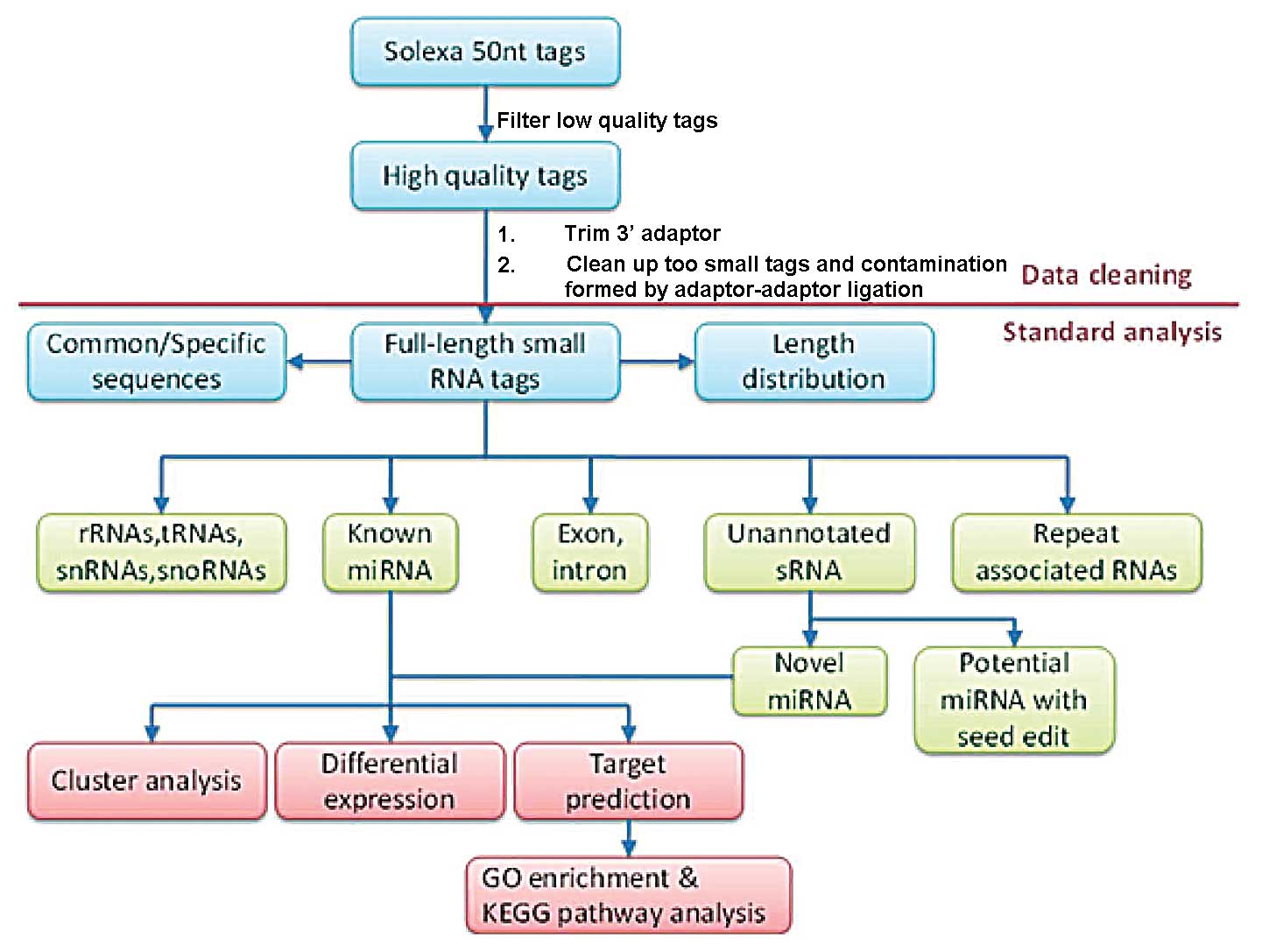
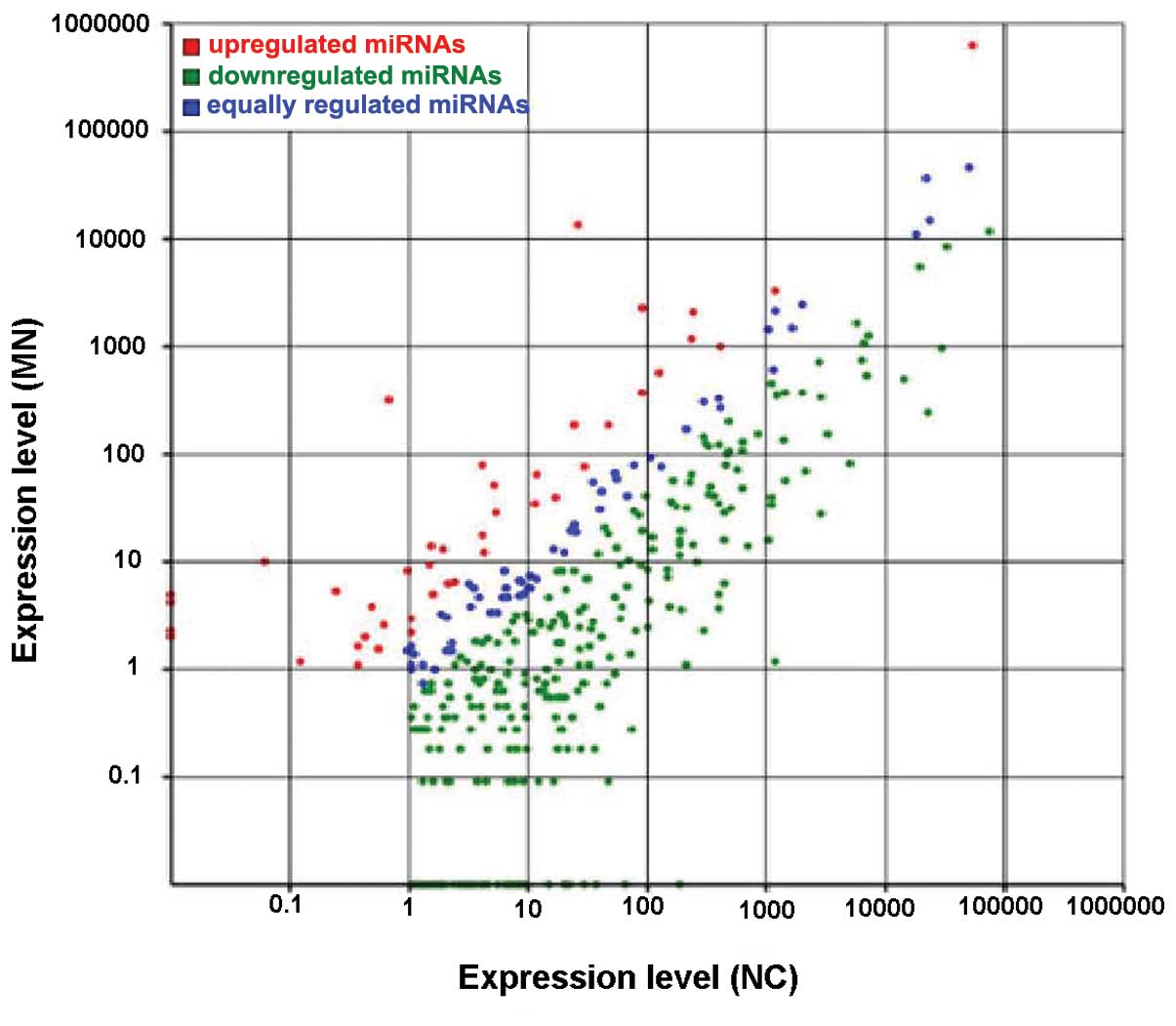
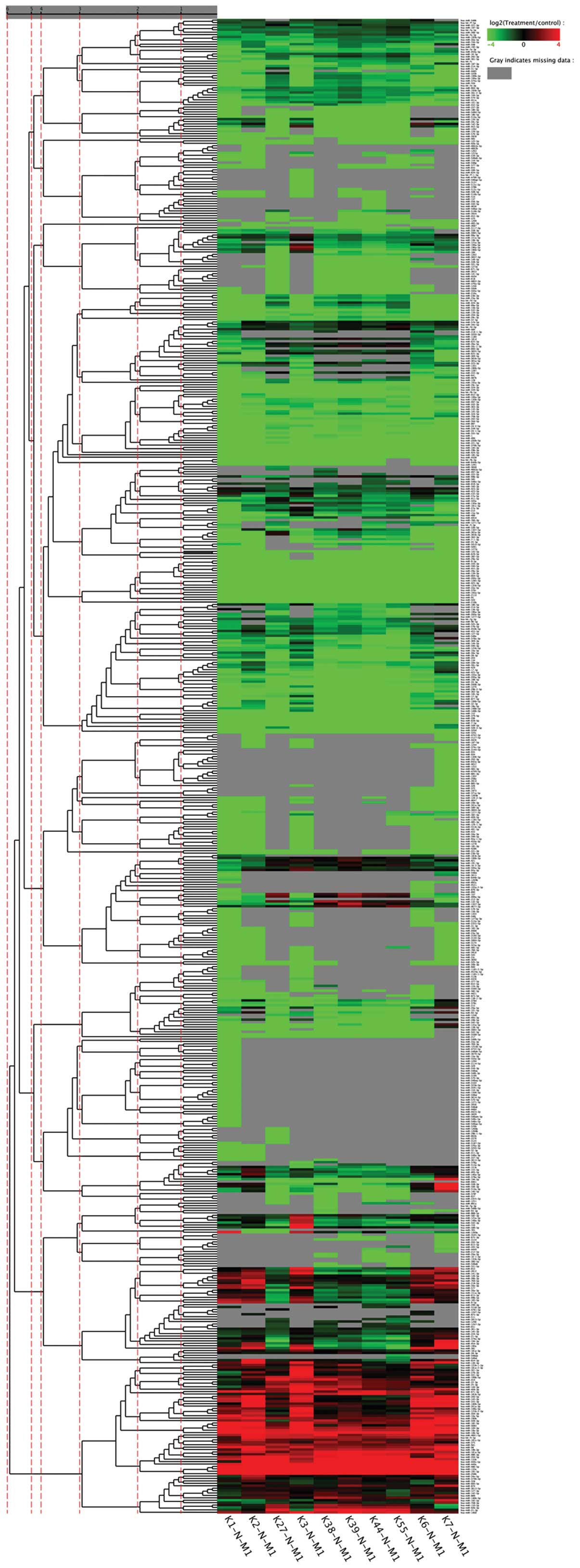
|
1
|
Ponticelli C and Passerini P: Can
prognostic factors assist therapeutic decisions in idiopathic
membranous nephropathy? J Nephrol. 23:156–163. 2010.PubMed/NCBI
|
|
2
|
Ponticelli C: Membranous nephropathy. J
Nephrol. 20:268–287. 2007.
|
|
3
|
Cybulsky AV, Quigg RJ and Salant DJ:
Experimental membranous nephropathy redux. Am J Physiol Renal
Physiol. 289:F660–F671. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shankland SJ: New insights into the
pathogenesis of membranous nephropathy. Kidney Int. 57:1204–1205.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Couser WG: Membranous nephropathy: a long
road but well traveled. J Am Soc Nephrol. 16:1184–1187. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fervenza FC, Sethi S and Specks U:
Idiopathic membranous nephropathy: diagnosis and treatment. Clin J
Am Soc Nephrol. 3:905–919. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Praga M and Rojas-Rivera J: Glomerular
disease: predicting outcomes in idiopathic membranous nephropathy.
Nat Rev Nephrol. 8:496–498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bataille S, Jourde N, Daniel L, et al:
Comparative safety and efficiency of five percutaneous kidney
biopsy approaches of native kidneys: a multicenter study. Am J
Nephrol. 35:387–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim VN and Nam JW: Genomics of microRNA.
Trends Genet. 22:165–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang B, Wang Q and Pan X: MicroRNAs and
their regulatory roles in animals and plants. J Cell Physiol.
210:279–289. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai Y, Sui W, Lan H, Yan Q, Huang H and
Huang Y: Comprehensive analysis of microRNA expression patterns in
renal biopsies of lupus nephritis patients. Rheumatol Int.
29:749–754. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saal S and Harvey SJ: MicroRNAs and the
kidney: coming of age. Curr Opin Nephrol Hypertens. 18:317–323.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li JY, Yong TY, Michael MZ and Gleadle JM:
Review: The role of microRNAs in kidney disease. Nephrology
(Carlton). 15:599–608. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morozova O and Marra MA: Applications of
next-generation sequencing technologies in functional genomics.
Genomics. 92:255–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li R, Li Y, Kristiansen K and Wang J:
SOAP: short oligonucleotide alignment program. Bioinformatics.
24:713–714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tino P: Basic properties and information
theory of Audic-Claverie statistic for analyzing cDNA arrays. BMC
Bioinformatics. 10:3102009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Minoche AE, Dohm JC and Himmelbauer H:
Evaluation of genomic high-throughput sequencing data generated on
Illumina HiSeq and genome analyzer systems. Genome Biol.
12:R1122011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huss M: Introduction into the analysis of
high-throughput-sequencing based epigenome data. Brief Bioinform.
11:512–523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang G, Kwan BC, Lai FM, Chow KM, Kam-Tao
Li P and Szeto CC: Expression of microRNAs in the urinary sediment
of patients with IgA nephropathy. Dis Markers. 28:79–86. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang G, Kwan BC, Lai FM, et al: Intrarenal
expression of miRNAs in patients with hypertensive nephrosclerosis.
Am J Hypertens. 23:78–84. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ichii O, Otsuka S, Sasaki N, Namiki Y,
Hashimoto Y and Kon Y: Altered expression of microRNA miR-146a
correlates with the development of chronic renal inflammation.
Kidney Int. 81:280–292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martinez NJ, Ow MC, Barrasa MI, et al: A
C. elegans genome-scale microRNA network contains composite
feedback motifs with high flux capacity. Genes Dev. 22:2535–2549.
2008.
|
|
26
|
Rajewsky N and Socci ND: Computational
identification of microRNA targets. Dev Biol. 267:529–535. 2004.
View Article : Google Scholar
|
|
27
|
Ulitsky I, Laurent LC and Shamir R:
Towards computational prediction of microRNA function and activity.
Nucleic Acids Res. 38:e1602010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang B, Stellwag EJ and Pan X:
Large-scale genome analysis reveals unique features of microRNAs.
Gene. 443:100–109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakagawa S, Niimura Y, Gojobori T, Tanaka
H and Miura K: Diversity of preferred nucleotide sequences around
the translation initiation codon in eukaryote genomes. Nucleic
Acids Res. 36:861–871. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Joshi CP, Zhou H, Huang X and Chiang VL:
Context sequences of translation initiation codon in plants. Plant
Mol Biol. 35:993–1001. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sui W, Dai Y, Huang Y, Lan H, Yan Q and
Huang H: Microarray analysis of MicroRNA expression in acute
rejection after renal transplantation. Transpl Immunol. 19:81–85.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sui W, Ou M, Chen J, et al: MicroRNA
expression profile of peripheral blood mononuclear cells of
Klinefelter syndrome. Exp Ther Med. 4:825–831. 2012.PubMed/NCBI
|
|
33
|
Patnaik SK, Yendamuri S, Kannisto E,
Kucharczuk JC, Singhal S and Vachani A: MicroRNA expression
profiles of whole blood in lung adenocarcinoma. PLoS One.
7:e460452012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dai Y, Sui W, Lan H, Yan Q, Huang H and
Huang Y: Microarray analysis of micro-ribonucleic acid expression
in primary immunoglobulin A nephropathy. Saudi Med J. 29:1388–1393.
2008.PubMed/NCBI
|
|
35
|
Chen YH, Wang SQ, Wu XL, et al:
Characterization of microRNAs expression profiling in one group of
Chinese urothelial cell carcinoma identified by Solexa sequencing.
Urol Oncol. 31:219–227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Osanto S, Qin Y, Buermans HP, et al:
Genome-wide microRNA expression analysis of clear cell renal cell
carcinoma by next generation deep sequencing. PLoS One.
7:e382982012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tian Z, Greene AS, Pietrusz JL, Matus IR
and Liang M: MicroRNA-target pairs in the rat kidney identified by
microRNA microarray, proteomic, and bioinformatic analysis. Genome
Res. 18:404–411. 2008. View Article : Google Scholar
|
|
38
|
Duisters RF, Tijsen AJ, Schroen B, et al:
miR-133 and miR-30 regulate connective tissue growth factor:
implications for a role of microRNAs in myocardial matrix
remodeling. Circ Res. 104:170–178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mezzano SA, Droguett MA, Burgos ME, et al:
Overexpression of chemokines, fibrogenic cytokines, and
myofibroblasts in human membranous nephropathy. Kidney Int.
57:147–158. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ho J, Ng KH, Rosen S, Dostal A, Gregory RI
and Kreidberg JA: Podocyte-specific loss of functional microRNAs
leads to rapid glomerular and tubular injury. J Am Soc Nephrol.
19:2069–2075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu G, Wu J, Zhou L, et al:
Characterization of the small RNA transcriptomes of androgen
dependent and independent prostate cancer cell line by deep
sequencing. PLoS One. 5:e155192010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Oulas A, Boutla A, Gkirtzou K, Reczko M,
Kalantidis K and Poirazi P: Prediction of novel microRNA genes in
cancer-associated genomic regions - a combined computational and
experimental approach. Nucleic Acids Res. 37:3276–3287. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dhahbi JM, Atamna H, Boffelli D, Magis W,
Spindler SR and Martin DI: Deep sequencing reveals novel microRNAs
and regulation of microRNA expression during cell senescence. PLoS
One. 6:e205092011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li G, Li Y, Li X, Ning X, Li M and Yang G:
MicroRNA identity and abundance in developing swine adipose tissue
as determined by Solexa sequencing. J Cell Biochem. 112:1318–1328.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Blow MJ, Grocock RJ, van Dongen S, et al:
RNA editing of human microRNAs. Genome Biol. 7:R272006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kawahara Y, Zinshteyn B, Sethupathy P,
Iizasa H, Hatzigeorgiou AG and Nishikura K: Redirection of
silencing targets by adenosine-to-inosine editing of miRNAs.
Science. 315:1137–1140. 2007. View Article : Google Scholar : PubMed/NCBI
|