Introduction
Local cancer control is an important objective of
primary treatment, and decreasing the rate of recurrence is also
important for patients. Hyperthermia (HT) induced by heat stress is
a promising approach for the treatment of various types of
malignant tumor, and is mainly used in combination with radiation
therapy and/or chemotherapy (1).
The great advantage of HT therapy is that it is tolerable for the
majority of patients without severe toxicity. A better
understanding of the mechanisms underlying its effects may provide
important information for clinical HT therapy; thus, the biological
effects have also been investigated for more than a decade.
However, the heat sensitivities of cancer cells vary widely because
of the differences in intrinsic heat sensitivity and resistance
development. Since thermal-resistant cancer cells reduce the
therapeutic effects of HT treatment, control of thermal resistance
is a substantial clinical problem.
The mechanism of thermal resistance in most cancer
cells involves the elevation of heat shock proteins (HSPs).
Mammalian HSPs have been classified according to their molecular
weights: Hsp110, Hsp90, Hsp70 and small HSPs (15–30 kDa) (2). HSPs are conserved proteins that are
produced to protect cells from stress-induced damage by assisting
the correct folding of nascent and stress-accumulated misfolded
proteins, and by preventing their aggregation (3,4).
Cancer cells must expand their metabolic and signal transduction
pathways, thereby becoming dependent on proteins, including
stress-inducible HSPs, that are dispensable for the survival of
normal cells. Therefore, the cytoprotective functions of HSPs
mentioned above are necessary to maintain the survival of cancer
cells (5). The expression and/or
activity of HSPs are abnormally high in cancer cells and further
increased after a variety of death stimuli, including HT (6).
microRNAs (miRNAs) are endogenous, evolutionarily
conserved small (18–22 nucleotides) non-coding RNAs that have been
shown to regulate gene expression post-transcriptionally (7). Currently, miRNAs are known to
regulate the expression of their target genes by suppressing mRNA
translation and/or degrading target mRNA transcription (8,9).
Due to their highly pleiotropic nature, each miRNA has the
potential to regulate hundreds or even thousands of protein-coding
RNA transcripts, and thus miRNAs are potentially capable of
influencing many different molecules. Previous studies (8–10)
have revealed that hundreds of miRNAs are found in the human genome
and are critical in various important biological processes,
including cell growth, proliferation, apoptosis and tumorigenesis.
However, because of their complex mechanisms, the functions of
miRNAs are not fully understood.
It has been suggested that miRNAs can functionally
interact with a variety of environmental factors (11), including radiation (12), and HT (13). Results of recent studies have also
shown that miRNAs play a role in the radiosensitivity of cancer
cells (14,15). However, few studies have been
conducted specifically to investigate the involvement of miRNAs in
thermal resistance in cancer cells. The aim of this study was to
examine the effect of miRNAs on thermal resistance in human oral
squamous cell carcinoma (OSCC) cell lines. The results showed that
elevation of the expression level of miR-27a reinforces HT-induced
cell death in thermal-resistant cells via a decrease in the
expression levels of HSPs, especially Hsp110 and Hsp90.
Materials and methods
Cell culture and HT treatment
The human OSCC cell lines HSC-2, HSC-3 and HSC-4
were obtained from the Human Science Research Resources Bank of the
Japan Health Sciences Foundation (Tokyo, Japan). The cell lines
were cultured in E-MEM medium (Wako Pure Chemical Industries, Ltd.,
Osaka, Japan) supplemented with 10% fetal bovine serum (FBS) at
37°C in humidified air with 5% CO2. For the HT
treatment, cells were cultured in 6- or 12-well plates for 72 h
prior to HT treatment. The plates were sealed with
Parafilm® and heated at 44°C for 60 min in a water bath.
After HT treatment, the cells were incubated at 37°C for the
indicated time periods until analysis (16).
Analysis of cell death
Cells were collected 24 h after HT treatment. For
the detection of total cell death (apoptotic and necrotic cell
death), the cells were washed with ice-cold phosphate-buffered
saline (PBS) and treated with 2.5 μg/ml propidium iodide (PI)
solution. For the detection of chromatin condensation, the cells
were stained using a Nuclear-ID Green Chromatin Condensation kit
(Enzo Life Sciences Inc., Farmingdale, NY, USA) according to the
manufacturer’s instructions. To obtain the distribution of cells in
a sub-G1 phase of the cell cycle, the cells were fixed with 70%
ice-cold ethanol, and subsequently treated with 0.25 μg/ml RNase A
and 50 μg/ml PI. The samples were then run on an Epics XL flow
cytometer (Beckman Coulter, Fullerton, CA, USA) (17).
Western blot analysis
Whole-cell extracts were prepared in RIPA lysis
buffer containing a cocktail of protease inhibitors (Nacalai
Tesque, Kyoto, Japan). The polyvinylidene difluoride membranes were
incubated with the primary antibody at 4°C for 18 h, and exposed to
the peroxidase-conjugated secondary antibody at room temperature
for 1 h. Immunoreactive proteins were visualized by a luminescent
image analyzer using a chemiluminescence detection system. Primary
antibodies used were as follows: a rabbit polyclonal anti-Hsp110
antibody, a rat monoclonal anti-Hsp90 antibody, a mouse monoclonal
anti-Hsp70 antibody, a mouse monoclonal anti-Hsp40 antibody (all
from MBL, Nagoya, Japan), a rabbit polyclonal anti-caspase-3
antibody (Cell Signaling Technology, Danvers, MA, USA) or a mouse
monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
antibody (Millipore Co., Temecula, CA, USA).
Detection of miRNAs
Total RNA containing small RNAs was extracted from
the cultured cells using a miRNeasy Mini kit (Qiagen, Valencia, CA,
USA) according to the manufacture’s instructions and used for the
global miRNA expression analysis. The quality of RNA was determined
with an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa
Clara, CA, USA). The miRNA microarray profiling was performed using
two miRNA array systems. One was the GeneChip® miRNA 2.0
array, which has unique probes for 15,644 mature miRNAs from
miRBase and 2,202 probes for pre-miRNA hairpin sequences
(Affymetrix, Inc., Santa Clara, CA, USA). Total RNA (1 μg) was
labeled by poly(A) polymerase addition using a Genisphere FlashTag
HSR kit following the manufacturer’s instructions (Genisphere,
Hatfield, PA, USA). Labeled RNA was hybridized to the miRNA 2.0
arrays. The arrays were washed and stained in a Fluidics Station
450, and image scanning was performed using an Affymetrix scanner.
The other array system was Toray’s 3D-Gene™ human miRNA array,
which contains ~1,700 probes selected from miRBase (Toray
Industries, Inc., Tokyo, Japan). Labeling, scanning and data mining
were performed at the Toray Research Center (Toray Industries,
Inc.).
Quantitative polymerase chain reaction
(qPCR) assay
Total RNA was extracted from cells using an RNeasy
total RNA extraction kit (Qiagen). qPCR was performed on a
Real-Time qPCR system (Mx3000P; Agilent Technologies, Inc.) using
SYBR Premix DimerEraser™ (Takara Bio, Inc., Shiga, Japan) according
to the manufacturer’s instructions. The reverse transcriptase
reaction was carried out with total RNA by using a random 6 mers
and an oligo(dT) primer (PrimeScript RT reagent kit; Takara Bio,
Inc.). PCR primers were designed based on the database. GAPDH was
used as a control for the normalization (17,18).
miRNA transfection
The pre-miRNAs, hsa-miR-23a mimic, hsa-miR-27a-3p
mimic/antisense, hsa-miR-30a antisense, has-miR-30c antisense,
hsa-miR-203 antisense, and miScript inhibitor negative control were
obtained from Qiagen. miRNA was transfected to cells using
Lipofectamine™ RNAiMAX (Life Technologies Co., Grand Island, NY,
USA) according to the manufacturer’s instructions. Briefly, the
cells were plated in 12-well plates in E-MEM medium supplemented
with 10% FBS. After growth for 20–24 h, the cells were replaced in
serum-reduced Opti-MEM (Life Technologies Co.) containing the
Lipofectamine™ RNAiMax/miRNA complexes that were prepared 20 min
before addition to the cells (forward transfection protocol). To
decrease the cellular toxicity of the complexes, Opti-MEM
containing the complexes was replaced with the complete culture
medium 6 h after transfection. Forty-eight hours after
transfection, the cells were exposed to HT.
Statistical analysis
Data are presented as the means ± standard
deviations (SDs). Differences between pairs of data sets were
analyzed using the Student’s t-test, with values of P<0.05
considered to indicate statistically significant differences.
Results
Effects of HT on the cell death in OSCC
cells
To evaluate the thermo-sensitivity of the HSC-2,
HSC-3 and HSC-4 OSCC cell lines, the effects of HT on the cell
death were examined using flow cytometry. Twenty-four hours after
HT treatment (44°C, 60 min), the percentages of total cell death
(apoptotic and necrotic dead cells) and condensed chromatin
condensation (apoptotic cell death) were significantly increased in
the HSC-2 and HSC-3 cells but not in the HSC-4 cells (Fig. 1A and B). The measurement of the
distribution of cells in a sub-G1 phase of the cell cycle, a marker
for apoptosis, also supported the idea that HSC-2 and HSC-3 were
more sensitive to HT than HSC-4 cells (Fig. 1C). The results suggested that HT
induces cell death in HSC-2 and HSC-3 but not in HSC-4 cells.
Therefore, HSC-4 cells were less sensitive to HT than the other two
cell lines.
Induction of HSPs in OSCC cells by
HT
Having established that the OSCC cell lines showed
different levels of thermal sensitivity, we examined the expression
patterns of HSPs in the OSCC cell lines by means of western blot
analysis. The basal expression level of the Hsp70 protein was
higher in HSC-3 and HSC-4 cells than in the HSC-2 cells (Fig. 2). On the other hand, the basal
expression level of the Hsp40 protein was higher in the HSC-2 and
HSC-4 cells than in the HSC-3 cells. In terms of the Hsp90 and
Hsp110 proteins, HSC-4 cells showed the highest expression levels
of these HSPs among the three cell lines examined. After HT
treatment, the protein expression levels of HSPs were increased in
a time-dependent manner, and the induction rates of Hsp70 and Hsp40
were marked in all the cell lines. Notably, 6 h after HT exposure,
the protein expression levels of Hsp110 and Hsp90 in HSC-4 cells
were further increased and were the highest among the three cell
lines (Fig. 2). Therefore, we
hypothesized that the different protein expression patterns of HSPs
may explain the differences in thermal sensitivity of the OSCC
cells lines, especially the thermo-resistance of HSC-4 cells.
Expression profiles of miRNAs in OSCC
cells
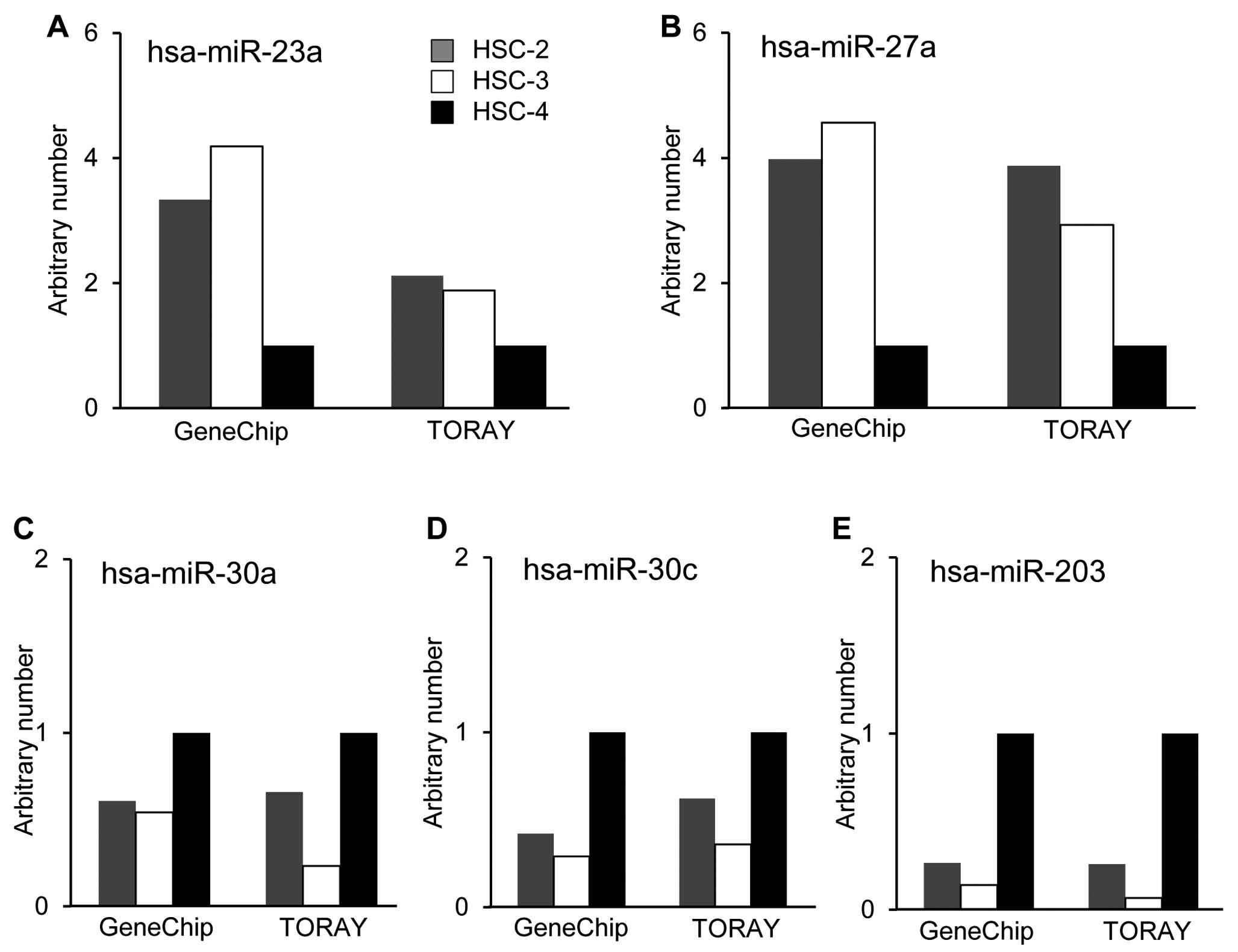
To identify miRNAs that were differentially
expressed and associated with the thermal sensitivity, we utilized
two different miRNA microarray systems provided by Affymetrix, Inc.
and Toray Industries, Inc. The results showed marked differences in
the miRNA expression patterns between the thermo-sensitive cell
lines, HSC-2 and HSC-3 cells, and the thermo-resistant cell line,
HSC-4, in the two systems. The basal expression levels of several
miRNAs in HSC-4 cells were significantly different compared with
those in HSC-2 and HSC-3 cells. The expression levels of
hsa-miR-23a and hsa-miR-27a in HSC-4 cells were lower than those of
other cells (Fig. 3A and B). On
the other hand, hsa-miR-30a, hsa-miR-30c and hsa-miR-203 were
preferentially expressed in HSC-4 cells compared with HSC-2 and
HSC-3 cells (Fig. 3C–E).
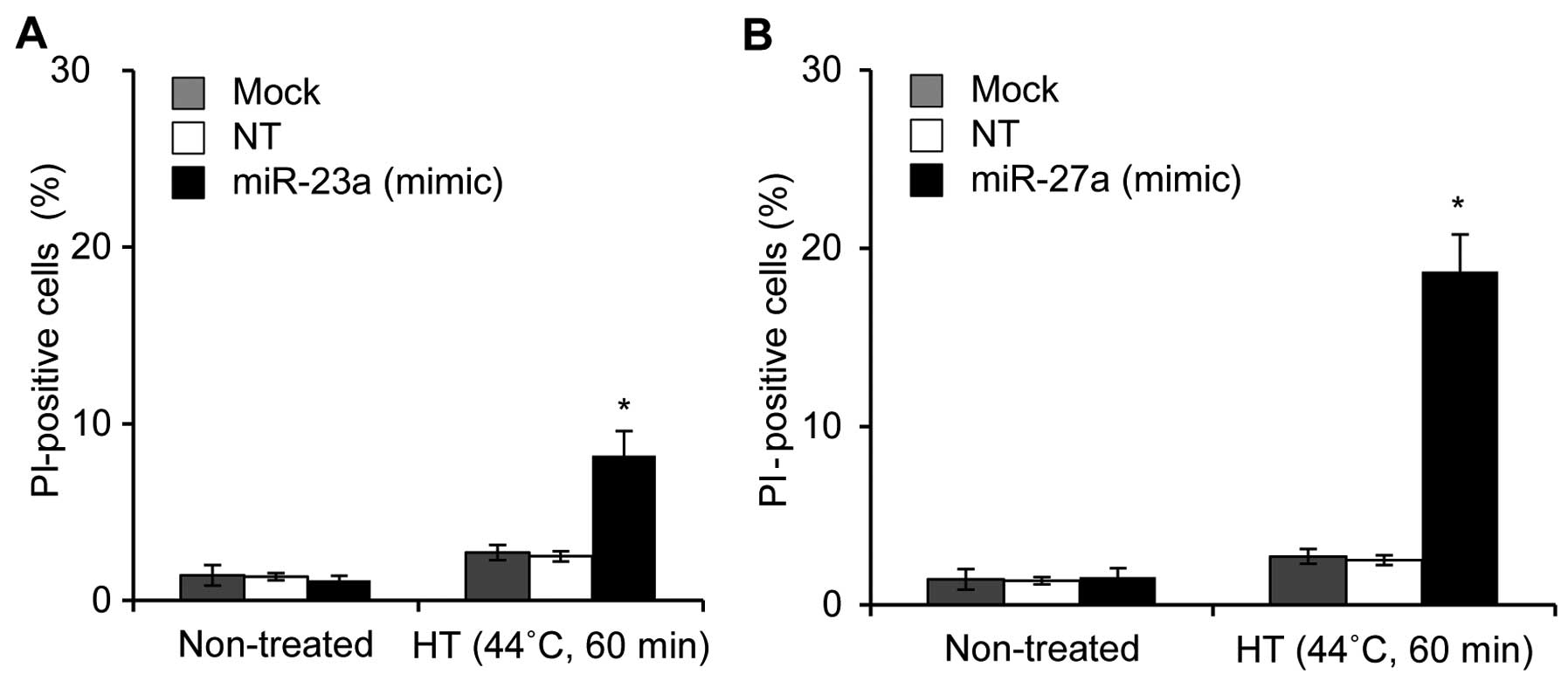
Effects of miRNA mimic or antisense
oligonucleotides on HT-induced cell death in HSC-4 cells
The two independent microarray systems clearly
demonstrated that five miRNAs (miR-23a, miR-27a, miR-30a, miR-30c
and miR-203) were candidate miRNAs for thermal sensitivity in OSCC
cells. Therefore, we examined the effects of miRNA mimic or
antisense oligonucleotides on HT-induced cell death. HSC-4 cells
were treated with HT 48 h after being transfected with the
oligonucleotide, and then cell death was evaluated 24 h after HT
treatment. Treatments of HSC-4 cells with mimic oligonucleotides
for miR-23a (20 nM) and miR-27a (20 nM) followed by HT
significantly elevated cell death, with the mean percentages of
cell death being 8.5 and 18.0%, respectively (Fig. 4A and B). However, cell death was
hardly enhanced by the treatment with a high concentration (100 nM)
of antisense oligonucleotide for miR-30, miR-30c or miR-203 in
HT-exposed cells (data not shown). These results demonstrated that
miR-23a and miR-27a may be involved in thermal sensitivity in HSC-4
cells.
Effects of the miR-27a mimic
oligonucleotide on the protein expression of HSPs and cleavage of
caspase-3 in HSC-4 cells under HT-treated conditions
We examined whether the protein expression of HSPs
was influenced by treatment with the miR-27a mimic oligonucleotide
in HSC-4 cells. The protein expression levels of Hsp110 and Hsp90
were significantly decreased in HSC-4 cells treated with the
miR-27a mimic oligonucleotide (20 nM) under HT conditions (Fig. 5). Simultaneously, the
oligonucleotide slightly decreased the expression levels of Hsp70
and Hsp40. A significant increase in caspase-3 cleavage, a marker
of apoptosis, was observed in the cells following combination
treatment with HT and oligonucleotide transfection. These results
suggested that the increased cleavage of caspase-3 may be due to
the decreased expression levels of HSPs, especially Hsp110 and
Hsp90, which promote cancer cell survival (19,20).
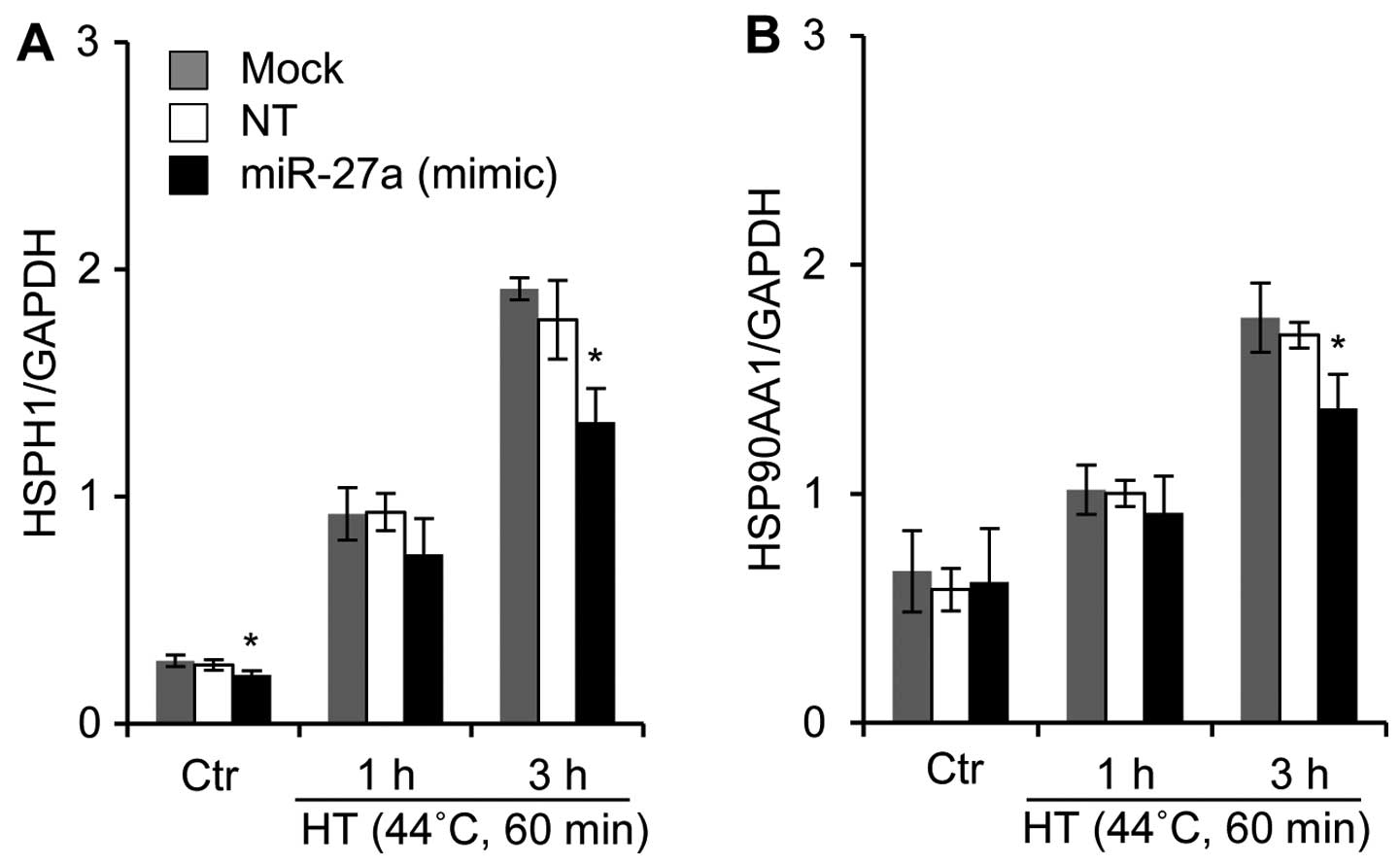
Effects of the miR-27a mimic
oligonucleotide on the mRNA expression of HSPs in HSC-4 cells under
HT-treated conditions
A bioinformatics-based approach was employed to
predict the putative targets using the TargetScan program hosted by
the Wellcome Trust Sanger Institute (21) and GGRNA hosted by DBCLS (22). We noted that one potential binding
site was found in the 3′-untranslated region (3′-UTR) or amino acid
coding sequences (CDS) of the human HSPH1 (Hsp110), HSP90AA1
(Hsp90), HSPA1A (Hsp70), HSPA1B (Hsp70), DNAJA1 (Hsp40), and DNAJB1
(HSp40) genes for human miR-27a. We also investigated whether a
miR-27a mimic oligonucleotide would affect the mRNA expression
levels of HSPs in HSC-4 cells. In the miR-27a mimic
oligonucleotide-transfected cells, the mRNA expression levels of
HSPH1 and HSP90AA1 were significantly reduced when the cells were
exposed to HT (Fig. 6). By
contrast, the oligonucleotide did not affect the mRNA expression of
HSPA1A, HSPA1B, DNAJA1 and DNAJB1 under either regular or HT
conditions (data not shown).
Discussion
Control of thermal resistance is one of the most
important issues in HT therapy. It has been reported that miRNAs
can play a critical role in cancer cells (23) by regulating cancer-related
pathways such as the cell cycle control, DNA damage response and
stress-sensitivity pathways (23,24). Microarray expression data from a
wide spectrum of cancers have provided evidence that aberrant miRNA
expression is the rule rather than the exception in cancer
(25). However, the resistance to
heat stress in cancer cells remains largely unexplored. In the
present study, we revealed that OSCC HSC-4 cells, the model of
thermally resistant cells used in the present study, had
characteristic patterns of miRNA expression compared with the
thermal-sensitive HSC-2 and HSC-3 OSCC cell lines. Of the miRNAs
expressed in this manner, miR-27a was most likely to be involved in
the thermal resistance in HSC-4 cells, because its constitutive
expression level was lower than the levels in thermal-sensitive
OSCC cells. Notably, transfection of HSC-4 cells with a miR-27a
mimic oligonucleotide elicited cell death under the HT conditions.
These results suggest that the low expression level of miR-27a
likely contributes to the mechanism of thermal resistance in OSCC
HSC-4 cells.
As for the targets of miR-27a, we focused on the HSP
family genes. miR-27a partially shares the seed sequence (26) to bind to the 3′-UTR or the amino
acid CDS region of HSP family genes. In general, miRNAs modulate
the gene expression in mammalian cells by base pairing to
complementary sites in the 3′-UTR of their target miRNAs. However,
results of a recent study showed that miRNAs can regulate target
miRNAs by binding to the CDS region (27). In the present study, the
expression levels of Hsp110 and Hsp90 significantly decreased when
HSC-4 cells were transfected with the miRNA-27a mimic
oligonucleotide under the HT conditions (Fig. 5), despite the low homology of the
seed sequence to these 3′-UTR regions. Additionally, these CDS
regions have partial homology to the miR-27a seed. Notably, the
miR-27a mimic oligonucleotide significantly suppressed the
expression of Hsp110 and Hsp90, not only at the levels of
translation, but also transcription. The detailed mechanism
underlying the control of gene expression via the CDS regions has
not yet been identified, although the homology between the miR-27a
seed sequence and CDS regions of Hsp110 and Hsp90 genes may be
important for the regulation of those expressions.
It is generally recognized that HSPs confer
substantial thermal resistance to cancer cells (6). Hsp110, one of the earliest HSPs
described in mammalian cells, plays an important role as a
chaperone under stress conditions (19,28) and participates in cellular thermal
resistance (29). In addition, it
has been recently demonstrated that the expression level of Hsp110
is elevated in highly metastatic colon cancer cell lines, and is
correlated with advanced clinical stages (30). Hsp90 is also a molecular chaperone
protein and is crucially involved in the function and stability of
many oncogene products and cell-signaling molecules (31,32). Functional inhibition of Hsp90 also
appears to enhance the sensitivity to HT on the incidence of cell
death (33). Overexpression of
Hsp90 was frequently observed in cancer cells, and, therefore, the
pharmacological inhibition of Hsp90 is an attractive strategy for
cancer therapy (34).
In conclusion, miR-27a may contribute to the thermal
sensitivity of OSCCs, presumably through regulation of the
expression of Hsp110 and Hsp90. Our data supports a model in which
miR-27a may overcome thermal resistance and serve as a predictive
marker of thermal sensitivity.
Acknowledgements
This study was supported in part by a Grant-in-Aid
for the Challenging Exploratory Research (23650303) and a
Grant-in-Aid for Scientific Research B (24310046) from the Japan
Society for the Promotion of Science. We would like to thank Dr
Ryohei Ogawa for helpful advice.
Abbreviations:
|
CDS
|
coding sequences
|
|
FBS
|
fetal bovine serum
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
HSPs
|
heat shock proteins
|
|
HT
|
hyperthermia
|
|
miRNAs
|
microRNAs
|
|
OSCC
|
oral squamous cell carcinoma
|
|
PBS
|
phosphate-buffered saline
|
|
PI
|
propidium iodide
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
3′-UTR
|
3′-untranslated region
|
References
|
1
|
Hildebrandt B, Wust P, Ahlers O, et al:
The cellular and molecular basis of hyperthermia. Crit Rev Oncol
Hematol. 43:33–56. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vos MJ, Hageman J, Carra S and Kampinga
HH: Structural and functional diversities between members of the
human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry.
47:7001–7011. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Minton KW, Karmin P, Hahn GM and Minton
AP: Nonspecific stabilization of stress-susceptible proteins by
stress-resistant proteins: a model for the biological role of heat
shock proteins. Proc Natl Acad Sci USA. 79:7107–7111. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lanneau D, Brunet M, Frisan E, Solary E,
Fontenay M and Garrido C: Heat shock proteins: essential proteins
for apoptosis regulation. J Cell Mol Med. 12:743–761. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ciocca DR and Calderwood SK: Heat shock
proteins in cancer: diagnostic, prognostic, predictive, and
treatment implications. Cell Stress Chaperones. 10:86–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calderwood SK, Khaleque MA, Sawyer DB and
Ciocca DR: Heat shock proteins in cancer: chaperones of
tumorigenesis. Trends Biochem Sci. 31:164–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meltzer PS: Cancer genomics: small RNAs
with big impacts. Nature. 435:745–746. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
10
|
Moss EG: MicroRNAs: hidden in the genome.
Curr Biol. 12:R138–R140. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J and Cui Q: Specific roles of
microRNAs in their interactions with environmental factors. J
Nucleic Acids. 2012:9783842012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Niemoeller OM, Niyazi M, Corradini S, et
al: MicroRNA expression profiles in human cancer cells after
ionizing radiation. Radiat Oncol. 6:292011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wilmink GJ, Roth CL, Ibey BL, et al:
Identification of microRNAs associated with hyperthermia-induced
cellular stress response. Cell Stress Chaperones. 15:1027–1038.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gwak HS, Kim TH, Jo GH, et al: Silencing
of microRNA-21 confers radio-sensitivity through inhibition of the
PI3K/AKT pathway and enhancing autophagy in malignant glioma cell
lines. PLoS One. 7:e474492012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chistiakov DA and Chekhonin VP:
Contribution of microRNAs to radio- and chemoresistance of brain
tumors and their therapeutic potential. Eur J Pharmacol. 684:8–18.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tabuchi Y, Wada S, Furusawa Y, Ohtsuka K
and Kondo T: Gene networks related to the cell death elicited by
hyperthermia in human oral squamous cell carcinoma HSC-3 cells. Int
J Mol Med. 29:380–386. 2012.
|
|
17
|
Kariya A, Tabuchi Y, Yunoki T and Kondo T:
Identification of common gene networks responsive to mild
hyperthermia in human cancer cells. Int J Mol Med. 32:195–202.
2013.PubMed/NCBI
|
|
18
|
Tabuchi Y, Furusawa Y, Kariya A, Wada S,
Ohtsuka K and Kondo T: Common gene expression patterns responsive
to mild temperature hyperthermia in normal human fibroblastic
cells. Int J Hyperthermia. 29:38–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hosaka S, Nakatsura T, Tsukamoto H,
Hatayama T, Baba H and Nishimura Y: Synthetic small interfering RNA
targeting heat shock protein 105 induces apoptosis of various
cancer cells both in vitro and in vivo. Cancer Sci. 97:623–632.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu W and Neckers L: Targeting the
molecular chaperone heat shock protein 90 provides a multifaceted
effect on diverse cell signaling pathways of cancer cells. Clin
Cancer Res. 13:1625–1629. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Plaisance-Bonstaff K and Renne R: Viral
miRNAs. Methods Mol Biol. 721:43–66. 2011. View Article : Google Scholar
|
|
22
|
Naito Y and Bono H: GGRNA: an ultrafast,
transcript-oriented search engine for genes and transcripts.
Nucleic Acids Res. 40:W592–W596. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cho WS: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: MicroRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo J, Miao Y, Xiao B, et al: Differential
expression of microRNA species in human gastric cancer versus
non-tumorous tissues. J Gastroenterol Hepatol. 24:652–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ott CE, Grünhagen J, Jäger M, et al:
MicroRNAs differentially expressed in postnatal aortic development
downregulate elastin via 3′ UTR and coding-sequence binding sites.
PLoS One. 6:e162502011.PubMed/NCBI
|
|
28
|
Oh HJ, Chen X and Subjeck JR: Hsp110
protects heat-denatured proteins and confers cellular
thermoresistance. J Biol Chem. 272:31636–31640. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oh HJ, Easton D, Murawski M, Kaneko Y and
Subjeck JR: The chaperoning activity of hsp110. Identification of
functional domains by use of targeted deletions. J Biol Chem.
274:15712–15718. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dorard C, de Thonel A, Collura A, et al:
Expression of a mutant HSP110 sensitizes colorectal cancer cells to
chemotherapy and improves disease prognosis. Nat Med. 17:1283–1289.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Den RB and Lu B: Heat shock protein 90
inhibition: rationale and clinical potential. Ther Adv Med Oncol.
4:211–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bagatell R and Whitesell L: Altered Hsp90
function in cancer: a unique therapeutic opportunity. Mol Cancer
Ther. 3:1021–1030. 2004.PubMed/NCBI
|
|
33
|
Ito A, Saito H, Mitobe K, et al:
Inhibition of heat shock protein 90 sensitizes melanoma cells to
thermosensitive ferromagnetic particle-mediated hyperthermia with
low Curie temperature. Cancer Sci. 100:558–564. 2009. View Article : Google Scholar
|
|
34
|
Workman P, Burrows F, Neckers L and Rosen
N: Drugging the cancer chaperone HSP90: combinatorial therapeutic
exploitation of oncogene addiction and tumor stress. Ann NY Acad
Sci. 1113:202–216. 2007. View Article : Google Scholar : PubMed/NCBI
|