Introduction
Human dermal fibroblasts (HDFs) are mesenchymal
cells specialized in extracellular matrix synthesis, including
collagen, elastin and hyaluronic acid, in the dermis (1,2).
Dermal fibroblast proliferation is important in wound healing and
skin structure homeostasis (3).
Therefore, a growth factor was used for proliferating HDFs in
cosmetics (4).
Epidermal growth factor (EGF), one of various growth
factors, is a small polypeptide first purified by Cohen from the
submaxillary gland of adult male mice (5). EGF is important in cell
proliferation, migration and differentiation (5). These roles are mediated via
activation of the EGF receptor (EGFR), a transmembrane glycoprotein
with tyrosine kinase activity (6). Activated EGFR regulates
proliferation and migration via activation of intrinsic signaling
molecules (7,8). EGF is added to serum-free media in
in vitro cell culture systems as EGF is essential to cell
growth (9). Furthermore, EGF
exerts cytoprotective effects from cell damage, such as senescence
(10). Therefore, EGF is used as
an inducer of dermal fibroblast proliferation in cosmetics and
medicine (4).
Phytosphingosine-1-phosphate (PhS1P) is derived from
fungi and plants, and is structurally similar to
sphingosine-1-phosphate (S1P), an endogenous signal lipid in
mammalian cells (11,12). PhS1P is an agonist for S1P
receptors, with a particularly high affinity for S1P4
(13). S1P receptors include the
isomers, S1P1, S1P2, S1P3 and
S1P4, in mammalian cells (13). Since each receptor activates
different downstream signals, the effects of these S1Ps are
slightly different (14,15). Therefore, we aimed to evaluate the
PhS1P function in HDFs. The results showed that PhS1P altered gene
expression and induced EGF-dependent proliferation as a synergistic
effector.
Materials and methods
Cell culture and materials
HDFs were purchased from Lonza (Basel, Switzerland)
and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco,
Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum (Sigma-Aldrich, St. Louis, MO, USA). The cells were
incubated at 37°C in a humidified incubator containing 5%
CO2. PhS1P was obtained from Phytos Co., Ltd. (Suwon,
Korea).
RNA extraction and microarray
Total RNA was isolated using RiboEX (GeneAll, Seoul,
Korea) and quantified based on the optical density ratio (280/260
nm) using a Bioanalyzer 2100 (28S RNA/18S RNA ratio; Agilent
Technologies, Santa Clara, CA, USA). Equal amounts of RNA were used
to synthesize cDNA and label it with biotin using an RNA
amplification kit (Ambion, Austin, TX, USA). After labeling, the
microarray was hybridized with biotin-labeled RNA and
streptavidin-Cy3 (Invitrogen Life Technologies, Carlsbad, CA, USA).
Following hybridization, the microarray was washed using wash E1BC
buffer and scanned using the iScan system (both from Illumina,
Hayward, CA, USA).
Microarray analyses
Microarray data were analyzed using Genespring GX
software version 11 (Agilent Technologies). mRNAs flagged ‘present’
in at least one sample were analyzed using fold-change. The
threshold cut-off was 1.3-fold for fold-change between non-treated
HDFs and PhS1P-treated HDFs. Significantly altered mRNAs were
sorted using the gene ontology (GO) tool.
Quantitative polymerase chain reaction
(qPCR)
cDNA was synthesized using MMLV-reverse
transcriptase (Invitrogen Life Technologies) according to the
manufacturer’s instructions. Synthesized cDNA was used for qPCR
(Line gene K; Bioer Technology, Co., Ltd., China) using specific
primers for cyclin A1, B1 and B2. Primers were designed by primer 3
(http://frodo.wi.mit.edu) (Table I). Expression was normalized to
β-actin.
 | Table IQuantitative PCR primer sequences. |
Table I
Quantitative PCR primer sequences.
| Gene | Primer sequences |
|---|
| Cyclin A2 | Forward
5′-TTATTGCTGGAGCTGCCTTT-3′ |
| Reverse
5′-CTCTGGTGGGTTGAGGAGAG-3′ |
| Cyclin B1 | Forward
5′-CGGGAAGTCACTGGAAACAT-3′ |
| Reverse
5′-AAACATGGCAGTGACACCAA-3′ |
| Cyclin B2 | Forward
5′-TTGCAGTCCATAAACCCACA-3′ |
| Reverse
5′-GAAGCCAAGAGCAGAGCAGT-3′ |
MTT assay
Cell viability was assessed using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. HDFs were cultured for 24 h in 96-well plates with PhS1P and
EGF. MTT tetrazolium salt (0.5 mg/ml; Sigma) was added to cells for
4 h. After incubation, the medium was replaced with dimethyl
sulfoxide in each well. The absorbance of each sample was measured
at 595 nm using a plate reader (Bio-Rad Laboratories, Hercules, CA,
USA).
Results
PhS1P cytotoxicity in HDFs
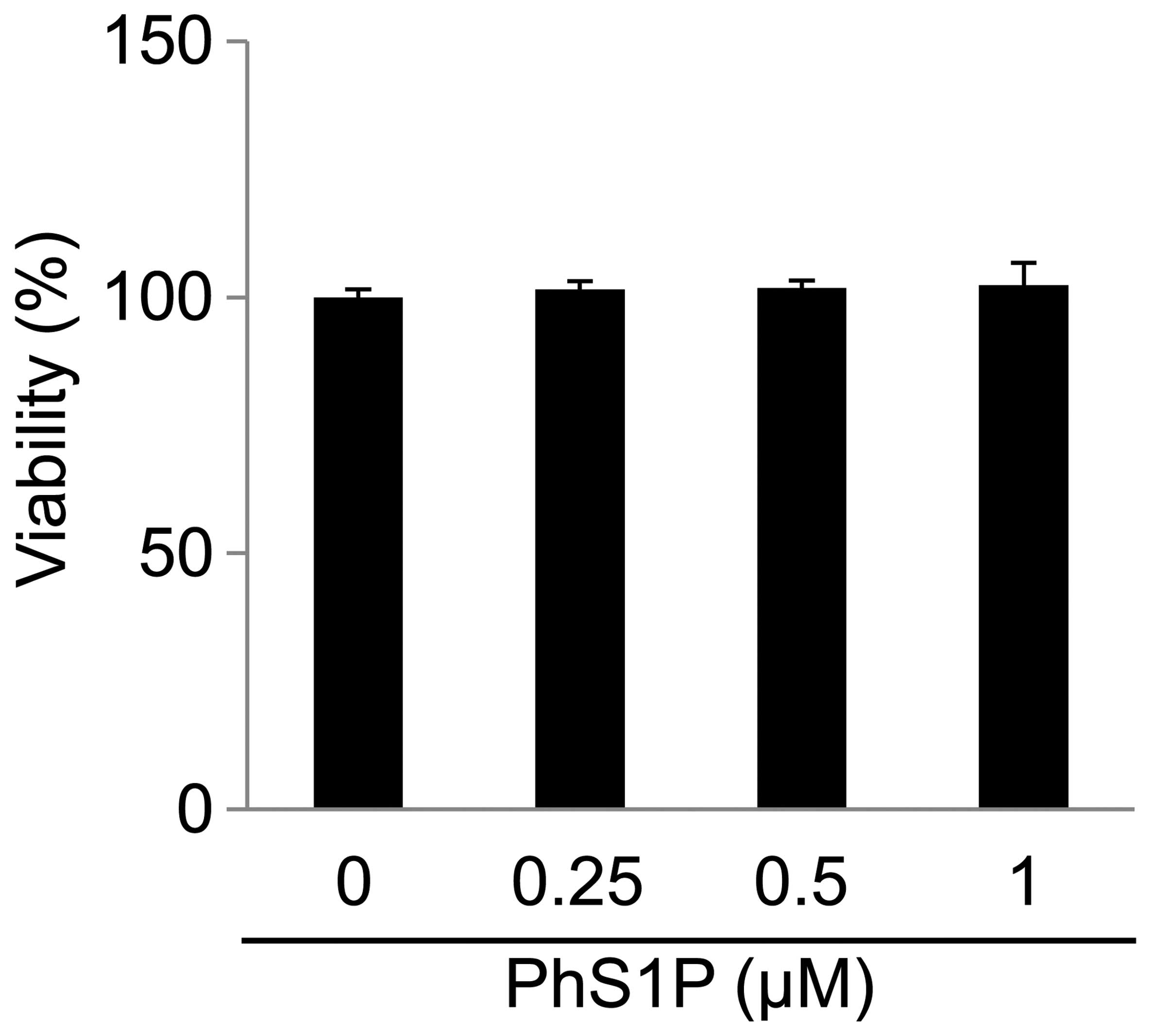
To assess the effect of PhS1P on cell viability,
HDFs were treated with various concentrations of PhS1P (0, 0.25,
0.5 and 1 μM) for 24 h (Fig. 1).
We determined that PhS1P at concentrations of ≤1 μM had no
cytotoxicity.
PhS1P alters the mRNA expression
profile
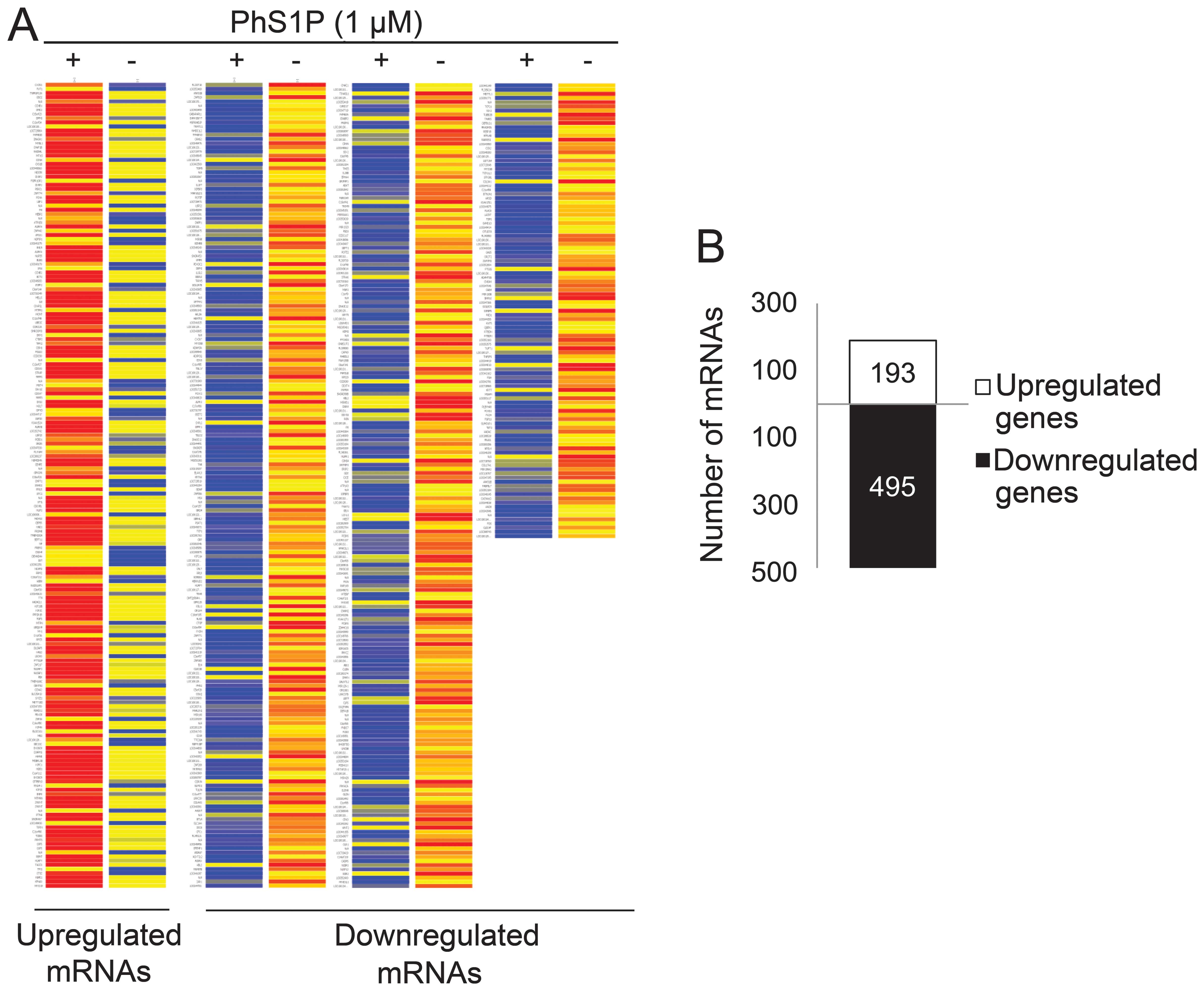
To identify genes that may play a PhS1P-dependent
role, we compared the expression profiles of non-treated and 1 μM
PhS1P-treated HDFs using the Illumina bead chip HumanHT-12. In
47,207 whole genes, we first filtered 35,973 genes that were
flagged ‘present’ with a frequency higher than the sensitivity of
detection in a minimum of one array. Fold-change was then analyzed
in these flag-filtered genes. Genes that changed by 1.3-fold
between non-treated and PhS1P-treated HDFs were presented on the
heat map (Fig. 2A). The analyses
identified 193 upregulated genes and 495 downregulated genes
(Fig. 2B).
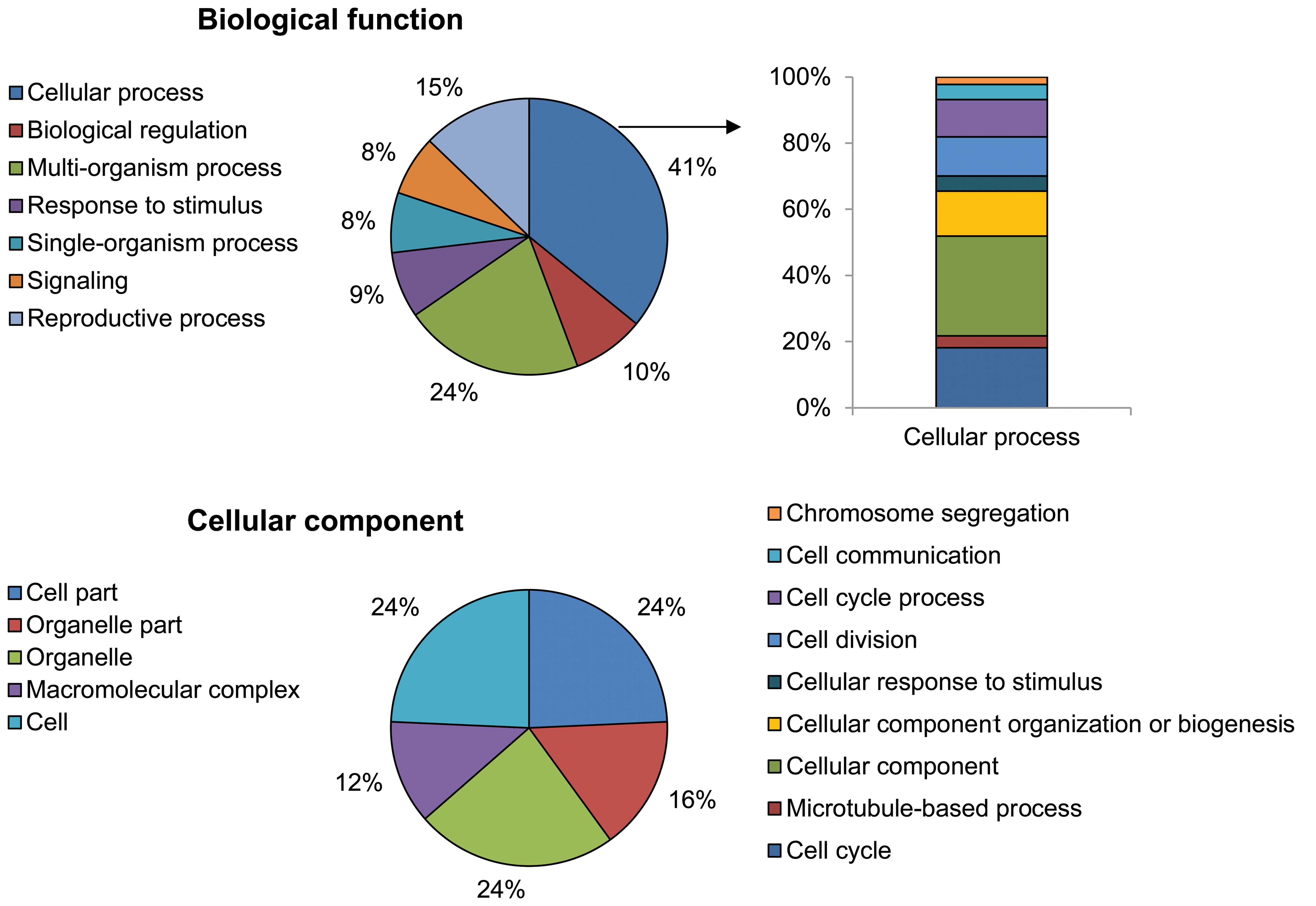
Using the GO analytical tool, the genes were sorted
based on their roles in biological processes (Fig. 3). Genes upregulated in
PhS1P-treated HDFs appeared to be involved in cell processes such
as cell cycle, cell division, microtubule-based processes,
chromosome segregation, cell communication, cellular responses to
stimuli, and cellular component organization or biogenesis. In
particular, cell cycle-related ontology (cell cycle, cell division
and chromosome segregation) was significantly enriched by
PhS1P.
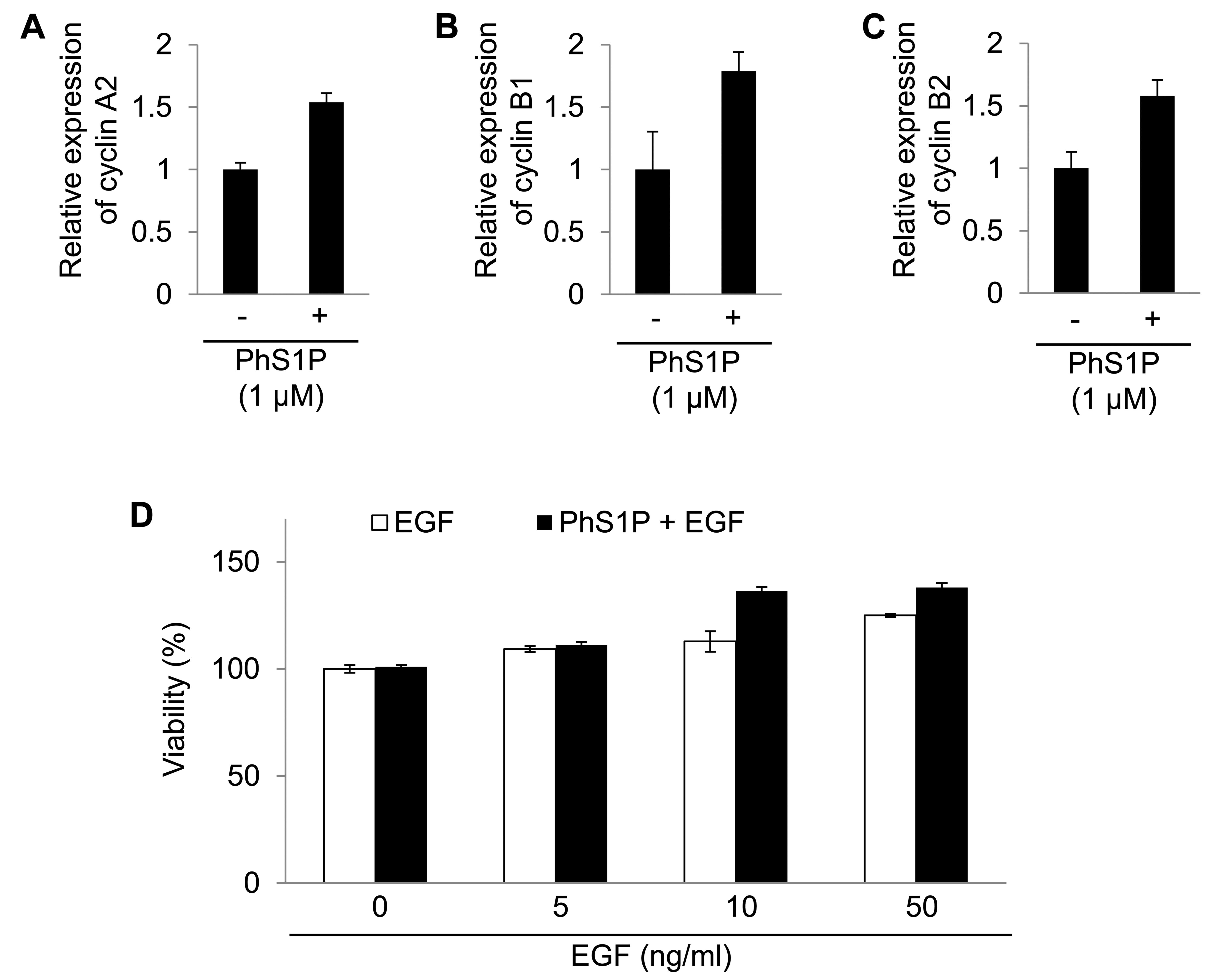
Cyclins A2, B1 and B2 were regulated by PhS1P.
Cyclins are well-known regulators in cells (16). Therefore, the analyses revealed
that PhS1P affected cell proliferation by altering specific mRNAs.
In addition, we confirmed the mRNA expression change of cyclin A2,
B1 and B2 using qPCR (Fig.
4).
We also examined the synergistic effect of PhS1P and
EGF on HDF viability. Compared with PhS1P-treated HDFs in the
absence of EGF, the viability of PhS1P-treated HDFs with EGF was
markedly increased (Fig. 4D).
These data indicate that PhS1P is a co-effector in the induction of
EGF-dependent cell proliferation.
Discussion
The aim of the study was to evaluate the effect of
PhS1P on EGF-induced proliferation in HDFs. PhS1P is
phytochemically derived from fungi, plants, and even mammalian
cells (12). In a previous study,
it was shown that PhS1P protects against hydrogen
peroxide-dependent growth arrest (17). Recent data from Lee et al
(17) suggested that PhS1P had no
significant effect on the proliferation at concentrations <1 μM
in HDFs (Fig. 1). However, PhS1P
(1 μM) regulates various cell cycle-related genes in HDFs (Fig. 2). In particular, the cyclins
(cyclin A2, B1 and B2), master regulators of the cell cycle, were
upregulated by PhS1P. Cyclin A2 regulates S-phase progression and
entry into mitosis (18,19). During S phase, cyclin A2 initiates
DNA synthesis (20). During the
G/M phase, cyclin A2 triggers entry into mitosis by activating
cyclin B1-Cdk1 (21).
In the progressing cell cycle, a sustained high
expression of cyclins is essential (16). However, overexpression of cyclins
is insufficient to induce cell cycle progression (22,23). As shown in Fig. 2, PhS1P upregulated mRNA expression
of cyclin A2, B1 and B2 although there was no change in cell
viability (Figs. 1 and 2).
Combining the present data with our previous data
(17), we determined that <1
μM PhS1P increases cyclin expression, but does not affect
viability. Moreover, EGF triggered proliferation in PhS1P-treated
HDFs (Fig. 4D). Therefore,
together with the results from Ikezawa et al (22), our data suggest that a
cyclin-enriched condition results in synergistic growth following
treatment with growth factors, such as EGF (24).
In summary, results of the present study have shown
that PhS1P regulates cell cycle-related genes. In addition, the
changes in gene expression synergistically trigger EGF-induced
proliferation.
Acknowledgements
We would like to thank all other members of Damy
Chemical Co., Ltd. for their support. This study was supported by
the Konkuk University in 2012.
References
|
1
|
Crigler L, Kazhanie A, Yoon TJ, Zakhari J,
Anders J, Taylor B and Virador VM: Isolation of a mesenchymal cell
population from murine dermis that contains progenitors of multiple
cell lineages. FASEB J. 21:2050–2063. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Giro MG, Oikarinen AI, Oikarinen H, Sephel
G, Uitto J and Davidson JM: Demonstration of elastin gene
expression in human skin fibroblast cultures and reduced
tropoelastin production by cells from a patient with atrophoderma.
J Clin Invest. 75:672–628. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schreier T, Degen E and Baschong W:
Fibroblast migration and proliferation during in vitro wound
healing. A quantitative comparison between various growth factors
and a low molecular weight blood dialysate used in the clinic to
normalize impaired wound healing. Res Exp Med (Berl). 193:195–205.
1993.
|
|
4
|
Allen G: Cosmetics - chemical technology
or biotechnology? Int J Cosmet Sci. 6:61–69. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carpenter G and Cohen S: Epidermal growth
factor. J Biol Chem. 265:7709–7712. 1990.
|
|
6
|
Cohen S: The receptor for epidermal growth
factor functions as a tyrosyl-specific kinase. Prog Nucleic Acid
Res Mol Biol. 29:245–247. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dittmar T, Husemann A, Schewe Y, Nofer JR,
Niggemann B, Zänker KS and Brandt BH: Induction of cancer cell
migration by epidermal growth factor is initiated by specific
phosphorylation of tyrosine 1248 of c-erbB-2 receptor via EGFR.
FASEB J. 16:1823–1825. 2002.PubMed/NCBI
|
|
8
|
Andl CD, Mizushima T, Nakagawa H, Oyama K,
Harada H, Chruma K, Herlyn M and Rustgi AK: Epidermal growth factor
receptor mediates increased cell proliferation, migration, and
aggregation in esophageal keratinocytes in vitro and in vivo. J
Biol Chem. 278:1824–1830. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Halleux C and Schneider YJ: Iron
absorption by CaCo2 cells cultivated in serum-free
medium as in vitro model of the human intestinal epithelial
barrier. J Cell Physiol. 158:17–28. 1994.
|
|
10
|
Shiraha H, Gupta K, Drabik K and Wells A:
Aging fibroblasts present reduced epidermal growth factor (EGF)
responsiveness due to preferential loss of EGF receptors. J Biol
Chem. 275:19343–19351. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim MK, Park KS, Lee H, Kim YD, Yun J and
Bae YS: Phytosphingosine-1-phosphate stimulates chemotactic
migration of L2071 mouse fibroblasts via pertussis toxin-sensitive
G-proteins. Exp Mol Med. 39:185–194. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pata MO, Hannun YA and Ng CK: Plant
sphingolipids: decoding the enigma of the Sphinx. New Phytol.
185:611–630. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Inagaki Y, Pham TT, Fujiwara Y, Kohno T,
Osborne DA, Igarashi Y, Tigyi G and Parrill AL: Sphingosine
1-phosphate analogue recognition and selectivity at S1P4 within the
endothelial differentiation gene family of receptors. Biochem J.
389:187–195. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Takuwa N, Du W, Kaneko E, Okamoto Y,
Yoshioka K and Takuwa Y: Tumor-suppressive sphingosine-1-phosphate
receptor-2 counteracting tumor-promoting sphingosine-1-phosphate
receptor-1 and sphingosine kinase 1 - Jekyll Hidden behind Hyde. Am
J Cancer Res. 1:460–481. 2011.PubMed/NCBI
|
|
15
|
Takuwa Y, Du W, Qi X, Okamoto Y, Takuwa N
and Yoshioka K: Roles of sphingosine-1-phosphate signaling in
angiogenesis. World J Biol Chem. 1:298–306. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: a changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JP, Cha HJ, Lee KS, Lee KK, Son JH,
Kim KN, Lee DK and An S: Phytosphingosine-1-phosphate represses the
hydrogen peroxide-induced activation of c-Jun N-terminal kinase in
human dermal fibroblasts through the phosphatidylinositol
3-kinase/Akt pathway. Arch Dermatol Res. 304:673–678. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ohashi A, Imai H and Minami N: Cyclin A2
is phosphorylated during the G2/M transition in mouse two-cell
embryos. Mol Reprod Dev. 66:343–348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alexandrow MG and Hamlin JL: Cdc6
chromatin affinity is unaffected by serine-54 phosphorylation,
S-phase progression, and overexpression of cyclin A. Mol Cell Biol.
24:1614–1627. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yam CH, Fung TK and Poon RY: Cyclin A in
cell cycle control and cancer. Cell Mol Life Sci. 59:1317–1326.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fung TK, Ma HT and Poon RY: Specialized
roles of the two mitotic cyclins in somatic cells: cyclin A as an
activator of M phase-promoting factor. Mol Biol Cell. 18:1861–1873.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ikezawa K, Ohtsubo M, Norwood TH and
Narayanan AS: Role of cyclin E and cyclin E-dependent kinase in
mitogenic stimulation by cementum-derived growth factor in human
fibroblasts. FASEB J. 12:1233–1239. 1998.PubMed/NCBI
|
|
23
|
Chou JL, Fan Z, DeBlasio T, Koff A, Rosen
N and Mendelsohn J: Constitutive overexpression of cyclin D1 in
human breast epithelial cells does not prevent G1 arrest induced by
deprivation of epidermal growth factor. Breast Cancer Res Treat.
55:267–283. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fisher D, Krasinska L, Coudreuse D and
Novák B: Phosphorylation network dynamics in the control of cell
cycle transitions. J Cell Sci. 125:4703–4711. 2012. View Article : Google Scholar : PubMed/NCBI
|