Introduction
Cytokines are glycosylated proteins that allow
communication among various cell types involved in immune response.
Interleukins (ILs) are cytokines mainly produced by T-cells, as
well by monocytes, macrophages and endothelial cells (1,2).
The different ILs share special biochemical or functional
characteristics and are numbered in order of their identification.
The emergence of new technologies is translating into a steady
increase in the number of known molecules (3). In 2008, Lin et al (4) produced 3,400 recombinant secreted
proteins that encode secreted proteins and extracellular domains of
transmembrane proteins in 293T cells and examined their activities
based on human monocyte screening assays. Subsequently, the authors
(4) discovered a novel cytokine,
IL-34. The human IL-34 protein is composed of 222 amino acids, has
a molecular mass of 39 kDa and forms homodimers. It binds to the
macrophage colony-stimulating factor (M-CSF) receptor, c-FMS (also
known as CSF-1 receptor), expressed on the cell surface of human
monocytes and has a stronger, although short-lived effect compared
to M-CSF. IL-34 has been shown to be involved in the process of
osteoclastogenesis and rheumatoid arthritis (RA) (5–8).
IL-34 has been shown to promote the proliferation, survival and
differentiation of monocytes and macrophages, the release of
pro-inflammatory chemokines, and thereby plays an important role in
innate immunity and inflammatory processes. It also plays an
important role in the regulation of osteoclast proliferation and
differentiation, and in the regulation of bone resorption (5–8).
IL-34 and M-CSF both signal via the same receptor,
the M-CSF receptor. Although IL-34 and M-CSF show no appreciable
similarity in their primary structure, they are evolutionally
distant ligands, but are structurally related (9). There is evidence indicating that the
M-CSF-IL-34-c-FMS axis is involved in the initiation, growth and
metastasis of tumors (10,11).
M-CSF levels may constitute a useful biomarker for a number of
types of cancer, as it is expressed at high levels in a number of
types of cancer, including breast cancer, ovarian cancer and
colorectal carcinoma and its expression correlates with a poor
prognosis (12). The direct
inhibition of M-CSF or the inhibition of c-FMS kinase activity can
lead to significant changes in the growth of grafted tumors
(13.14). Tumor-associated macrophages are the most abundant
component of the leukocyte infiltrate of solid tumors. In
M-CSF-deficient mice (M-CSFop/op or
M-CSF−/−), the growth of the primary tumor and the
metastatic spread of tumor cells has been shown to be significantly
reduced due to the inability of angiogenesis to feed the tumors
(12–15).
However, studies on the role of IL-34 in
tumorigenesis. In the present study, we identified the IL-34 gene
in various mammalian genomes by comparative genomic analyses. The
conserved transcription factor-binding sites within the promoter
region of the human IL-34 gene were then searched. Analyses of the
expression data, functional relevant single nucleotide
polymorphisms (SNPs) and comparative proteomic analysis were also
conducted. Furthermore, a meta-analysis of the prognostic value of
the IL-34 gene in various types of cancer was performed.
Materials and methods
Identification of the novel IL-34 gene in
vertebrate genomes and integrative genomic analyses
All the IL-34 gene and amino acid sequences were
obtained from the Ensembl database (http://www.ensembl.org/index.html), based on
orthologous and paralogous relationships. The gained IL-34
sequences were applied as queries to search the IL-34 gene using
BLAST at the National Center for Biotechnology Information (NCBI),
in order to confirm whether their best hit was an IL-34 gene
(16–18). The number and length of IL-34
exons and introns in all competent sequences were investigated for
exon-intron conservation analyses. The number, length and
structures of the exons and introns in IL-34 in all species were
also collected from the Ensembl database (http://www.ensembl.org/index.html). Conserved
transcription factor-binding sites within the promoter region of
the human IL-34 gene were obtained from SABiosciences’ proprietary
database which combines Text Mining Application and data from the
UCSC Genome Browser (19–21).
Comparative proteomic analysis of IL-34
protein
The protein coding sequences of IL-34 were aligned
using ClustalW software implemented in MEGA 5.05. We constructed a
maximum likelihood (ML) tree of IL-34 amino acid sequences using
MEGA 5.05 with the optimal model (Kimura 2-parameter model). For
the relative support of the internal node, bootstrap analysis was
performed with 1,000 replications for ML reconstructions (22). The program CodeML implemented in
the PAML 4.7 software package was used to investigate whether the
IL-34 protein is under positive selection (23). The site-specific model was exerted
using likelihood ratio tests (LRTs) to compare the M7 (null model)
with the M8 model. M7 is a null model that does not allow for any
codons with ω >1, whereas the M8 model allows for positively
selective sites (ω >1). When the M8 model was fitted to the data
more efficiently (P-value <0.05) than the null model (M7), the
presence of sites with ω >1 was suggested. On the contrary, the
results of P-value >0.05 proved the absence of sites with ω
>1. Twice the log likelihood difference between the two compared
models (2Δl) was compared against χ2 with critical
values of 5.99 and 9.21 at 0.05 and 0.01 significance levels,
respectively, as previously described (24).
Functionally relevant SNP evaluation of
the human IL-34 gene and identification of somatic mutations in
human cancer
Functionally relevant SNPs of the human IL-34 gene
were identified as previously described (16–21). The SNPs were extracted from the
Ensembl (http://www.ensembl.org) and the NCBI
SNPdb (http://www.ncbi.nlm.nih.gov)
databases. The SNPs that disrupted exonic splicing enhancer/exonic
splicing silencer (ESE/ESS) motifs and cause missence mutations
were also identified. The identification of somatic mutations of
the human IL-34 gene in human cancer was conducted in the Catalogue
of Somatic Mutations in Cancer (COSMIC), a database for mining
complete cancer genomes in the catalogue of somatic mutations in
cancer (25).
In silico expression analyses of the
human IL-34 gene
Expressed sequence tags (ESTs) derived from the
human IL-34 gene were searched for using the BLAST programs as
previously described (26–29).
The human IL-34 gene (NM_152456) was used as query sequences for
the BLAST programs. The expression profiles for normal human
tissues were obtained from GeneAnnot (30) and ArrayExpress (31) databases. Northern analysis of the
NCBI uniGene dataset was also performed (19–21).
Meta-analysis of the prognostic value of
the IL-34 gene in cancer
A database termed ‘PrognoScan’ has been previously
developed (32). This database
includes a large collection of publicly available cancer microarray
datasets with clinical annotation, as well as a tool for assessing
the biological association between gene expression and prognosis.
PrognoScan employs the minimum P-value approach for grouping
patients for survival analysis. PrognoScan provides a powerful
platform for evaluating potential tumor markers and therapeutic
targets and is publicly accessible at http://www.sabiosciences.com. The human IL-34 gene was
used as an input source as a query and the data were collected for
analysis.
Results
Comparative proteomic analysis of IL-34
protein identified in vertebrate genomes
All the IL-34 gene and protein sequences were
collected from the Ensembl database and confirmed by BLAST at NCBI.
The complete IL-34 gene was identified in the human, chimpanzee,
gibbon, macaque, orangutan, marmoset, bushbaby, pika, squirrel,
rat, mouse, kangaroo rat, elephant, cat, dog, panda, ferret, pig,
horse, cow, flycatcher, chicken, zebrafish, platyfish and tilapia.
The sequence and structural alignment of IL-34 is illustrated in
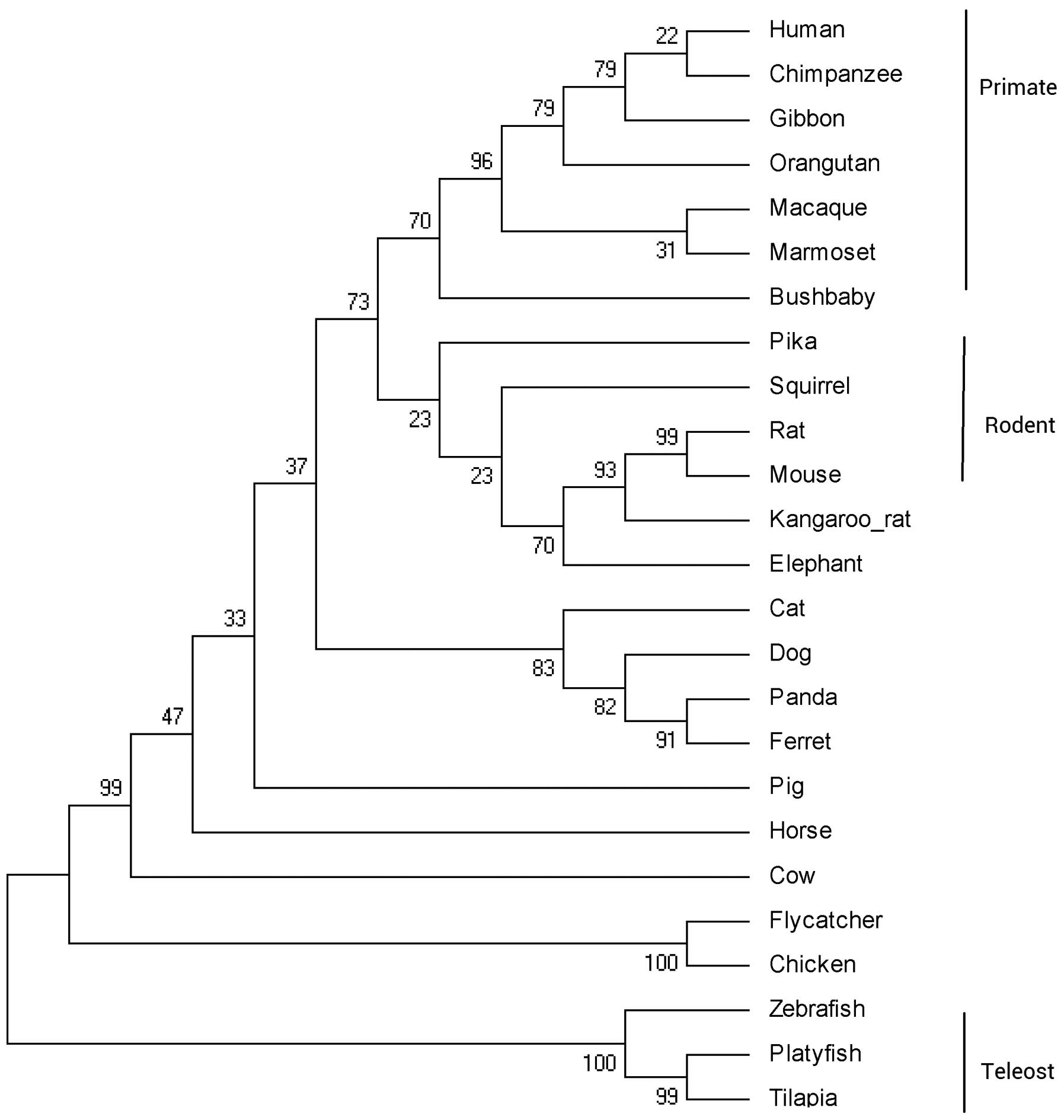
Fig. 1. The phylogenetic tree was
constructed according to the protein coding sequences of IL-34
using the ML method (Fig. 2). The
IL-34 gene from the primate lineage, rodent lineage and teleost
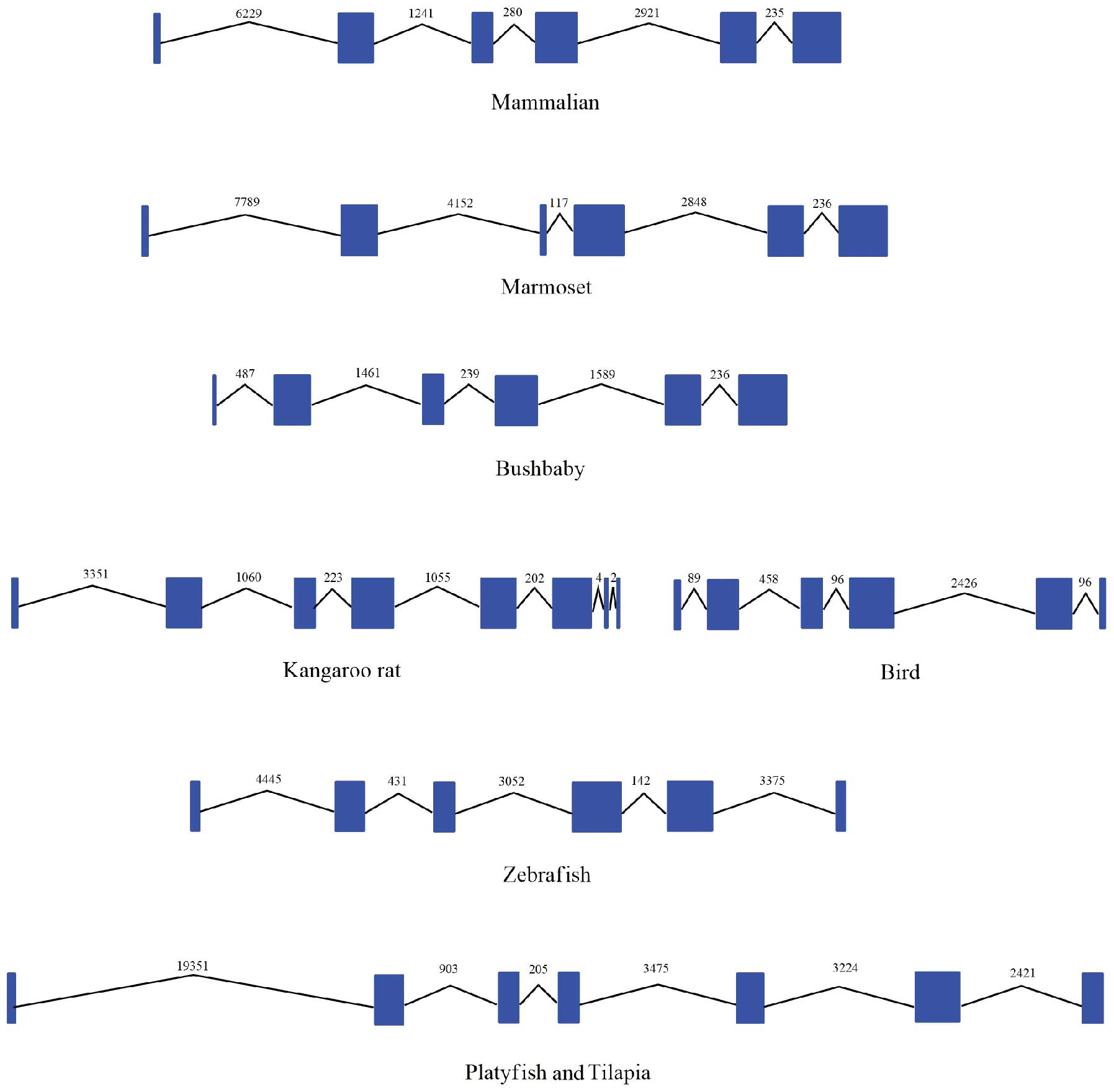
lineage forms a species-specific cluster. The exon-intron
information collected from the Ensembl database is presented in
Table I and Fig. 3. In the majority of genomes, the
IL-34 gene has 6 exons with similar lengths in different species
(Table I). In the majority of
vertebrates, the IL-34 gene shows exon-intron conservation with 5
introns and similar sizes of each intron. With exception, there are
8 exons and 7 introns in the IL-34 gene in the kangaroo rat.
Moreover, the IL-34 gene in the platyfish and tilapia contains 7
exons and 6 introns. Thus, the intron deletions of the IL-34 gene
may occur during the evolutionary process in fish. Furthermore,
site-specific tests for positive selection were performed for
vertebrate, mammalian, primate and mammalian excluding primate,
rodent and teleost lineages. Although some positive selection sites
were computed, only the 2Δl of M7 and M8 of mammalian IL-34 was
>5.99, indicating that the M8 model was more efficient than the
M7 model in fitting the data. It seemed that mammalian IL-34 was
under positive selecetion pressure with the identified positively
selected site, 196Val (Table
II).
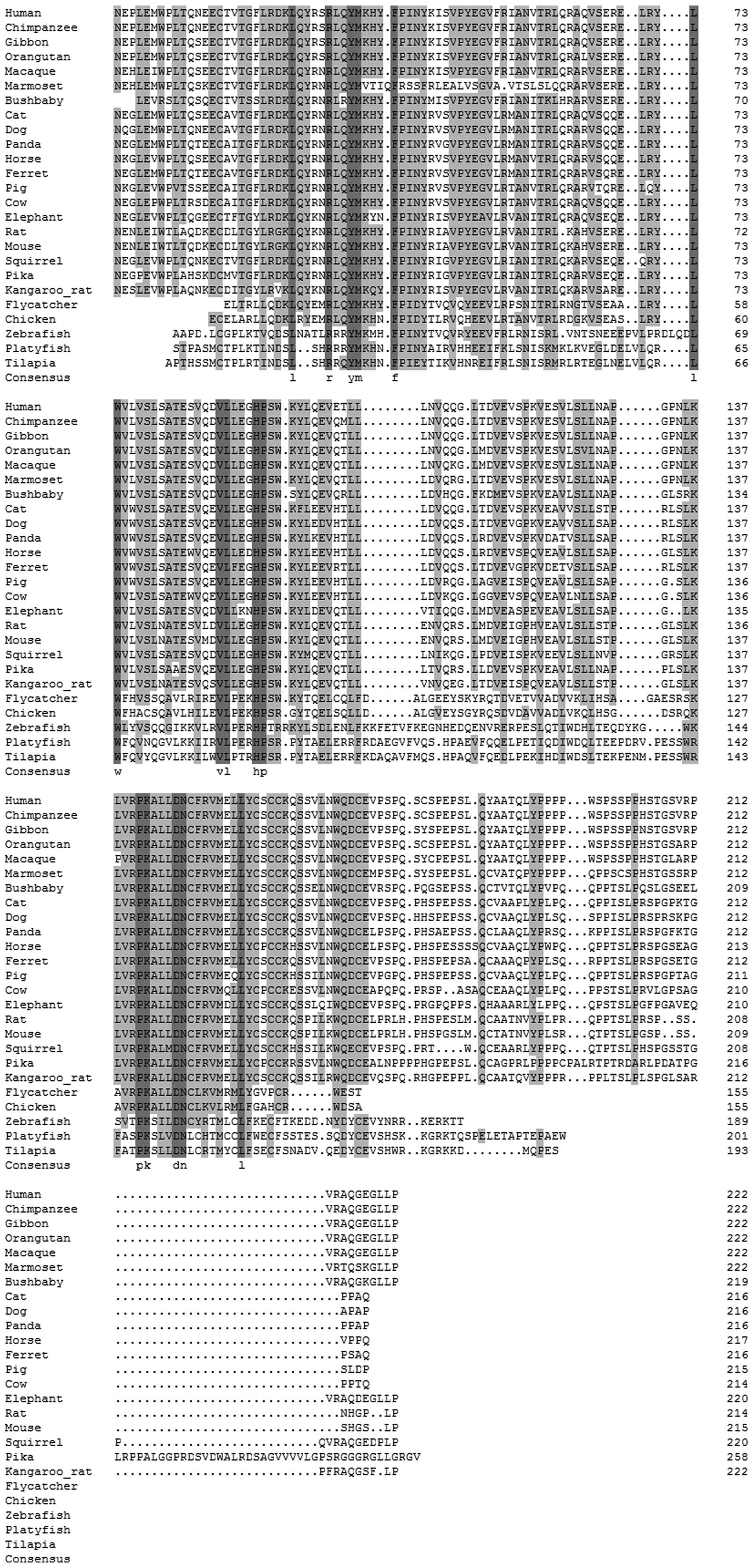 | Figure 1Sequence and structural alignment of
vertebrate interleukin-34 (IL-34). All the IL-34 gene and protein
sequences were collected from the Ensembl database and confirmed by
BLAST at the National Center for Biotechnology Information (NCBI).
The complete IL-34 gene was identified in 25 various mammalian
genomes, such as the human, chimpanzee, gibbon, macaque, orangutan,
marmoset, bushbaby, pika, squirrel, rat, mouse, kangaroo rat,
elephant, cat, dog, panda, ferret, pig, horse, cow, flycatcher,
chicken, zebrafish, platyfish and tilapia genomes. |
 | Table IExon and intron lengths of IL-34. |
Table I
Exon and intron lengths of IL-34.
|
Length
(bp) |
|---|
|
|
|---|
| Species | Exon 1 | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | Exon 4 | Intron 4 | Exon 5 | Intron 5 | Exon 6 | Intron 6 | Exon 7 | Intron 7 | Exon 8 | Total exons |
|---|
| Human | 28 | 7,562 | 134 | 1,930 | 78 | 280 | 162 | 2,496 | 136 | 243 | 191 | - | - | - | - | 729 |
| Chimpanzee | 28 | 8,179 | 134 | 1,926 | 78 | 280 | 162 | 2,495 | 136 | 250 | 191 | - | - | - | - | 729 |
| Gibbon | 28 | 6,472 | 134 | 1,930 | 78 | 280 | 162 | 2,538 | 136 | 250 | 191 | - | - | - | - | 729 |
| Orangutan | 28 | 7,643 | 134 | 2,042 | 78 | 280 | 162 | 2,563 | 136 | 258 | 191 | - | - | - | - | 729 |
| Macaque | 28 | 7,369 | 134 | 2,064 | 78 | 288 | 162 | 2,531 | 136 | 239 | 191 | - | - | - | - | 729 |
| Marmoset | 28 | 7,789 | 142 | 4,152 | 27 | 117 | 205 | 2,848 | 136 | 236 | 191 | - | - | - | - | 729 |
| Bushbaby | 15 | 487 | 147 | 1,461 | 78 | 239 | 162 | 1,589 | 136 | 236 | 170 | - | - | - | - | 711 |
| Cat | 28 | 6,405 | 134 | 787 | 78 | 285 | 162 | 2,902 | 136 | 230 | 173 | - | - | - | - | 711 |
| Dog | 28 | 6,254 | 134 | 940 | 78 | 281 | 162 | 2,366 | 136 | 238 | 173 | - | - | - | - | 711 |
| Panda | 28 | 5,018 | 134 | 797 | 78 | 284 | 162 | 2,066 | 136 | 230 | 173 | - | - | - | - | 711 |
| Horse | 28 | 5,227 | 134 | 841 | 78 | 284 | 162 | 2,162 | 136 | 217 | 176 | - | - | - | - | 714 |
| Ferret | 28 | 4,504 | 134 | 771 | 78 | 306 | 162 | 2,043 | 136 | 234 | 173 | - | - | - | - | 711 |
| Cow | 28 | 5,849 | 134 | 751 | 78 | 261 | 162 | 2,879 | 133 | 229 | 170 | - | - | - | - | 705 |
| Rat | 28 | 5,484 | 134 | 1,125 | 78 | 269 | 159 | 5,154 | 136 | 264 | 170 | - | - | - | - | 705 |
| Mouse | 28 | 5,672 | 134 | 684 | 78 | 258 | 162 | 5,463 | 136 | 195 | 170 | - | - | - | - | 708 |
| Squirrel | 28 | 5,565 | 134 | 789 | 78 | 277 | 162 | 3,231 | 136 | 210 | 185 | - | - | - | - | 723 |
| Kangaroo rat | 28 | 3,351 | 134 | 1,060 | 78 | 223 | 162 | 1,055 | 136 | 202 | 160 | 4 | 16 | 2 | 15 | 729 |
| Flycatcher | 28 | 92 | 104 | 293 | 78 | 84 | 174 | 3,627 | 139 | 99 | 20 | - | - | - | - | 543 |
| Chicken | 28 | 86 | 104 | 623 | 78 | 108 | 174 | 1,225 | 133 | 82 | 20 | - | - | - | - | 537 |
| Zebrafish | 40 | 4,445 | 107 | 431 | 81 | 3,052 | 201 | 142 | 169 | 3,375 | 44 | - | - | - | - | 642 |
| Platyfish | 40 | 26,738 | 104 | 1,344 | 78 | 256 | 78 | 4,479 | 108 | 2,376 | 184 | 4,042 | 86 | - | - | 678 |
| Tilapia | 37 | 11,964 | 104 | 461 | 78 | 154 | 78 | 2,170 | 108 | 4,072 | 184 | 799 | 59 | - | - | 648 |
 | Table IISite-specific tests for positive
selection on IL-34. |
Table II
Site-specific tests for positive
selection on IL-34.
| Species | Models | Estimates of
parameters | lnL | 2Δl | Positively selected
sites |
|---|
| Vertebrate | M7 | P=1.03903,
Q=5.39585 | −4659.912970 | 2.057682 | NS |
| M8 | P0=0.98039,
P=1.20110, Q=7.10944 (P1=0.01961) w=1.00000 | −4657.855288 | | |
| Mammalian | M7 | P=0.48941,
Q=1.45186 | −4490.139825 | 6.824409 | 196 Va |
| M8 | P0=0.96625,
P=0.63368, Q=2.34086 (P1=0.03375), w=1.81330 | −4483.315416 | | |
| Primate | M7 | P=0.01895,
Q=0.02238 | −1715.925824 | 0.033788 | NS |
| M8 | P0=0.56073,
P=0.00997, Q=0.16501 (P1=0.43927) w=1.00000 | −1715.959612 | | |
| Mammalian excluding
primate | M7 | P=0.30844,
Q=1.06769 | −3530.180011 | 2.160234 | 200 Qa |
| M8 | P0=0.98088,
P=0.35332, Q=1.40393 (P1=0.01912) w=2.08098 | −3528.019777 | | |
| Rodent | M7 | P=0.33256,
Q=1.26857 | −1743.972997 | 0.000104 | NS |
| M8 | P0=0.99999,
P=0.33225, Q=1.26708 (P1=0.00001) w=3.17132 | −1743.973101 | | |
| Teleost | M7 | P=0.55893,
Q=1.64564 | −1656.009963 | 0.088367 | NS |
| M8 | P0=0.99243,
P=0.58684, Q=1.81573 (P1=0.00757) w=8.77551 | −1655.921596 | | |
Expression profile of the human IL-34
gene
By EST sequence searching, the human IL-34 gene was
found to be expressed in the adult and fetal brain, the
hippocampus, spleen, embryonic stem cells, heart, medulla, lung,
testes, ovaries, metastatic chondrosarcoma, epidermis,
keratinocytes, osteoarthritic cartilage, adipose tissue, choroid,
eyes, amygdala, kidneys, thymus, small intestine, hypothalamus,
islets of Langerhans, glioblastoma and the retinal pigment
epithelium. The investigation of available microarray analyses and
‘virtual northern blot analysis’ revealed a predominant expression
of IL-34 in the lymph nodes, brain, heart, skeletal muscle, colon,
adipocyte, kidneys, liver, lungs, thyroid, adrenal gland, ovaries,
prostate and testes. When performing a search in the PrognoScan
database, the human IL-34 gene was also found to be expressed in
various types of cancer, such as blood, brain, breast, colorectal,
eye, head and neck, lung, ovarian and skin cancer.
Comparative genomic anlaysis of the human
IL-34 gene
The upstream transcription factor 1 (USF1),
regulatory factor X-1 (RFX1), the Sp1 transcription factor 1, POU
class 3 homeobox 2 (POU3F2) and the forkhead box L1 (FOXL1)
regulatory transcription factor binding sites were identified in
the IL-34 gene upstream (promoter) region.
Functionally relevant SNP evaluation of
the human IL-34 gene and identification of somatic mutations in
human cancer
A total of 2,141 available SNPs were identified in
the human IL-34 gene. Among these SNPs, a total of 55 SNPs were
functionally relevant; these included 32 SNPs causing missense
mutations, 3 exonic splicing enhancer SNPs and 20 SNPs causing
nonsense mutations (Table III).
As presented in Table IV, by
performing a search of the COSMIC database, we identified 18
somatic mutations of the IL-34 gene in cancer.
 | Table IIIFunctionally relevant SNP evaluation
of the human IL-34 gene. |
Table III
Functionally relevant SNP evaluation
of the human IL-34 gene.
| SNP ID | Chr 16 position
sequence | Sequence | Type | Amino acid
change |
|---|
| rs200158701 | 70680854(+) | CCATGC/TCCCGG | mis | PS |
| rs192337001 | 70680866(+) | GCTTCA/CCCTGG | mis | TP |
| rs139133476 | 70688459(+) | CCTTGG/CCGTGG | mis | AG |
| rs142890682 | 70688461(+) | TTGGCG/ATGGCC | mis | MV |
| rs118062333 | 70690511(+) | AACACT/CACTTC | mis | HY |
| rs200597979 | 70690960(+) | GGGCCA/GCCCAT | mis | HR |
| rs8046424 | 70690989(+) | AGGTGC/GAGACG | mis | QE |
| rs187166563 | 70693576(+) | CCCAGA/GGCCAA | mis | EG |
| rs142214904 | 70693626(+) | GCTTCC/TGGGTC | mis | RW |
| rs7206509 | 70693945(+) | GCCAAG/CTCCTC | mis | TS |
| rs201277640 | 70693984(+) | GTATGC/TGGCCA | mis | AV |
| rs202122982 | 70694001(+) | TGTACC/TCTCCG | mis | PS |
| rs148286339 | 70694011(+) | GCCCCC/TGTGGT | mis | PL |
| rs141513638 | 70694056(+) | GAGGCC/TGGTCA | mis | PL |
| rs112639369 | 70694073(+) | AGGGCG/AAGGGC | mis | KE |
| rs1444643201 | 70694076(+) | GCGAGG/AGCCTC | mis | SG |
| rs367851338 | 70693627(+) | CTTCCA/GGGTCA | mis | QR |
| rs368143418 | 70690933(+) | TGAGTC/TGGTGC | mis | SL |
| rs374665339 | 70690963(+) | CCACCC/TATCCT | mis | PL |
| rs368923655 | 70691023(+) | CCTCAC/TGGTGA | mis | TM |
| rs368367274 | 70693597(+) | GGTGCA/GGCCCA | mis | QR |
| rs200891924 | 70693560(+) | TGTCCC/ATCTTG | mis | IL |
| rs372998917 | 70694041(+) | CTCCAC/TGGGCT | mis | TM |
| rs370436386 | 70690927(+) | TGCCAC/TTGAGT | mis | TL |
| rs144427482 | 70690571(+) | CCAACG/ATCACC | mis | IV |
| rs201108464 | 70693569(+) | TGAATG/ACCCCA | mis | TA |
| rs144144426 | 70690541(+) | GTGTGC/TCTTAC | mis | PS |
| rs377411431 | 70690885(+) | CGAGCG/TGGAGC | mis | RL |
| rs369011177 | 70680875(+) | GGCTGC/TGCTGT | mis | RC |
| rs145782768 | 70693979(+) | TTGCAG/CTATGC | mis | HQ |
| rs201784459 | 70694005(+) | CCCTCC/TGCCCC | mis | PL |
| rs200488835 | 70693655(+) | TCCTGC/GTGTAA | mis | CW |
| rs3813904 | 70680744(+) | TGACTG/CAGTGA | ese | |
| rs3813905 | 70680850(+) | ACCACC/GATGCC | ese | |
| rs4985556 | 70694000(+) | CTGTAC/ACCTCC | ese | |
 | Table IVSomatic mutations of IL-34 in cancer
tissue. |
Table IV
Somatic mutations of IL-34 in cancer
tissue.
| Position (AA) | Mutation (CDS) | Mutation (amino
acid) | Mutation ID
(COSM) | Count | Mutation type |
|---|
| 4 | c.11G>A | p.G4D | COSM973055 | 1 | Substitution -
missense |
| 9 | c.25C>T | p.R9C | COSM3691133 | 1 | Substitution -
missense |
| 33 | c.99G>A | p.E33E | COSM704311 | 1 | Substitution -
coding silent |
| 38 | c.114G>A | p.T38T | COSM973057 | 1 | Substitution -
coding silent |
| 42 |
c.125_126GG>AA | p.R42Q | COSM143555 | 1 | Substitution -
missense |
| 59 | c.176C>T | p.P59L | COSM108032 | 1 | Substitution -
missense |
| 61 | c.182A>G | p.N61S | COSM3387573 | 1 | Substitution -
missense |
| 104 | c.311C>T | p.S104L | COSM435667 | 1 | Substitution -
missense |
| 155 | c.465C>T | p.N155N | COSM328594 | 9 | Substitution -
coding silent |
| 170 | c.508C>T | p.R170W | COSM194870 | 1 | Substitution -
missense |
| 183 | c.549C>A | p.S183R | COSM3402448 | 1 | Substitution -
missense |
| 197 | c.589C>T | p.Q197* | COSM1379321 | 1 | Substitution -
nonsense |
| 197 | c.590A>G | p.Q197R | COSM1379322 | 1 | Substitution -
missense |
| 197 | c.591G>A | p.Q197Q | COSM40324 | 1 | Substitution -
coding silent |
| 208 | c.623C>T | p.A208V | COSM417292 | 2 | Substitution -
missense |
| 208 | c.624G>A | p.A208A | COSM1177412 | 1 | Substitution -
coding silent |
| 217 | c.651G>A | p.P217P | COSM1379323 | 1 | Substitution -
coding silent |
| 229 | c.686C>T | p.S229L | COSM417291 | 3 | Substitution -
missense |
Meta-analysis of the prognostic value of
the human IL-34 gene in cancer
When the name of a gene is submitted, PrognoScan
displays a summary in table format of tests for the gene with
columns for dataset, cancer type, subtype, endpoint, cohort,
contributor, array type, probe ID, number of patients, optimal
cutpoint, Pmin and Pcor. Among the databases which detected the
expression of the IL-34 gene, 5 out of 40 tests revealed an
association between the expression of the IL-34 gene and cancer
prognosis (blood cancer, 1/4; brain cancer, 1/4; breast cancer,
0/11; colorectal cancer, 1/7; eye cancer, 0/1; head and neck
cancer, 0/3; lung cancer, 2/6; ovarian cancer, 0/3; and skin
cancer, 0/1) with a 5% significance level (Table V). Among the two types of lung
cancer, the lower expression of the IL-34 gene was related to poor
survival and was found in non-small cell lung cancer (NSCLC) case
(GSE8894). However, a higher expression of the IL-34 gene was
related to poor survival in a case of adenocarcinoma (GSE31210). As
for blood cancer cases and colorectal cancer, we found that a lower
expression of the IL-34 gene was associated with poor survival.
However, in the brain cancer cases, a higher expression of the
IL-34 gene was related to poor survival.
 | Table VDataset content from the PrognoScan
database demonstrating an association between microarray analyses
in IL-34 and cancer prognosis. |
Table V
Dataset content from the PrognoScan
database demonstrating an association between microarray analyses
in IL-34 and cancer prognosis.
| Database | Case type | Subsyte | No. of
patients | Endpoint | Cutpoint | P-value | Prognosis | (Refs.) |
|---|
|
GSE12417-GPL570 | Blood cancer | AML | 79 | Overall
survival | 0.18 | 0.028 | 1 | (45) |
| GSE4412-GPL97 | Brain cancer | Glioma | 74 | Overall
survival | 0.72 | 0.003 | 2 | (46) |
| GSE17537 | Colorectal
cancer | | 55 | Overall
survival | 0.38 | 0.04 | 1 | (47) |
| GSE31210 | Lung cancer | Adenocarcinoma | 204 | Relapse-free
survival | 0.89 | 0.03 | 2 | (48) |
| GSE8894 | Lung cancer | NSCLC | 138 | Relapse-free
survival | 0.4 | 0.0002 | 1 | (49) |
Discussion
IL-34 was identified by functional screening of a
library of secreted proteins, based on its ability to support human
monocyte survival and to promote, with the same efficiency as
M-CSF, the formation of the colony forming unit-macrophage (CFU-M)
in human bone marrow cell cultures (4).
In the present study, we identified the complete
IL-34 gene in 25 various mammalian genomes, including the human,
chimpanzee, gibbon, macaque, orangutan, marmoset, bushbaby, pika,
squirrel, rat, mouse, kangaroo rat, elephant, cat, dog, panda,
ferret, pig, horse, cow, flycatcher, chicken, zebrafish, platyfish
and tilapia genomes. In addition, we found that IL-34 existed in
all types of vertebrates, including fish, amphibians, birds and
mammals. The IL-34 gene has a similar 7 exon/6 intron gene
organization in various species, and genes in the IL-34 loci were
syntenically conserved (33,34). The phylogenetic tree demonstrated
that the IL-34 gene from the primate lineage, rodent lineage and
teleost lineage formed a species-specific cluster. From the
alignment and phylogenetic tree, mammalian IL-34 was conversed
among vertebrate genomes, suggesting that the function of the IL-34
gene plays an important physiological role in all vertebrates in
the long evolutionary process. It seemed that the mammalian IL-34
gene was under positive selection pressure with the identified
positively selected site, 196Val. This is in accordance the with
multiple biological functions of a cytokine, which plays a key role
in the immune system.
IL-34 mRNA is widely expressed in various types of
tissue, including tissue of the heart, brain, lung, liver, kidneys,
thymus and spleen (4).
Accordingly, by EST sequence searching, the IL-34 gene was also
found to be expressed in various other types of tissues and cells,
including the hippocampus, embryonic stem cells, medulla, testes,
ovaries, metastatic chondrosarcoma, epidermis, keratinocytes,
osteoarthritic cartilage, adipose tissue, choroid, eyes, amygdala
thymus, small intestine, hypothalamus, islets of Langerhans,
glioblastoma and the retinal pigment epithelium. This suggests that
the IL-34 gene is widely expressed in many types of tissues and
organs. The investigation of available microarray analyses and
‘virtual northern blot analysis’ confirmed the predominant
expression of IL-34 in the lymph nodes, brain, heart, skeletal
muscle, colon, adipocyte, kidneys, liver, lung, thyroid, adrenal
gland, ovaries, prostate and testes. A total of 55 functionally
relevant SNPs, including 32 SNPs causing missense mutations, 3
exonic splicing enhancer SNPs and 20 SNPs causing nonsense
mutations were identified from 2,141 available SNPs in the human
IL-34 gene, which may affect the multiple functions of IL-34.
However, the effects of these SNPs on the physiological and
pathological function of IL-34 require further investigation.
IL-34 and M-CSF both signal via the same receptor,
the M-CSF receptor, c-FMS. It has been shown that M-CSF is
expressed at high levels in many types of tumor, including breast
cancer, ovarian cancer and colorectal carcinoma and correlates with
a poor prognosis (10–15). However, studies on the role of
IL-34 in tumor development are limited. In the present study, we
firstly found that IL-34 was indeed expressed in various types of
cancer, such as blood, brain, breast, colorectal, eye, head and
neck, lung, ovarian and skin cancer. A total of 5 out of 40 tests
(1 blood cancer, 1 brain cancer, 1 colorectal cancer and 2 lung
cancer) revealed an association between IL-34 gene expression and
cancer prognosis. The mechanisms responsible for the involvement of
IL-34 in the progression of these tumors require further
investigation. It should be noted that the association between the
expression of IL-34 and prognosis varies in different types of
cancer, even in the same type of cancer from different databases.
This suggests that the function of IL-34 in these tumors may be
multidimensional, not only functioning as a tumor inhibitor or
promoter. Moreover, we identified 18 somatic mutations of IL-34 in
cancer tissue in the present study. The mechanisms through which
these mutations affect tumor formation require further
investigation. These data suggest that IL-34, similar to M-CSF, is
involved in tumor formation.
USF1, RFX1, Sp1, POU3F2 and FOXL1 regulatory
transcription factor binding sites were identified in the IL-34
gene upstream (promoter) region. USF-1 is an important
transcription factor that participates in glucose metabolism and
tumorigenesis. It has a negative effect on cell proliferation in
some cell types and stabilizes the p53 protein and promotes a
transient cell cycle arrest, in the presence of DNA damage
(34,35). RFX1 is unique transcription factor
that contains a highly conserved 76-amino-acid DNA binding domain.
RFX1 can directly regulate CD44 expression (36,37). This mechanism may contribute to
the effects of RFX1 on the proliferation, survival and invasion of
glioblastoma cells. Sp1 is a member of the Sp/Krüppel-like factor
(KLF) family of transcription factors that play a critical role in
embryonic and early postnatal development, differentiation, cell
cycle regulation and in multiple diseases, including cancer
(38–41). POU domain transcription factors
are present in a number of cell lineages where they perform various
functions, either as ubiquitous regulators of ‘housekeeping’ genes,
or as developmental- and lineage-specific coordinators of cell fate
decisions (42). POU3F2 has been
shown to be responsive to MAPK pathway activation and to modulate
the levels of microphthalmia-associated transcription factor (MITF)
so as to suppress the differentiated melanocytic phenotype and to
enhance tumor metastasis (29).
FOXL1 is located at the junction of multiple signaling pathways and
plays critical roles in a variety of physiological and pathological
processes, including cancer development. These tumor-related
transcriptional factors may be involved in the effects of IL-34 in
tumors (28,43,44).
Acknowledgements
This study was sponsored by grants from the Chinese
National Natural Science Foundation (81202077 and 810001329), the
National Major Scientific and Technological Special Project for
‘Significant New Drugs Development’ (2011ZX09302-003-02), the
Jiangsu Province Major Scientific and Technological Special Project
(BM2011017) and a Project Funded by the Priority Academic Program
Development of Jiangsu Higher Education Institutions.
References
|
1
|
Wakabayashi S, Yamaguchi K, Kumakura S, et
al: Effects of anesthesia with sevoflurane and propofol on the
cytokine/chemokine production at the airway epithelium during
esophagectomy. Int J Mol Med. 34:137–144. 2014.PubMed/NCBI
|
|
2
|
Signorelli SS, Fiore V and Malaponte G:
Inflammation and peripheral arterial disease: the value of
circulating biomarkers (Review). Int J Mol Med. 33:777–783.
2014.PubMed/NCBI
|
|
3
|
Clavel G, Thiolat A and Boissier MC:
Interleukin newcomers creating new numbers in rheumatology: IL-34
to IL-38. Joint Bone Spine. 80:449–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin H, Lee E, Hestir K, Leo C, et al:
Discovery of a cytokine and its receptor by functional screening of
the extracellular proteome. Science. 320:807–811. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boström EA and Lundberg P: The newly
discovered cytokine IL-34 is expressed in gingival fibroblasts,
shows enhanced expression by pro-inflammatory cytokines, and
stimulates osteoclast differentiation. PLoS One. 8:e816652013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moon SJ, Hong YS, Ju JH, Kwok SK, Park SH
and Min JK: Increased levels of interleukin 34 in serum and
synovial fluid are associated with rheumatoid factor and anticyclic
citrullinated peptide antibody titers in patients with rheumatoid
arthritis. J Rheumatol. 40:1842–1849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hwang SJ, Choi B, Kang SS, et al:
Interleukin-34 produced by human fibroblast-like synovial cells in
rheumatoid arthritis supports osteoclastogenesis. Arthritis Res
Ther. 14:R142012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z, Buki K, Vääräniemi J, Gu G and
Väänänen HK: The critical role of IL-34 in osteoclastogenesis. PLoS
One. 6:e186892011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
El-Gamal MI, Anbar HS, Yoo KH and Oh CH:
FMS kinase inhibitors: Current status and future prospects. Med Res
Rev. 33:599–636. 2013. View Article : Google Scholar
|
|
10
|
Burns CJ and Wilks AF: c-FMS inhibitors: a
patent review. Expert Opin Ther Pat. 21:147–165. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baud’huin M, Renault R, Charrier C, et al:
Interleukin-34 is expressed by giant cell tumours of bone and plays
a key role in RANKL-induced osteoclastogenesis. J Pathol.
221:77–86. 2010. View Article : Google Scholar
|
|
12
|
Petráčková M, Staněk L, Mandys V, Dundr P
and Vonka V: Properties of bcr-abl-transformed mouse 12B1 cells
secreting interleukin-2 and granulocyte-macrophage colony
stimulating factor (GM-CSF): II. Adverse effects of GM-CSF. Int J
Oncol. 40:1915–1922. 2012.
|
|
13
|
Fournier P, Aigner M and Schirrmacher V:
Targeting of IL-2 and GM-CSF immunocytokines to a tumor vaccine
leads to increased anti-tumor activity. Int J Oncol. 38:1719–1729.
2011.PubMed/NCBI
|
|
14
|
Kitoh Y, Saio M, Gotoh N, et al: Combined
GM-CSF treatment and M-CSF inhibition of tumor-associated
macrophages induces dendritic cell-like signaling in vitro. Int J
Oncol. 38:1409–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tada F, Abe M, Hirooka M, et al: Phase
I/II study of immunotherapy using tumor antigen-pulsed dendritic
cells in patients with hepatocellular carcinoma. Int J Oncol.
41:1601–1609. 2012.PubMed/NCBI
|
|
16
|
Yang L, Luo Y and Wei J: Integrative
genomic analyses on Ikaros and its expression related to solid
cancer prognosis. Oncol Rep. 24:571–577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang L, Luo Y, Wei J and He S: Integrative
genomic analyses on IL28RA, the common receptor of interferon-λ1,
-λ2 and -λ3. Int J Mol Med. 25:807–812. 2010.PubMed/NCBI
|
|
18
|
Yang L, Wei J and He S: Integrative
genomic analyses on interferon-λs and their roles in cancer
prediction. Int J Mol Med. 25:299–304. 2010.PubMed/NCBI
|
|
19
|
Yu H, Yuan J, Xiao C and Qin Y:
Integrative genomic analyses of recepteur d’origine nantais and its
prognostic value in cancer. Int J Mol Med. 31:1248–1254.
2013.PubMed/NCBI
|
|
20
|
Wang M, Wei X, Shi L, Chen B, Zhao G and
Yang H: Integrative genomic analyses of the histamine H1 receptor
and its role in cancer prediction. Int J Mol Med. 33:1019–1026.
2014.PubMed/NCBI
|
|
21
|
Wang B, Chen K, Xu W, Chen D, Tang W and
Xia TS: Integrative genomic analyses of secreted protein acidic and
rich in cysteine and its role in cancer prediction. Mol Med Rep.
10:1461–1468. 2014.PubMed/NCBI
|
|
22
|
Kumar S, Nei M, Dudley J and Tamura K:
MEGA: a biologist-centric software for evolutionary analysis of DNA
and protein sequences. Brief Bioinform. 9:299–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Z: PAML: a program package for
phylogenetic analysis by maximum likelihood. Comput Appl Biosci.
13:555–556. 1997.PubMed/NCBI
|
|
24
|
Yang Z, Nielsen R, Goldman N and Pedersen
AM: Codon-substitution models for heterogeneous selection pressure
at amino acid sites. Genetics. 155:431–449. 2000.PubMed/NCBI
|
|
25
|
Forbes SA, Bindal N, Bamford S, et al:
COSMIC: mining complete cancer genomes in the Catalogue of Somatic
Mutations in Cancer. Nucleic Acids Res. 39:D945–D950. 2011.
View Article : Google Scholar :
|
|
26
|
Katoh Y and Katoh M: Integrative genomic
analyses on GLI1: positive regulation of GLI1 by Hedgehog-GLI,
TGFβ-Smads, and RTK-PI3K-AKT signals, and negative regulation of
GLI1 by Notch-CSL-HES/HEY, and GPCR-Gs-PKA signals. Int J Oncol.
35:187–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Katoh M and Katoh M: Integrative genomic
analyses of WNT11: transcriptional mechanisms based on canonical
WNT signals and GATA transcription factors signaling. Int J Mol
Med. 24:247–251. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Katoh M and Katoh M: Transcriptional
mechanisms of WNT5A based on NF-κB, Hedgehog, TGFβ, and Notch
signaling cascades. Int J Mol Med. 23:763–769. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Katoh M and Katoh M: Integrative genomic
analyses of ZEB2: Transcriptional regulation of ZEB2 based on
SMADs, ETS1, HIF1α, POU/OCT, and NF-κB. Int J Oncol. 34:1737–1742.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chalifa-Caspi V, Yanai I, Ophir R, et al:
GeneAnnot: comprehensive two-way linking between oligonucleotide
array probesets and GeneCards genes. Bioinformatics. 20:1457–1458.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Parkinson H, Sarkans U, Shojatalab M, et
al: ArrayExpress - a public repository for microarray gene
expression data at the EBI. Nucleic Acids Res. 33:D553–D555. 2005.
View Article : Google Scholar
|
|
32
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: a new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang T, Kono T, Monte MM, et al:
Identification of IL-34 in teleost fish: differential expression of
rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor.
Mol Immunol. 53:398–409. 2013. View Article : Google Scholar
|
|
34
|
Bouafia A, Corre S, Gilot D, Mouchet N,
Prince S and Galibert MD: p53 requires the stress sensor USF1 to
direct appropriate cell fate decision. PLoS Genet. 10:e10043092014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ikeda R, Nishizawa Y, Tajitsu Y, et al:
Regulation of major vault protein expression by upstream
stimulating factor 1 in SW620 human colon cancer cells. Oncol Rep.
31:197–201. 2014.
|
|
36
|
Katoh Y and Katoh M: Comparative genomics
on Vangl1 and Vangl2 genes. Int J Oncol. 26:1435–1440.
2005.PubMed/NCBI
|
|
37
|
Feng C, Zhang Y, Yin J, Li J, Abounader R
and Zuo Z: Regulatory factor X1 is a new tumor suppressive
transcription factor that acts via direct downregulation of CD44 in
glioblastoma. Neuro Oncol. 16:1078–1085. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu F, Zhu X, Han T, et al: The oncoprotein
hepatitis B X-interacting protein promotes the migration of ovarian
cancer cells through the upregulation of S-phase kinase-associated
protein 2 by Sp1. Int J Oncol. 45:255–263. 2014.PubMed/NCBI
|
|
39
|
Liang X, Li ZL, Jiang LL, Guo QQ, Liu MJ
and Nan KJ: Suppression of lung cancer cell invasion by LKB1 is due
to the downregulation of tissue factor and vascular endothelial
growth factor, partly dependent on SP1. Int J Oncol. 44:1989–1997.
2014.PubMed/NCBI
|
|
40
|
Chae JI, Cho JH, Lee KA, et al: Role of
transcription factor Sp1 in the quercetin-mediated inhibitory
effect on human malignant pleural mesothelioma. Int J Mol Med.
30:835–841. 2012.PubMed/NCBI
|
|
41
|
Lee KA, Lee YJ, Ban JO, et al: The
flavonoid resveratrol suppresses growth of human malignant pleural
mesothelioma cells through direct inhibition of specificity protein
1. Int J Mol Med. 30:21–27. 2012.PubMed/NCBI
|
|
42
|
Moritsugu R, Tamai K, Nakano H, et al:
Functional analysis of the nuclear localization signal of the POU
transcription factor Skn-1a in epidermal keratinocytes. Int J Mol
Med. 34:539–544. 2014.PubMed/NCBI
|
|
43
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
44
|
Katoh M and Katoh M: Transcriptional
regulation of WNT2B based on the balance of Hedgehog, Notch, BMP
and WNT signals. Int J Oncol. 34:1411–1415. 2009.PubMed/NCBI
|
|
45
|
Metzeler KH, Hummel M, Bloomfield CD, et
al: An 86-probe-set gene-expression signature predicts survival in
cytogenetically normal acute myeloid leukemia. Blood.
112:4193–4201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Freije WA, Castro-Vargas FE, Fang Z, et
al: Gene expression profiling of gliomas strongly predicts
survival. Cancer Res. 64:6503–6510. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Smith JJ, Deane NG, Wu F, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar
|
|
48
|
Okayama H, Kohno T, Ishii Y, et al:
Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar
|
|
49
|
Lee ES, Son DS, Kim SH, et al: Prediction
of recurrence-free survival in postoperative non-small cell lung
cancer patients by using an integrated model of clinical
information and gene expression. Clin Cancer Res. 14:7397–7404.
2008. View Article : Google Scholar : PubMed/NCBI
|