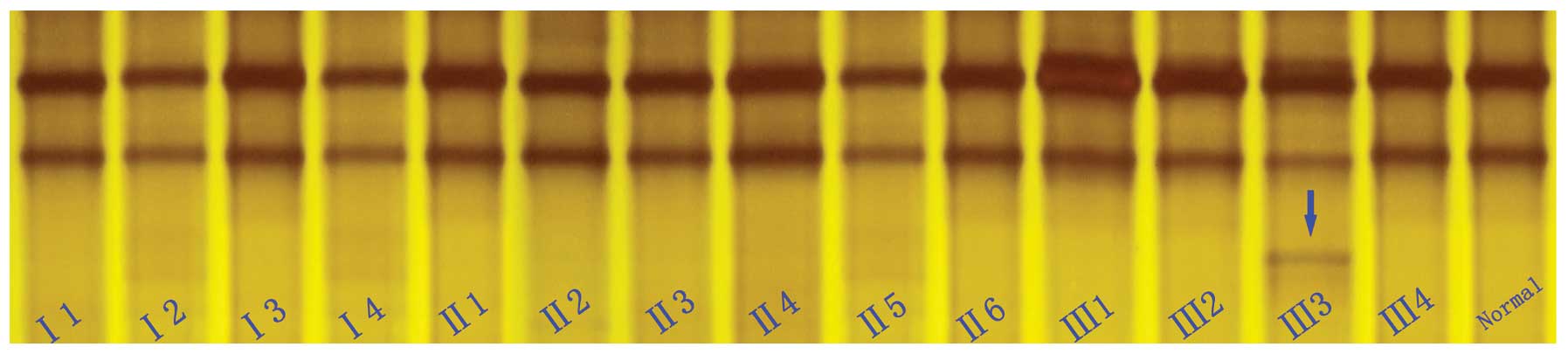
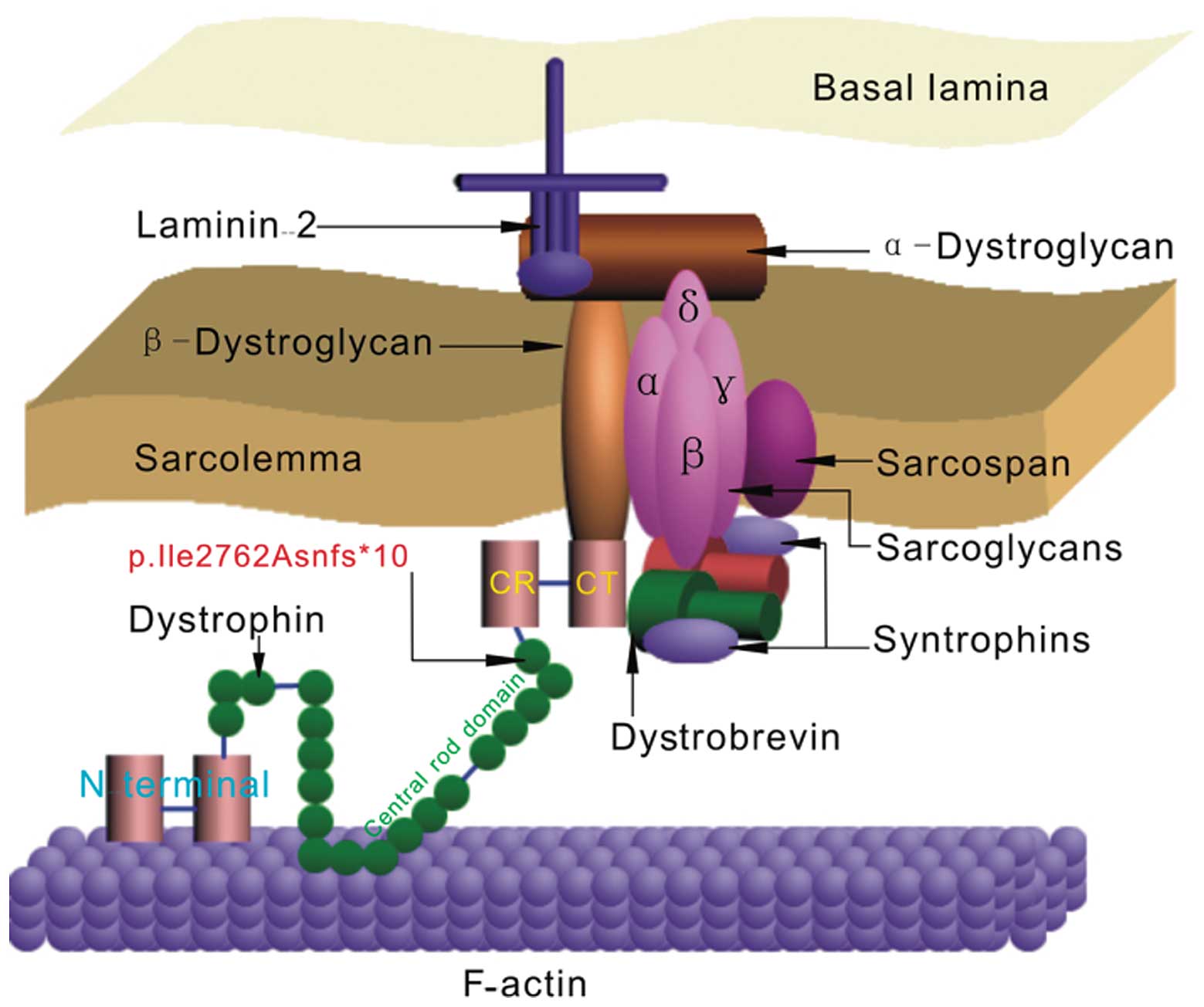
|
1
|
Emery AE: The muscular dystrophies.
Lancet. 359:687–695. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoffman EP, Brown RH and Kunkel LM:
Dystrophin: The protein product of the Duchenne Muscular Dystrophy
locus. Cell. 51:919–928. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Emery AE: Population frequencies of
inherited neuromuscular disorders - a world survey. Neuromuscul
Disord. 1:19–29. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blake DJ, Weir A, Newey SE and Davies KE:
Function and genetics of dystrophin and dystrophin-related proteins
in muscle. Physiol Rev. 82:291–329. 2002.PubMed/NCBI
|
|
5
|
van Essen AJ, Kneppers AL, van der Hout
AH, et al: The clinical and molecular genetic approach to Duchenne
and Becker muscular dystrophy: an updated protocol. J Med Genet.
34:805–812. 1997.PubMed/NCBI
|
|
6
|
Sura T, Eu-ahsunthornwattana J,
Pingsuthiwong S and Busabaratana M: Sensitivity and frequencies of
dystrophin gene mutations in Thai DMD/BMD patients as detected by
multiplex PCR. Dis Markers. 25:115–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Batchelor CL and Winder SJ: Sparks,
signals and shock absorbers: How dystrophin loss causes muscular
dystrophy. Trends Cell Biol. 16:198–205. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Koenig M, Hoffman EP, Bertelson CJ, et al:
Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and
preliminary genomic organization of the DMD gene in normal and
affected individuals. Cell. 50:509–517. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koenig M, Monaco AP and Kunkel LM: The
complete sequence of dystrophin predicts a rod-shaped cytoskeleton
protein. Cell. 53:219–228. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ahn AH and Kunkel LM: The structural and
functional diversity of dystrophin. Nat Genet. 3:283–291. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nishio H, Takeshima Y, Narita N, et al:
Identification of a novel first exon in the human dystrophin gene
and of a new promoter located more than 500 kb upstream of the
nearest known promoter. J Clin Invest. 94:1037–1042. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Magri F, Del Bo R, D’Angelo MG, et al:
Clinical and molecular characterization of a cohort of patients
with novel nucleotide alterations of the dystrophin gene detected
by direct sequencing. BMC Med Genet. 12:372011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dent KM, Dunn DM, von Niederhausern AC, et
al: Improved molecular diagnosis of dystrophinopathies in an
unselected clinical cohort. Am J Med Genet A. 134:295–298. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tuffery-Giraud S, Chambert S, Demaille J
and Claustres M: Point mutations in the dystrophin gene: evidence
for frequent use of cryptic splice sites as a result of splicing
defects. Hum Mutat. 14:359–368. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Q, Li-Ling J, Lin C, et al:
Characteristics of dystrophin gene mutations among Chinese patients
as revealed by multiplex ligation-dependent probe amplification.
Genet Test Mol Biomarkers. 13:23–30. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cai F, Zhu J, Chen W, et al: A novel PAX6
mutation in a large Chinese family with aniridia and congenital
cataract. Mol Vis. 16:1141–1145. 2010.PubMed/NCBI
|
|
17
|
Wang Q, Shen J, Splawski I, et al: SCN5A
mutations associated with an inherited cardiac arrhythmia, long QT
syndrome. Cell. 80:805–811. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sadoulet-Puccio HM and Kunkel LM:
Dystrophin and its isoforms. Brain Pathol. 6:25–35. 1996.
View Article : Google Scholar
|
|
19
|
Roberts RG: Dystrophins and dystrobrevins.
Genome Biol. 2:REVIEWS30062001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Davies KE, Tinsley JM and Blake DJ:
Molecular analysis of Duchenne muscular dystrophy: past, present,
and future. Ann NY Acad Sci. 758:287–296. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ozawa E: Our trails and trials in the
subsarcolemmal cytoskeleton network and muscular dystrophy
researches in the dystrophin era. Proc Jpn Acad Ser B Phys Biol
Sci. 86:798–821. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Wang W, Li R, et al: The diploid
genome sequence of an Asian individual. Nature. 456:60–65. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nishida A, Kataoka N, Takeshima Y, et al:
Chemical treatment enhances skipping of a mutated exon in the
dystrophin gene. Nat Commun. 2:3082011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hall N: Advanced sequencing technologies
and their wider impact in microbiology. J Exp Biol. 210:1518–1525.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Singh SM, Kongari N, Cabello-Villegas J
and Mallela KM: Missense mutations in dystrophin that trigger
muscular dystrophy decrease protein stability and lead to
cross-beta aggregates. Proc Natl Acad Sci USA. 107:15069–15074.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ishikawa-Sakurai M, Yoshida M, Imamura M,
et al: ZZ domain is essentially required for the physiological
binding of dystrophin and utrophin to beta-dystroglycan. Hum Mol
Genet. 13:693–702. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kemaladewi DU, Hoogaars WM, van Heiningen
SH, et al: Dual exons kipping in myostatin and dystrophin for
Duchenne muscular dystrophy. BMC Med Genomics. 4:362011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hwa HL, Chang YY, Huang CH, et al: Small
mutations of the DMD gene in Taiwanese families. J Formos Med
Assoc. 107:463–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sitnik R, Campiotto S, Vainzof M, et al:
Novel point mutations in the dystrophin gene. Hum Mutat.
10:217–222. 1997. View Article : Google Scholar : PubMed/NCBI
|