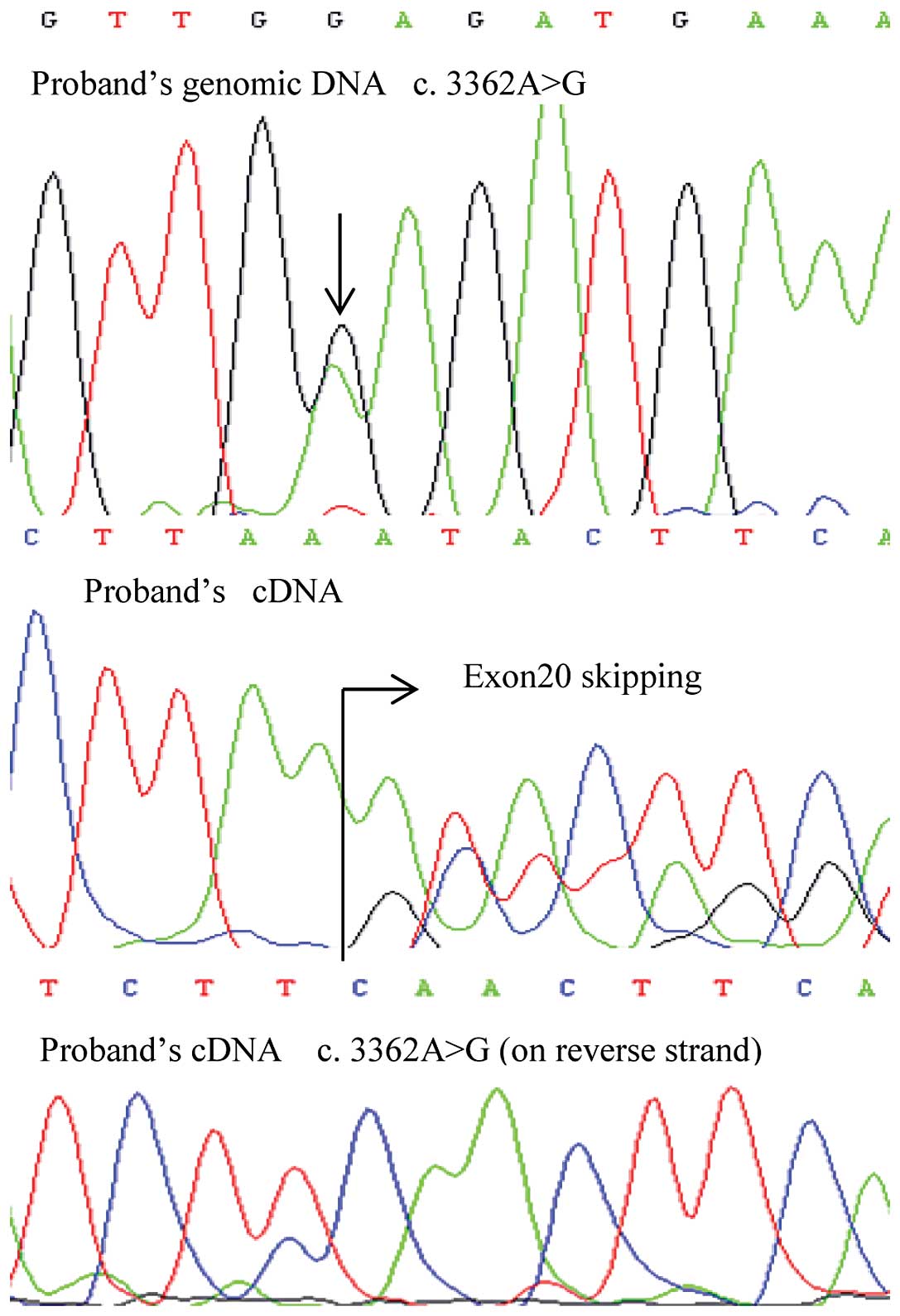
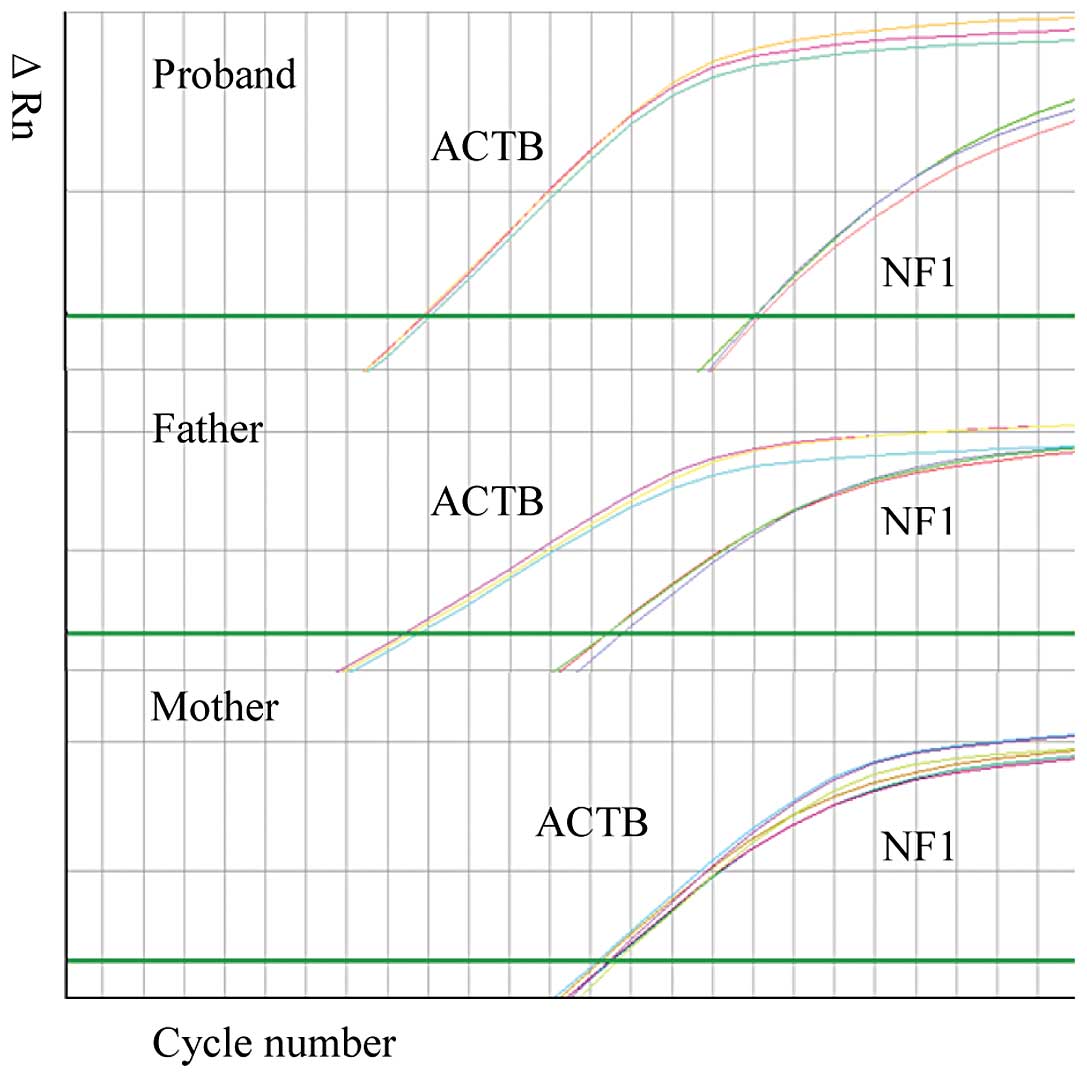
|
1
|
Williams VC, Lucas J, Babcock MA, Gutmann
DH, Korf B and Maria BL: Neurofibromatosis type 1 revisited.
Pediatrics. 123:124–133. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
No authors listed. Neurofibromatosis.
Conference statement. National Institutes of Health Consensus
Development Conference. Arch Neurol. 45:575–578. 1988.PubMed/NCBI
|
|
3
|
Griffiths S, Thompson P, Frayling I and
Upadhyaya M: Molecular diagnosis of neurofibromatosis type 1: 2
years experience. Fam Cancer. 6:21–34. 2007.PubMed/NCBI
|
|
4
|
Pasmant E, Sabbagh A, Spurlock G,
Laurendeau I, Grillo E, Hamel MJ, Martin L, Barbarot S, Leheup B,
Rodriguez D, Lacombe D, Dollfus H, Pasquier L, Isidor B, Ferkal S,
Soulier J, Sanson M, Dieux-Coeslier A, Bièche I, Parfait B, Vidaud
M, Wolkenstein P, Upadhyaya M and Vidaud D; members of the NF
France Network. NF1 microdeletions in neurofibromatosis type 1:
from genotype to phenotype. Hum Mutat. 31:1506–1518. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aoki Y, Niihori T, Narumi Y, Kure S and
Matsubara Y: The RAS/MAPK Syndromes: Novel roles of the RAS pathway
in human genetic disorders. Hum Mutat. 29:992–1006. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cichowski K and Jacks T: NF1 tumor
suppressor gene function: narrowing the GAP. Cell. 104:593–604.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yunoue S, Tokuo H, Fukunaga K, Feng L,
Ozawa T, Nishi T, Kikuchi A, Hattori S, Kuratsu J, Saya H and Araki
N: Neurofibromatosis type I tumor suppressor neurofibromin
regulates neuronal differentiation via its GTPase-activating
protein function toward Ras. J Biol Chem. 278:26958–26969. 2003.
View Article : Google Scholar
|
|
8
|
Schubbert S, Shannon K and Bollag G:
Hyperactive Ras in developmental disorders and cancer. Nat Rev
Cancer. 7:295–308. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Trovó-Marqui AB and Tajara EH:
Neurofibromin: a general outlook. Clin Genet. 70:1–13. 2006.
|
|
10
|
Nemethova M, Bolcekova A, Ilencikova D,
Durovcikova D, Hlinkova K, Hlavata A, Kovacs L, Kadasi L and
Zatkova A: Thirty-nine novel neurofibromatosis 1 (NF1) gene
mutations identified in Slovak patients. Ann Hum Genet. 77:364–379.
2013.PubMed/NCBI
|
|
11
|
Ko JM, Sohn YB, Jeong SY, Kim HJ and
Messiaen LM: Mutation spectrum of NF1 and clinical characteristics
in 78 Korean patients with neurofibromatosis type 1. Pediatr
Neurol. 48:447–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang C, Li WH, Krainer AR and Zhang MQ:
RNA landscape of evolution for optimal exon and intron
discrimination. Proc Natl Acad Sci USA. 105:5797–5802. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kolovos P, Knoch TA, Grosveld FG, Cook PR
and Papantonis A: Enhancers and silencers: an integrated and simple
model for their function. Epigenetics Chromatin. 5:12012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Messiaen LM, Callens T, Mortier G, Beysen
D, Vandenbroucke I, Van Roy N, Speleman F and Paepe AD: Exhaustive
mutation analysis of the NF1gene allows identification of 95% of
mutations and reveals a high frequency of unusual splicing defects.
Hum Mutat. 15:541–555. 2000.PubMed/NCBI
|
|
15
|
Ars E, Serra E, Garcia J, Kruyer H, Gaona
A, Lázaro C and Estivill X: Mutations affecting mRNA splicing are
the most common molecular defects in patients with
neurofibromatosis type 1. Hum Mol Genet. 9:237–247. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zatkova A, Messiaen L, Vandenbroucke I,
Wieser R, Fonatsch C, Krainer AR and Wimmer K: Disruption of exonic
splicing enhancer elements is the principal cause of exon skipping
associated with seven nonsense or missense alleles of NF1. Hum
Mutat. 24:491–501. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wimmer K, Roca X, Beiglböck H, Callens T,
Etzler J, Rao A, Krainer A, Fonatsch C and Messiaen L: Extensive in
silico analysis of NF1 splicing defects uncovers determinants for
splicing outcome upon 5′ splice-site disruption. Hum Mutat.
28:599–612. 2007.PubMed/NCBI
|
|
18
|
Pros E, Gómez C, Martín T, Fábregas P,
Serra E and Lázaro C: Nature and mRNA effect of 282 different NF1
point mutations: focus on splicing alterations. Hum Mutat.
29:E173–E193. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sironi M, Menozzi G, Riva L, Cagliani R,
Comi GP, Bresolin N, Giorda R and Pozzoli U: Silencer elements as
possible inhibitors of pseudoexon splicing. Nucleic Acids Res.
32:1783–1791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ars E, Kruyer H, Morell M, Pros E, Serra
E, Ravella A, Estivill X and Lázaro C: Recurrent mutations in the
NF1 gene are common among neurofibromatosis type 1 patients. J Med
Genet. 40:e822003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Castle B, Baser ME, Huson SM, Cooper DN
and Upadhyaya M: Evaluation of genotype-phenotype correlations in
neurofibromatosis type 1. J Med Genet. 40:e1092003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kluwe L, Friedrich R, Korf B, Fahsold R
and Mautner V: NF1 mutations in neurofibromatosis 1 patients with
plexiform neurofibromas. Hum Mutat. 19:3092002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van Minkelen R, van Bever Y, Kromosoeto J,
Withagen-Hermans C, Nieuwlaat A, Halley D and van den Ouweland A: A
clinical and genetic overview of 18 years neurofibromatosis type 1
molecular diagnosis in the Netherlands. Clin Genet. May
18–2013.(Epub ahead of print).
|
|
24
|
Upadhyaya M, Huson SM, Davies M, Thomas N,
Chuzhanova N, Giovannini S, Evans DG, Howard E, Kerr B, Griffiths
S, Consoli C, Side L, Adams D, Pierpont M, Hachen R, Barnicoat A,
Li H, Wallace P, Van Biervliet JP, Stevenson D, Viskochil D,
Baralle D, Haan E, Riccardi V, Turnpenny P, Lazaro C and Messiaen
L: An absence of cutaneous neurofibromas associated with a 3-bp
inframe deletion in exon 17 of the NF1 gene (c.2970–2972 delAAT):
evidence of a clinically significant NF1 genotype-phenotype
correlation. Am J Hum Genet. 80:140–151. 2007.PubMed/NCBI
|
|
25
|
Nelson KK and Green MR: Mechanism for
cryptic splice site activation during pre-mRNA splicing. Proc Natl
Acad Sci USA. 87:6253–6257. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blencowe BJ: Exonic splicing enhancers:
mechanism of action, diversity and role in human genetic diseases.
Trends Biochem Sci. 25:106–110. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Colapietro P, Gervasini C, Natacci F,
Rossi L, Riva P and Larizza L: NF1 exon 7 skipping and sequence
alterations in exonic splice enhancers(ESEs) in a
neurofibromatiosis 1 patient. Hum Genet. 113:551–554. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vandenbroucke I, Callens T, De Paepe A and
Messiaen L: Complex splicing pattern generates great diversity in
human NF1 transcripts. BMC Genomics. 3:132002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Z, Rolish ME, Yeo G, Tung V, Mawson M
and Burge CB: Systematic identification and analysis of exonic
spicing silencers. Cell. 119:831–845. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang XH and Chasin LA: Computational
definition of sequence motifs governing constitutive exon splicing.
Genes Dev. 18:1241–1250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baralle D and Baralle M: Splicing in
action: assessing disease causing sequence changes. J Med Genet.
42:737–748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Haque A, Buratti E and Baralle FE:
Functional properties and evolutionary splicing constraints on a
composite exonic regulatory element of splicing in CFTR exon 12.
Nucleic Acids Res. 38:647–659. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gottfried O, Viskochil D and Couldwell W:
Neurofibromatosis type 1 and tumorigenesis: molecular mechanisms
andtherapeutic implications. Neurosurg Focus. 28:E82010. View Article : Google Scholar : PubMed/NCBI
|