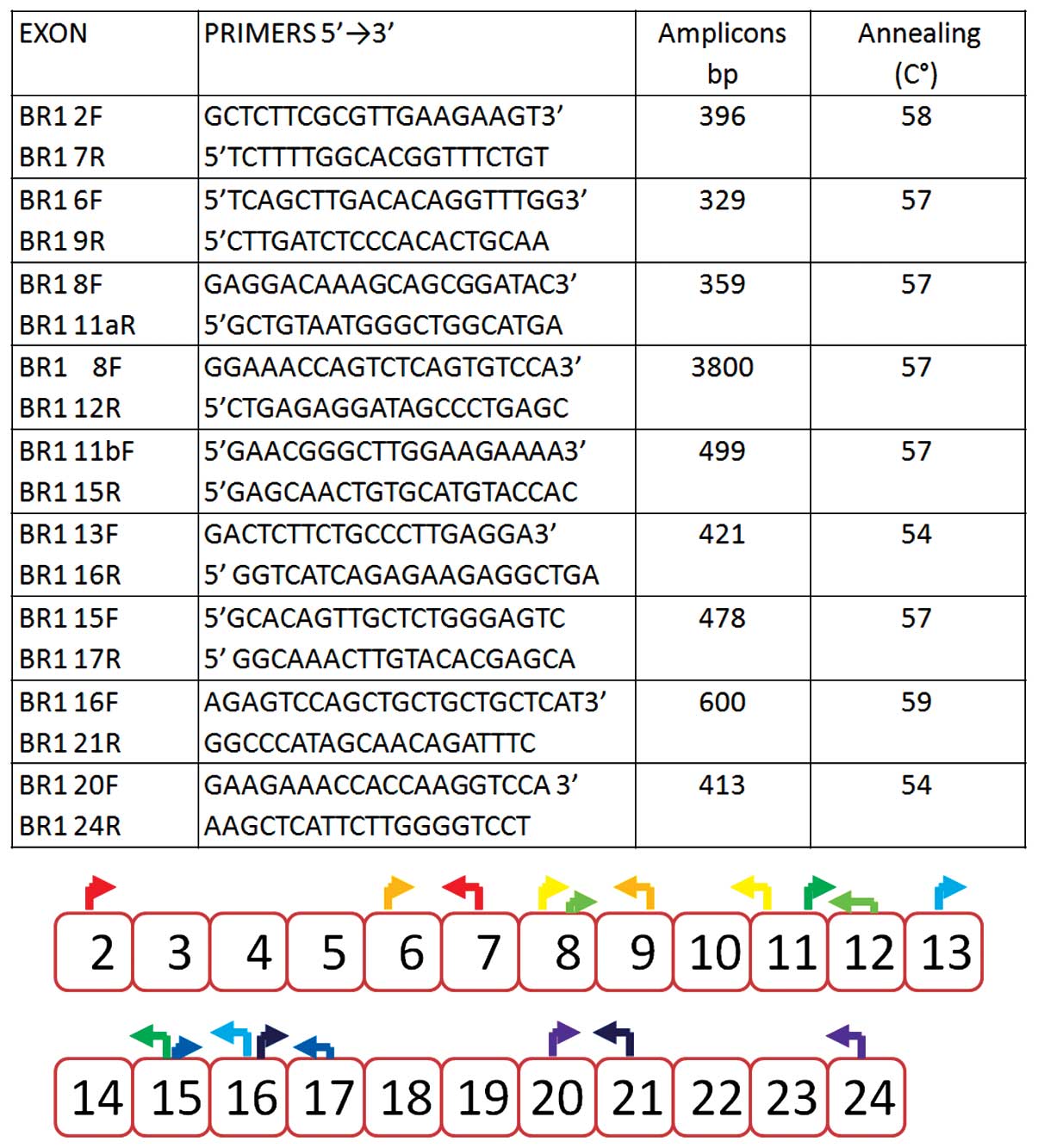
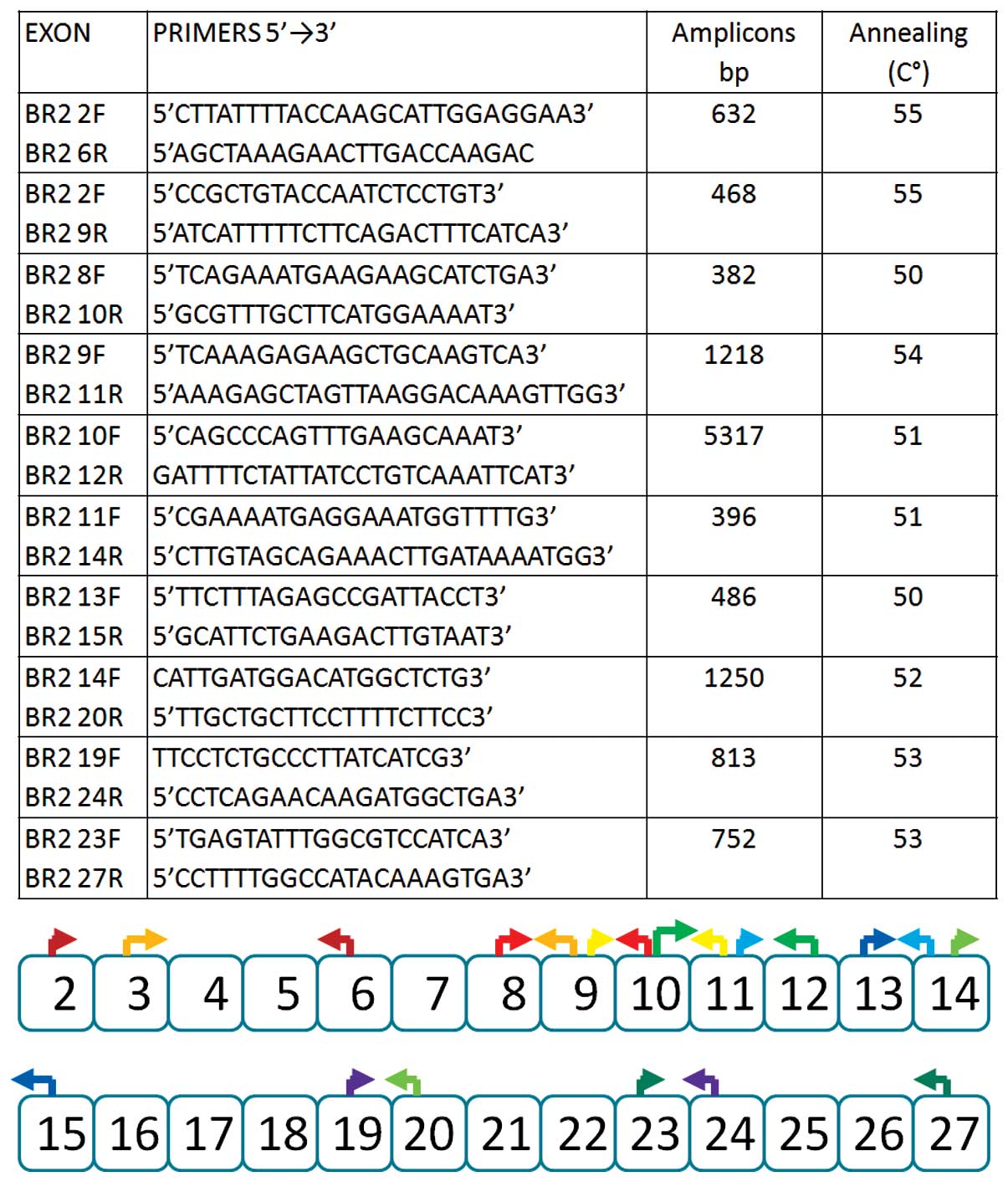
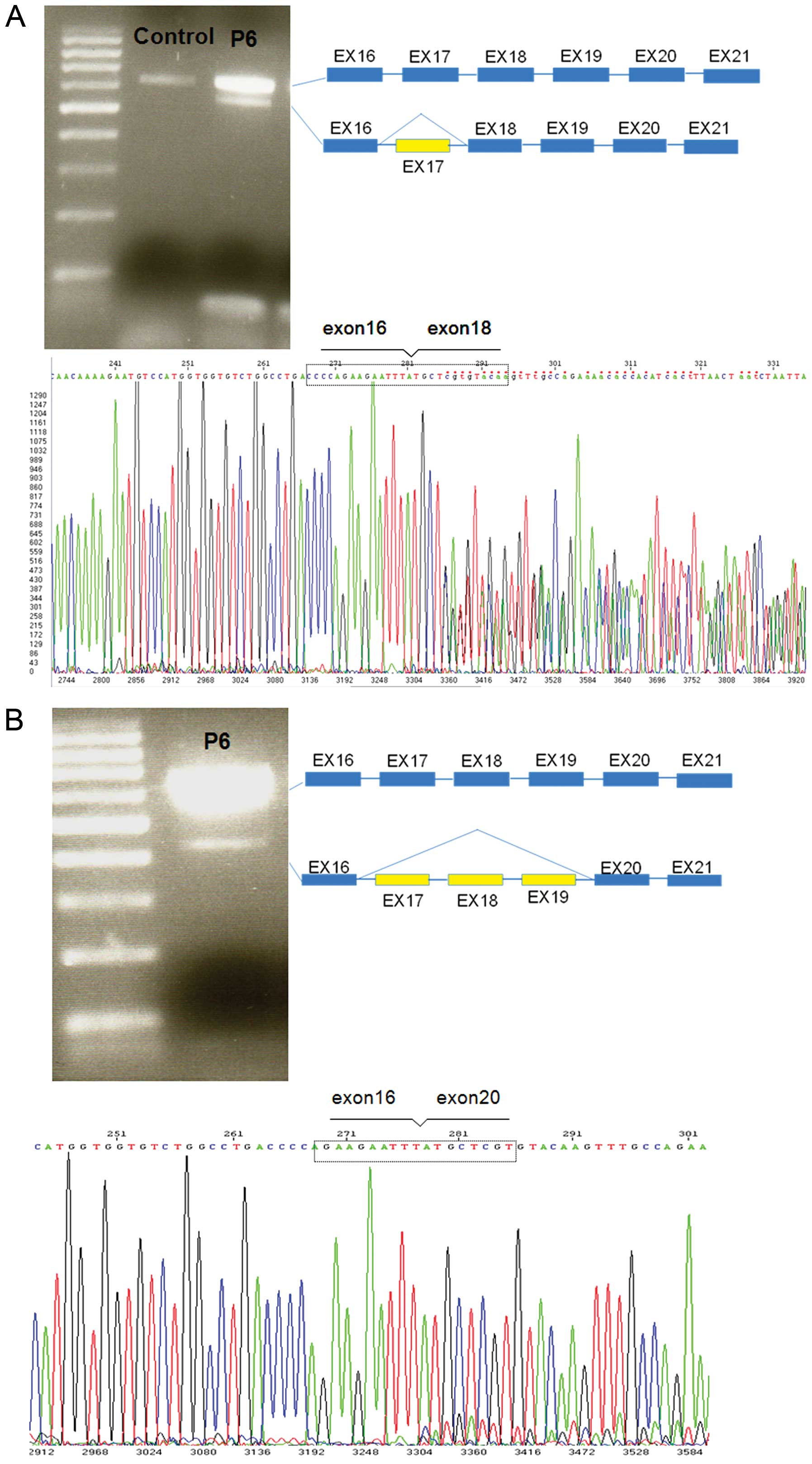
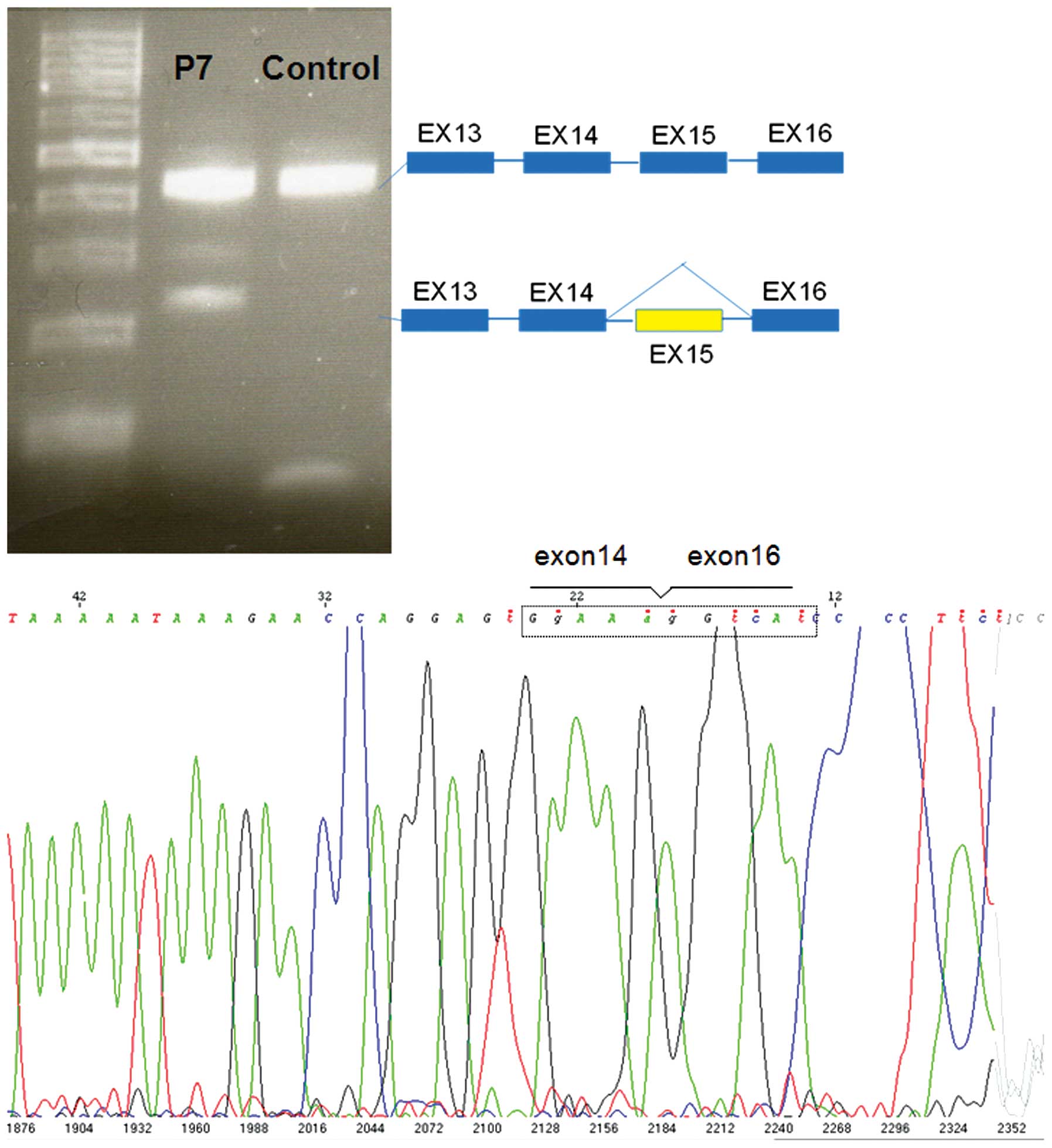
|
1
|
Miki Y, Swensen J, Shattuck-Eidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM,
Ding W, et al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wooster R, Bignell G, Lancaster J, Swift
S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C and Micklem G:
Identification of the breast-cancer susceptibility gene BRCA2.
Nature. 378:789–792. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stratton MR and Rahman N: The emerging
landscape of breast cancer susceptibility. Nat Genet. 40:17–22.
2008. View Article : Google Scholar
|
|
4
|
Sanz DJ, Acedo A, Infante M, Durán M,
Pérez-Cabornero L, Esteban-Cardeñosa E, Lastra E, Pagani F, Miner C
and Velasco EA: A high proportion of DNA variants of BRCA1 and
BRCA2 is associated with aberrant splicing in breast/ovarian cancer
patients. Clin Cancer Res. 16:1957–1967. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chatterjee S and Pal JK: Role of 5′- and
3′-untranslated regions of mRNAs in human diseases. Biol Cell.
101:251–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farrugia DJ, Agarwal MK, Pankratz VS, et
al: Functional assays for classification of BRCA2 variants of
uncertain significance. Cancer Res. 68:3523–3531. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Walker LC, Whiley PJ, Couch FJ, et al:
Detection of splicing aberrations caused by BRCA1 and BRCA2
sequence variants encoding missense substitutions: implications for
prediction of pathogenicity. Hum Mutat. 31:E1484–E1505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cartegni L, Chew SL and Krainer AR:
Listening to silence and understanding non-sense: exonic mutations
that affect splicing. Nat Rev Genet. 3:285–298. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spurdle AB, Couch FJ, Hogervorst FB,
Radice P and Sinilnikova OM: Prediction and assessment of splicing
alterations: implications for clinical testing. Hum Mutat.
29:1304–1313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu M, Conzen SD, Cole CN and Arrick BA:
Characterization of functional messenger RNA splice variants of
BRCA1 expressed in nonmalignant and tumor-derived breast cells.
Cancer Res. 56:4578–4581. 1996.PubMed/NCBI
|
|
11
|
Wang H, Shao N, Ding QM, Cui J, Reddy ES
and Rao VN: BRCA1 proteins are transported to the nucleus in the
absence of serum and splice variants BRCA1a, BRCA1b are tyrosine
phosphoproteins that associate with E2F, cyclins and cyclin
dependent kinases. Oncogene. 15:143–157. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilson CA, Payton MN, Elliott GS, Buaas
FW, Cajulis EE, Grosshans D, Ramos L, Reese DM, Slamon DJ and
Calzone FJ: Differential subcellular localization, expression and
biological toxicity of BRCA1 and the splice variant BRCA1-delta11b.
Oncogene. 14:1–16. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Orban TI and Olah E: Expression profiles
of BRCA1 splice variants in asynchronous and in G1/S synchronized
tumor cell lines. Biochem Biophys Res Commun. 280:32–38. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Orban TI and Olah E: Emerging roles of
BRCA1 alternative splicing. Mol Pathol. 56:191–197. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maniccia AW, Lewis C, Begum N, et al:
Mitochondrial localization, ELK-1 transcriptional regulation and
growth inhibitory functions of BRCA1, BRCA1a, and BRCA1b proteins.
J Cell Physiol. 219:634–641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Colombo M, Blok MJ, Whiley P, et al:
Comprehensive annotation of splice junctions supports pervasive
alternative splicing at the BRCA1 locus: a report from the ENIGMA
consortium. Hum Mol Genet. 23:3666–3680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen M and Manley JL: Mechanisms of
alternative splicing regulation: insights from molecular and
genomics approaches. Nat Rev Mol Cell Biol. 10:741–754.
2009.PubMed/NCBI
|
|
18
|
Tesoriero AA, Wong EM, Jenkins MA, Hopper
JL, Brown MA, Chenevix-Trench G, Spurdle AB and Southey MC;
kConFab: Molecular characterization and cancer risk associated with
BRCA1 and BRCA2 splice site variants identified in multiple-case
breast cancer families. Hum Mutat. 26:4952005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Antoniou AC, Durocher F, Smith P, Simard J
and Easton DF; INHERIT BRCAs program members: BRCA1 and BRCA2
mutation predictions using the BOADICEA and BRCAPRO models and
penetrance estimation in high-risk French-Canadian families. Breast
Cancer Res. 8:R32006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Antoniou AC, Cunningham AP, Peto J, et al:
The BOADICEA model of genetic susceptibility to breast and ovarian
cancers: updates and extensions. Br J Cancer. 98:1457–1466. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Garibay GR, Acedo A, García-Casado Z,
et al: Capillary electrophoresis analysis of conventional splicing
assays: IARC analytical and clinical classification of 31BRCA2
genetic variants. Hum Mutat. 35:53–57. 2014. View Article : Google Scholar
|
|
22
|
Houdayer C, Caux-Moncoutier V, Krieger S,
et al: Guidelines for analysis in molecular diagnosis derived from
a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2
variants. Hum Mutat. 33:1228–1238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Whiley PJ, de la Hoya M, Thomassen M, et
al: Comparison of mRNA splicing assay protocols across multiple
laboratories: recommendations for best practicein standardized
clinical testing. Clin Chem. 60:341–352. 2014. View Article : Google Scholar
|
|
24
|
Montagna M, Santacatterina M, Torri A,
Menin C, Zullato D, Chieco-Bianchi L and D’Andrea E: Identification
of a 3 kb Alu-mediated BRCA1 gene rearrangement in two
breast/ovarian cancer families. Oncogene. 18:4160–4165. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sevcik J, Falk M, Macurek L, et al:
Expression of human BRCA1Delta17-19 alternative splicing variant
with a truncated BRCT domain in MCF-7 cells results in impaired
assembly of DNA repair complexes and aberrant DNA damage response.
Cell Signal. 25:1186–1193. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mazoyer S: Genomic rearrangements in the
BRCA1 and BRCA2 genes. Hum Mutat. 25:415–422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bonnet C, Krieger S, Vezain M, et al:
Screening BRCA1 and BRCA2 unclassified variants for splicing
mutations using reverse transcription PCR on patient RNA and an ex
vivo assay based on a splicing reporter minigene. J Med Genet.
45:438–446. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gutiérrez-Enríquez S, Coderch V, Masas M,
Balmaña J and Diez O: The variants BRCA1 IVS6-1G>A and BRCA2
IVS15+1G>A lead to aberrant splicing of the transcripts. Breast
Cancer Res Treat. 117:461–465. 2009. View Article : Google Scholar
|
|
29
|
Perrin-Vidoz L, Sinilnikova OM,
Stoppa-Lyonnet D, Lenoir GM and Mazoyer S: The nonsense-mediated
mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing
premature termination codons. Hum Mol Genet. 11:2805–2814. 2002.
View Article : Google Scholar : PubMed/NCBI
|