|
1
|
Hildebrandt B, Wust P, Ahlers O, et al:
The cellular and molecular basis of hyperthermia. Crit Rev Oncol
Hematol. 43:33–56. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vos MJ, Hageman J, Carra S and Kampinga
HH: Structural and functional diversities between members of the
human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry.
47:7001–7011. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Minton KW, Karmin P, Hahn GM and Minton
AP: Nonspecific stabilization of stress-susceptible proteins by
stress-resistant proteins: a model for the biological role of heat
shock proteins. Proc Natl Acad Sci USA. 79:7107–7111. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lanneau D, Brunet M, Frisan E, Solary E,
Fontenay M and Garrido C: Heat shock proteins: essential proteins
for apoptosis regulation. J Cell Mol Med. 12:743–761. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ciocca DR and Calderwood SK: Heat shock
proteins in cancer: diagnostic, prognostic, predictive, and
treatment implications. Cell Stress Chaperones. 10:86–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calderwood SK, Khaleque MA, Sawyer DB and
Ciocca DR: Heat shock proteins in cancer: chaperones of
tumorigenesis. Trends Biochem Sci. 31:164–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meltzer PS: Cancer genomics: small RNAs
with big impacts. Nature. 435:745–746. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
10
|
Moss EG: MicroRNAs: hidden in the genome.
Curr Biol. 12:R138–R140. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J and Cui Q: Specific roles of
microRNAs in their interactions with environmental factors. J
Nucleic Acids. 2012:9783842012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Niemoeller OM, Niyazi M, Corradini S, et
al: MicroRNA expression profiles in human cancer cells after
ionizing radiation. Radiat Oncol. 6:292011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wilmink GJ, Roth CL, Ibey BL, et al:
Identification of microRNAs associated with hyperthermia-induced
cellular stress response. Cell Stress Chaperones. 15:1027–1038.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gwak HS, Kim TH, Jo GH, et al: Silencing
of microRNA-21 confers radio-sensitivity through inhibition of the
PI3K/AKT pathway and enhancing autophagy in malignant glioma cell
lines. PLoS One. 7:e474492012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chistiakov DA and Chekhonin VP:
Contribution of microRNAs to radio- and chemoresistance of brain
tumors and their therapeutic potential. Eur J Pharmacol. 684:8–18.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
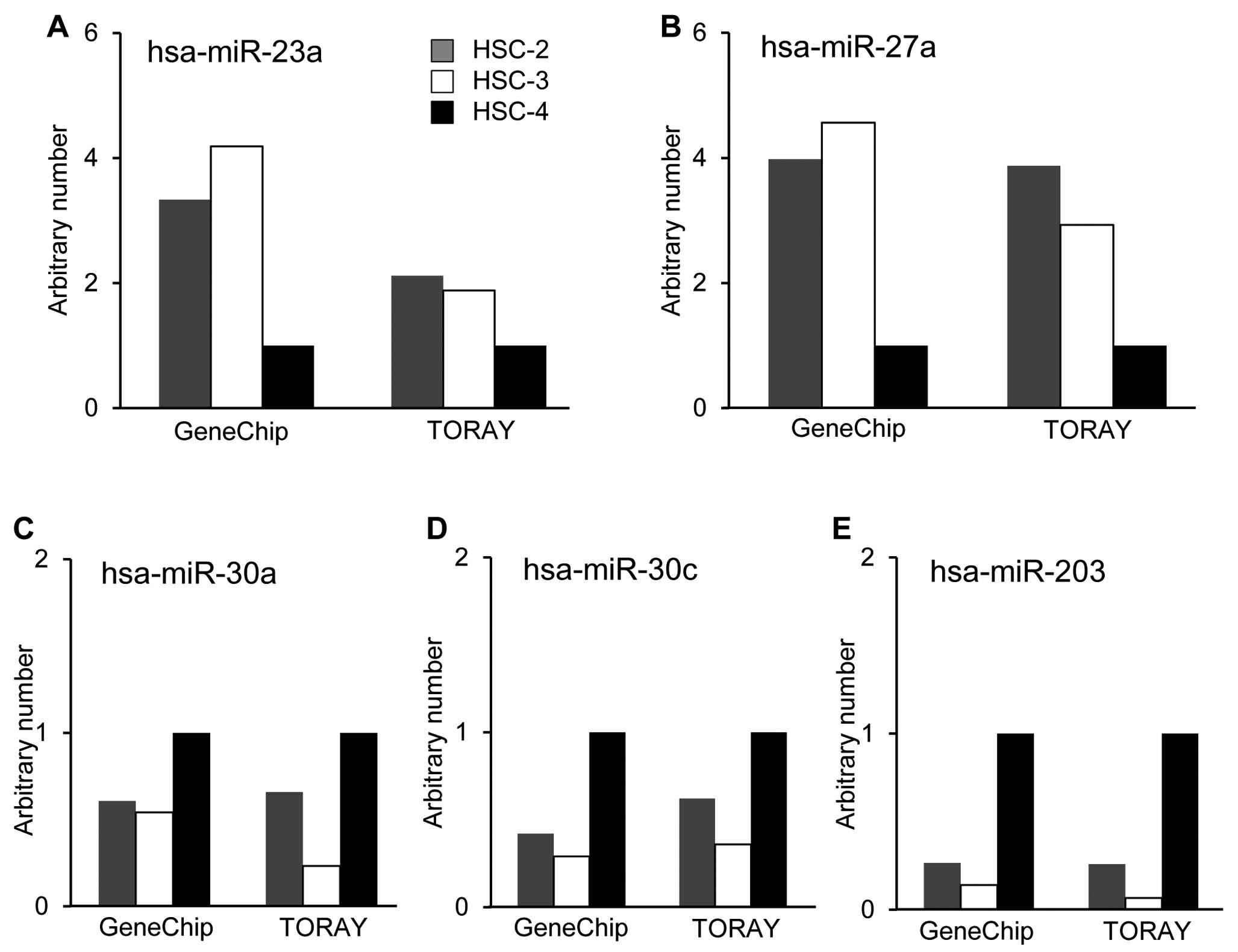
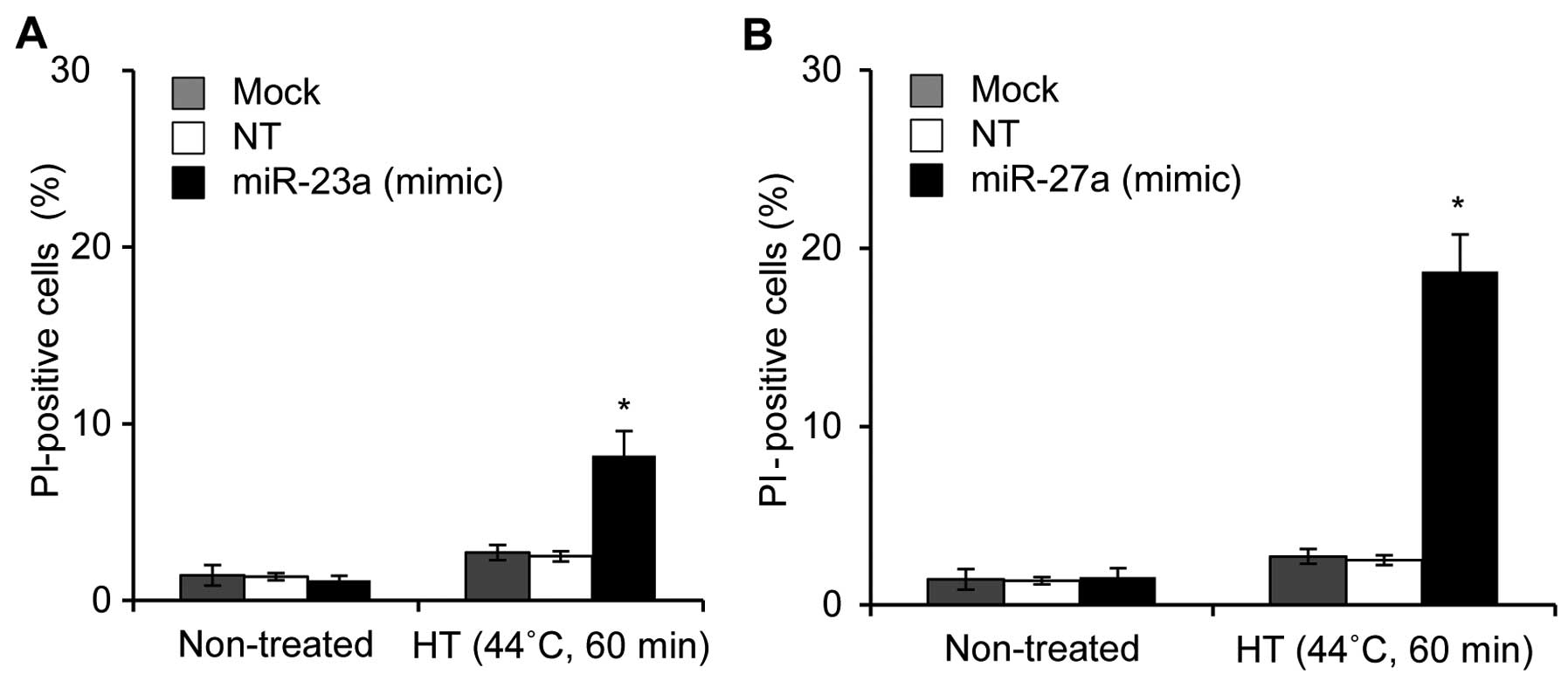
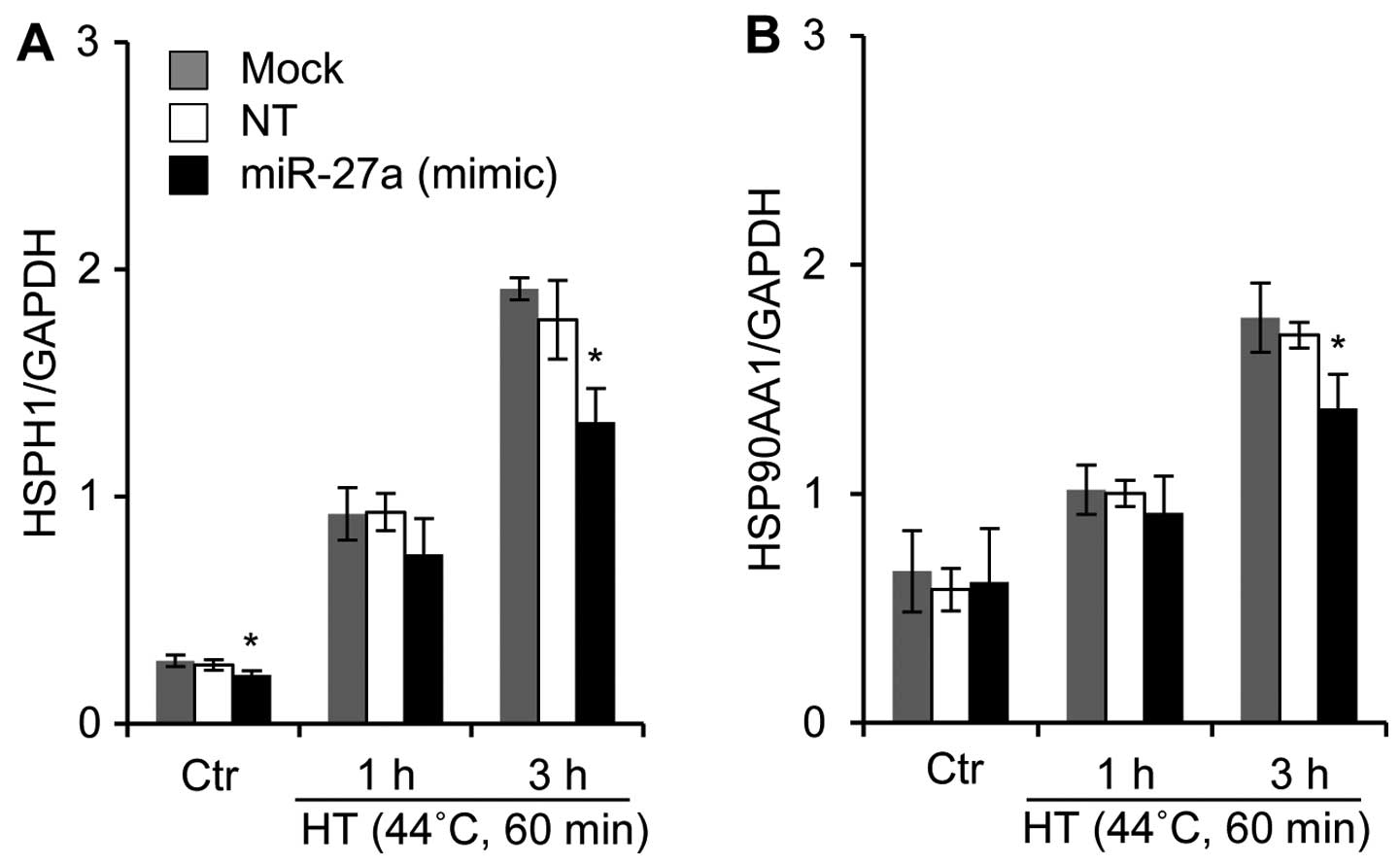
Tabuchi Y, Wada S, Furusawa Y, Ohtsuka K
and Kondo T: Gene networks related to the cell death elicited by
hyperthermia in human oral squamous cell carcinoma HSC-3 cells. Int
J Mol Med. 29:380–386. 2012.
|
|
17
|
Kariya A, Tabuchi Y, Yunoki T and Kondo T:
Identification of common gene networks responsive to mild
hyperthermia in human cancer cells. Int J Mol Med. 32:195–202.
2013.PubMed/NCBI
|
|
18
|
Tabuchi Y, Furusawa Y, Kariya A, Wada S,
Ohtsuka K and Kondo T: Common gene expression patterns responsive
to mild temperature hyperthermia in normal human fibroblastic
cells. Int J Hyperthermia. 29:38–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hosaka S, Nakatsura T, Tsukamoto H,
Hatayama T, Baba H and Nishimura Y: Synthetic small interfering RNA
targeting heat shock protein 105 induces apoptosis of various
cancer cells both in vitro and in vivo. Cancer Sci. 97:623–632.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu W and Neckers L: Targeting the
molecular chaperone heat shock protein 90 provides a multifaceted
effect on diverse cell signaling pathways of cancer cells. Clin
Cancer Res. 13:1625–1629. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Plaisance-Bonstaff K and Renne R: Viral
miRNAs. Methods Mol Biol. 721:43–66. 2011. View Article : Google Scholar
|
|
22
|
Naito Y and Bono H: GGRNA: an ultrafast,
transcript-oriented search engine for genes and transcripts.
Nucleic Acids Res. 40:W592–W596. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cho WS: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: MicroRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo J, Miao Y, Xiao B, et al: Differential
expression of microRNA species in human gastric cancer versus
non-tumorous tissues. J Gastroenterol Hepatol. 24:652–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ott CE, Grünhagen J, Jäger M, et al:
MicroRNAs differentially expressed in postnatal aortic development
downregulate elastin via 3′ UTR and coding-sequence binding sites.
PLoS One. 6:e162502011.PubMed/NCBI
|
|
28
|
Oh HJ, Chen X and Subjeck JR: Hsp110
protects heat-denatured proteins and confers cellular
thermoresistance. J Biol Chem. 272:31636–31640. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oh HJ, Easton D, Murawski M, Kaneko Y and
Subjeck JR: The chaperoning activity of hsp110. Identification of
functional domains by use of targeted deletions. J Biol Chem.
274:15712–15718. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dorard C, de Thonel A, Collura A, et al:
Expression of a mutant HSP110 sensitizes colorectal cancer cells to
chemotherapy and improves disease prognosis. Nat Med. 17:1283–1289.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Den RB and Lu B: Heat shock protein 90
inhibition: rationale and clinical potential. Ther Adv Med Oncol.
4:211–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bagatell R and Whitesell L: Altered Hsp90
function in cancer: a unique therapeutic opportunity. Mol Cancer
Ther. 3:1021–1030. 2004.PubMed/NCBI
|
|
33
|
Ito A, Saito H, Mitobe K, et al:
Inhibition of heat shock protein 90 sensitizes melanoma cells to
thermosensitive ferromagnetic particle-mediated hyperthermia with
low Curie temperature. Cancer Sci. 100:558–564. 2009. View Article : Google Scholar
|
|
34
|
Workman P, Burrows F, Neckers L and Rosen
N: Drugging the cancer chaperone HSP90: combinatorial therapeutic
exploitation of oncogene addiction and tumor stress. Ann NY Acad
Sci. 1113:202–216. 2007. View Article : Google Scholar : PubMed/NCBI
|