Introduction
MicroRNAs (miRNAs/miRs) are small, non-coding RNAs.
They were first identified as developmental mediators in
Caenorhabditis (C.) elegans (1,2).
It is well known that miRNAs have critical roles in the regulation
of gene expression. They are initially transcribed as long RNAs and
are then processed by two RNase complexes, Drosha and Dicer, into
22-nucleotide duplexes. These duplexes are loaded into RNA-induced
silencing complexes (3,4). The effect of miRNA-mediated
modulation of gene expression has been documented across the animal
kingdom during numerous steps of neuronal development, from early
neurogenesis to synaptogenesis (5–8).
Abundant and diverse miRNA expression has also been reported in the
central nervous system (9–11).
The presence of three copies of all or part of human
chromosome 21 (Hsa21) results in the constellation of physiological
traits known as the Down syndrome (DS), also called trisomy 21
(12). Bioinformatic analysis has
demonstrated that Hsa21 harbors five miRNA genes, miR-99a, let-7c,
miR-125b-2, miR-155 and miR-802 (12,13). miRNA expression profiling, miRNA
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR), and miRNA in situ hybridization experiments have
shown that the expression of these miRNAs is markedly higher in
fetal brain and heart specimens from individuals with DS than that
in samples from age- and gender-matched controls (12,14,15). miR-125b, a homolog of lin-4, was
first discovered in C. elegans, in which it regulates
developmental timing (1). Ectopic
expression of miR-125b can increase the relative number of
differentiated SH-SY5Y cells that show neurite outgrowth (16). miR-125b is upregulated during the
differentiation of human neural progenitor ReNcell VM cells, and
high levels of miR-125b have been shown to promote neurite
outgrowth in these cells (16).
miR-125b also affects dendritic spine morphology. NR2A, which is a
subunit of NMDA receptors and affects synaptic plasticity, is a
target of miR-125b (17). In
hippocampal neurons, NR2A expression is negatively regulated
through its 3′-untranslated region by fragile X mental retardation
1, miR-125b and argonaute 1 (17). In poorly differentiated cerebellar
granule cell progenitors (GCPs), miR-125b-2 is downregulated, but
it promotes GCP differentiation and antagonizes the effects induced
by sonic hedgehog (Shh) via targeting activating components of the
Hh signaling pathway (18). The
present study was performed to assess the association of miR-125b
with the nervous system.
Two recent studies have demonstrated the
contribution of miR-125b to early neuronal development in embryos
(19,20). These studies used the mouse
embryonic stem cell (mESC) lines R1 mESCs or E14Tg2a as a model to
demonstrate that miR-125b is associated with a specific step during
neural differentiation of mESCs. Ectopical expression of miR-125b
did not affect the self-renewal of undifferentiated ESCs. However,
the expression of a number of miRNAs changed significantly during
ESC differentiation, among which miR-125b showed a marked reduction
as compared with that in the control. Another study from 2012
showed that overexpression of miR-125b did not affect the ectoderm
and neuron differentiation in mESCs (19), which was in contrast with a study
from 2013, which reported that the ectopic expression of miR-125b
blocked ESC differentiation at the epiblast stage (20). Furthermore, exploration of the
targets of miR-125b led to the discovery of two distinct targets,
Lin28 and Dies1 (19,20).
The present study investigated the cellular function
of the overexpression of miR-125b-2 in mESCs. Stable
miR-125b-2-expressing mESC lines were established, and it was shown
that the ectopic expression of miR-125b-2 did not affect the
self-renewal and proliferation of mESCs. To elucidate the
underlying mechanism and the function of miR-125b-2 in the neuronal
differentiation of ESCs, ESC-specific germ layer markers
characteristic for endoderm, ectoderm and mesoderm were assessed in
embryoid bodies. The findings of the present study highlighted an
important role of miR-125b-2 in the regulation of ESC germ layer
differentiation and revealed a novel mechanism for cell lineage
determination and neuronal differentiation.
Materials and methods
Cell culture
The mouse ESC line (mESC), E14Tg2a (American Type
Culture Collection, Manassas, VA, USA), was kindly provided by
Professor Ping Li (Key Laboratory of Molecular Medicine, Fudan
University, Shanghai, China). Cells were maintained on feeder-free,
gelatin-coated plates (Gibco-BRL, Invitrogen Life Technologies,
Carlsbad, CA, USA) in the following medium: Dulbecco’s modified
Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA)
supplemented with 2 mM glutamine, 100 U/ml penicillin/streptomycin,
1 mM sodium pyruvate (all from Invitrogen Life Technologies), 1 mM
non-essential amino acids (Invitrogen Life Technologies), 0.1 mM
l-mercaptoethanol
(Sigma-Aldrich, St. Louis, MO, USA), 15% fetal bovine serum (FBS;
Thermo Fisher Scientific), and 103 U/ml leukemia
inhibitory factor (LIF; Millipore, Billerica, MA, USA). The 293T
cells were obtained from Professor Ping Li were cultured in
high-glucose DMEM supplemented with 10% FBS at 37°C, with 5%
CO2 for maintenance.
Plasmid constructs, viral packaging and
ESC transfection
Mouse genomic DNA was purified from the mESC line,
E14Tg2a, using GenElute™ Mammalian Genomic DNA Miniprep Kits
(Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s
insrtuctions. The coding regions of mouse miR-125b-2 were amplified
by polymerase chain reaction (PCR) of mouse genomic DNA. [PCR
reactions were performed in a total volume of 50 µl
consisting of 1 µl of mouse genomic DNA, 10 µl of 5X
Prime STAR™ Buffer, 4 µl dNTP Mixture (2.5 mM), 1 µl
of each primer (10 µM), and 0.5 µl Prime STAR™ HS DNA
Polymerase (2.5 U/µl) (Takara Bio, Inc., Dalian, China). PCR
amplifications were carried out on a ThermoHybaid PCR express
(Thermo Fisher Scientific, Waltham, MA, USA) and PCR products were
analyzed by electrophoresis on a 2.0% agarose gel (Biowest, Spain)
containing 0.5 µg/ml of ethidium bromide. Gel images were
captured and analyzed using the Quantity One System (Bio-Rad,
Hercules, USA)]. They were inserted into the AgeI and
EcoRI sites of the pLKO.1 vector. pLKO.1-miR-125b-2
lentiviral vectors combined with packaging plasmids, pMD2.G and
psPAX2, were co-transfected into 293T cells using Lipofectamine
2000 reagent (Invitrogen Life Technologies) according to the
manufacturer’s instructions. All plasmids, such as, pLKO.1, pMD2.G
and psPAX2 were kindly provided by Professor Ping Li (Key
Laboratory of Molecular Medicine, Fudan University).
Virus-containing supernatant was collected 48 h after transfection
and filtered through 0.45-µm filters (Millipore). ESCs were
incubated in the virus supernatant supplemented with 4 mg/ml
polybrene (Sigma-Aldrich, St. Louis, MO, USA) for 48 h and then the
cells were re-plated in fresh mESC culturing medium. Puromycine
(Sigma-Aldrich, St. Louis, MO, USA) was added at a final
concentration of 2 mg/ml and resistant colonies were selected after
1 week. Pure lentivirus served as a negative control.
Cell proliferation assays
Cell proliferation was evaluated using the Cell
Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan)
according to manufacturer’s instructions. Cells at 12 h
post-transfection were seeded into 96-well plates at 5,000
cells/well. Following 24, 48, 72, 96 and 120 h of transfection, 10
µl CCK-8 solution was added to each well. The plate was
incubated for 1–4 h in a humidified CO2 incubator at
37°C and the absorbance was measured at 450 nm using a Model 680
microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Embryoid body culture
mESC differentiation was induced by transferring
~1,000 cells in 15 µl differentiation medium onto the lid of
a 100-mm dish. The cells were cultured for nine days as a hanging
drop to facilitate the formation of embryoid bodies (EBs). Each
dish contained ~80 embryoid bodies. These were cultured in
differentiation medium containing DMEM supplemented with 2 mM
glutamine, 100 U/ml penicillin/streptomycin, 1 mM sodium pyruvate,
1 mM non-essential amino acids, 0.1 mM l-mercaptoethanol and 15% FBS. EB
medium was changed every other day.
Neuronal differentiation
The mESCs were grown for four days to form
unattached EBs in differentiation medium containing DMEM
supplemented with 2 mM glutamine, 100 U/ml penicillin/streptomycin,
1 mM sodium pyruvate, 1 mM nonessential amino acids, 0.1 mM
L-mercaptoethanol and 15% FBS. After 4 days of embryoid body
formation the cells were treated with 1 µM
all-trans-retinoic acid (Sigma-Aldrich, Buchs, Switzerland)
for an additional 4 days. These EBs were digested and transferred
to poly-d-lysine/laminin-coated tissue
culture dishes (Sigma-Aldrich, St. Louis, MO, USA). The cells were
then incubated in DMEM with 10% heat-inactivated FBS to induce
neuronal differentiation.
RNA extraction and reverse transcription
quantitative PCR (RT-qPCR)
Total RNA from mESCs was isolated using the TRIzol
reagent (Invitrogen Life Technologies) according to the
manufacturer’s instructions, and the concentration was determined
by the ratio of the absorbance at 260 to that at 280 nm using a
NanoDrop® ND-1000 spectrophotometer (Thermo Fisher
Scientific). To measure the content of miR-125b-2, 500 ng total RNA
was poly-A tailed and reverse transcribed to cDNA using an
All-in-One™ miRNA qRT-PCR Detection kit (Cat. no. AOMD-Q050;
GeneCopoeia Inc., Rockville, MD, USA) according to the
manufacturer’s instructions. Real-time PCR was then performed using
an ABI7300 Real-Time PCR System (Applied Biosystems, Foster City,
CA, USA) with miRNA-specific forward and reverse primers. Each
reaction was performed with 2 µl template cDNA, 10 µl
2X All-in-One qPCRMix, 2 µl of each primer (2 µM),
0.4 µl 50X ROX Reference Dye, and water to adjust to a final
volume of 20 µl. All reactions were incubated on a 96-well
plate at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec,
65°C for 20 sec and 72°C for 10 sec. Statistical analysis was
performed using SDS software version 1.4.1 (Applied Biosystems).
For the analyses of marker genes of mESCs, EBs and neurons, RNA (1
µg) of each sample was used for reverse transcription with
the Prime Script® RT reagent kit (Takara Bio, Inc.)
using Oligo(dT) Primer at 37°C for 15 min, followed by 85°C for 5
sec. The amplified cDNA was quantified using SYBR®
Premix Ex Taq™ (DRR041A; Takara Bio, Inc.) according to the
manufacturer’s instructions. Each reaction was performed with 2
µl template cDNA, 10 µl 2X SYBR Premix Ex Taq. 0.4
µl of each primer (10 µM), 0.4 µl 50X ROX
Reference Dye, and water to adjust to a final volume of 20
µl. All reactions were incubated on a 96-well plate at 95°C
for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 31
sec. Real-time PCR was then performed using the same qPCR apparatus
and statistical analysis was performed using the same software. The
resulting cDNA was then amplified by qPCR using the primers listed
in Table I. The housekeeping
genes U6 and the GAPDH were used to normalize the samples, using
the 2−∆∆Ct method. All primers were obtained from
GeneCopoeia.
 | Table IPrimers used for reverse
transcription quantitative polymerase chain reaction. |
Table I
Primers used for reverse
transcription quantitative polymerase chain reaction.
| Name | Forward primer
(5′→3′) | Reverse primer
(5′→3′) |
|---|
| Oct4 |
CTGAGGGCCAGGCAGGAGCACGAG |
CTGTAGGGAGGGCTTCGGGCACTT |
| Nanog |
AGGGTCTGCTACTGAGATGCTCTG |
CAACCACTGGTTTTTCTGCCACCG |
| Klf4 |
CAAGTCCCCTCTCTCCATTATCAAGAG |
CCACTACGTGGGATTTAAAAGTGCCTC |
| Rex1 |
AAGCCGTATCAGTGCACGTTCGAAGGCT |
ATGCGTGTATCCCCAGTGCCTCTGTCAT |
| Gata6 |
TTGCTCCGGTAACAGCAGTG |
GTGGTCGCTTGTGTAGAAGGA |
| Foxa2 |
CCCTACGCCAACATGAACTCG |
GTTCTGCCGGTAGAAAGGGA |
| Nestin |
CTGAGAACTCTCGCTTGCAGACA |
GGAAATGCAGCTTCAGCTTGG |
| Foxf1 |
CGGAGAAGCCGCCCTACT |
GCGCGCCTGAGAAACTG |
| Brachyury |
GCGGGAAAGAGCCTGCAGTA |
TTCCCCGTTCACGTACTTCC |
| Map2 |
CATCGCCAGCCTCGGAACAAACAG |
TGCGCAAATGGAACTGGAGGCAAC |
| U6 |
CTCGCTTCGGCAGCACA |
CTCGCTTCGGCAGCACA |
| GAPDH |
GTATGACTCCACTCACGGCAAA |
TTCCCATTCTCGGCCTTG |
Immunofluorescence analysis
Cells were permeabilized with 0.25% Triton X-100
(Sigma-Aldrich, St. Louis, MO, USA) for 10 min at 37°C and then
fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA)
in phosphate-buffered saline (PBS) for 15 min at room temperature.
The fixed cells were blocked for 20 min with PBS containing 5%
bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO, USA).
Next, the cells were incubated for 16 h at 4°C with mixtures
containing primary antibodies specific to the ESC markers, Nanog
(1:100; species raised in: rabbit; specificity: rat, human and
mouse; monoclonal antibody; Millipore, AB5731), octamer-binding
transcription factor 4 (Oct4; 1:100; species raised in: mouse;
specificity: human and mouse; monoclonal antibody; Millipore,
MABD76) and sex-determining region Y-box 2 (Sox2; 1:100; species
raised in: mouse; specificity: human and mouse; monoclonal
antibody; Millipore, MAB4343) in PBS containing 1% BSA. The cells
were washed three times with PBS containing 1% BSA. As the
secondary antibody, goat anti-mouse immunoglobulin G conjugated to
fluorescein isothiocycanate (Sigma-Aldrich, St. Louis, MO, USA) was
applied at dilutions of 1:500. The Hoechst 33342 reagent
(Sigma-Aldrich, St. Louis, MO, USA) was used to detect the nuclei
in cells. After washing with PBS for three times, the cells were
analyzed using a confocal scanning laser fluorescence microscope
(Model FV300; Olympus, Tokyo, Japan).
Statistical analysis
Values are expressed as the mean ± standard error of
three independent experiments. Statistical significance of
differences was calculated using Prism software (version 4.0a;
GraphPad Software, Inc., La Jolla, CA, USA) by one-way analysis of
variance. P<0.05 was considered to indicate a statistically
significant difference between values.
Results
miR-125b-2 overexpression does not affect
the pluripotency and self-renewal of mESCs
To determine the function of miR-125b-2 in the
maintenance of pluripotency and self-renewal, ESCs that
overexpressed miR-125b-2 were established by transfection with a
pLKO.1 lentiviral expression vector. According to addgene
(http://www.addgene.org), the plasmid psPAX2
produces a higher titer than pCMV-dR8.2 dvpr and contains a robust
CAG promoter for efficient expression of packaging proteins
(21–23). Therefore, a lentiviral pLKO.1
vector containing an miR-125b-2 expression vector, a psPAX2
packaging vector and a pMD2.G envelope vector were combined at a
ratio of 4:3:1. Twenty-four hours after transfection, the cells
were selected using puromycin. After one week, the overexpression
of miR-125b-2 was verified by RT-qPCR. The results showed that the
expression levels of miR-125b-2 in mESCs were 36 times higher than
those in the controls (Fig. 1A),
which indicated the successful establishment of stably
miR-125b-2-expressing mESCs. Next, to determine the effects of
miR-125b-2 on the self-renewal of mESCs, the expression of the mESC
markers Klf4, Nanog, Rex1 and Oct4 was detected by RT-qPCR. There
were no significant differences in the expression levels of
self-renewal markers between miR-125b-2-transfected cells and
control cells (Fig. 1B)
Furthermore, Nanog, Sox2 and Oct4 were detected by
immunohistochemistry. Fluorescence microscopy showed that all four
markers remained detectable in miR-125b-2-transfected cells.
Phase-contrast images of the morphology of the colonies in the
presence of LIF showed no obvious differences in morphology between
miR-125b-2-transfected cells and control cells (Fig. 1C). These results indicated that
the overexpression of miR-125b-2 had no influence on the
maintenance of mESCs.
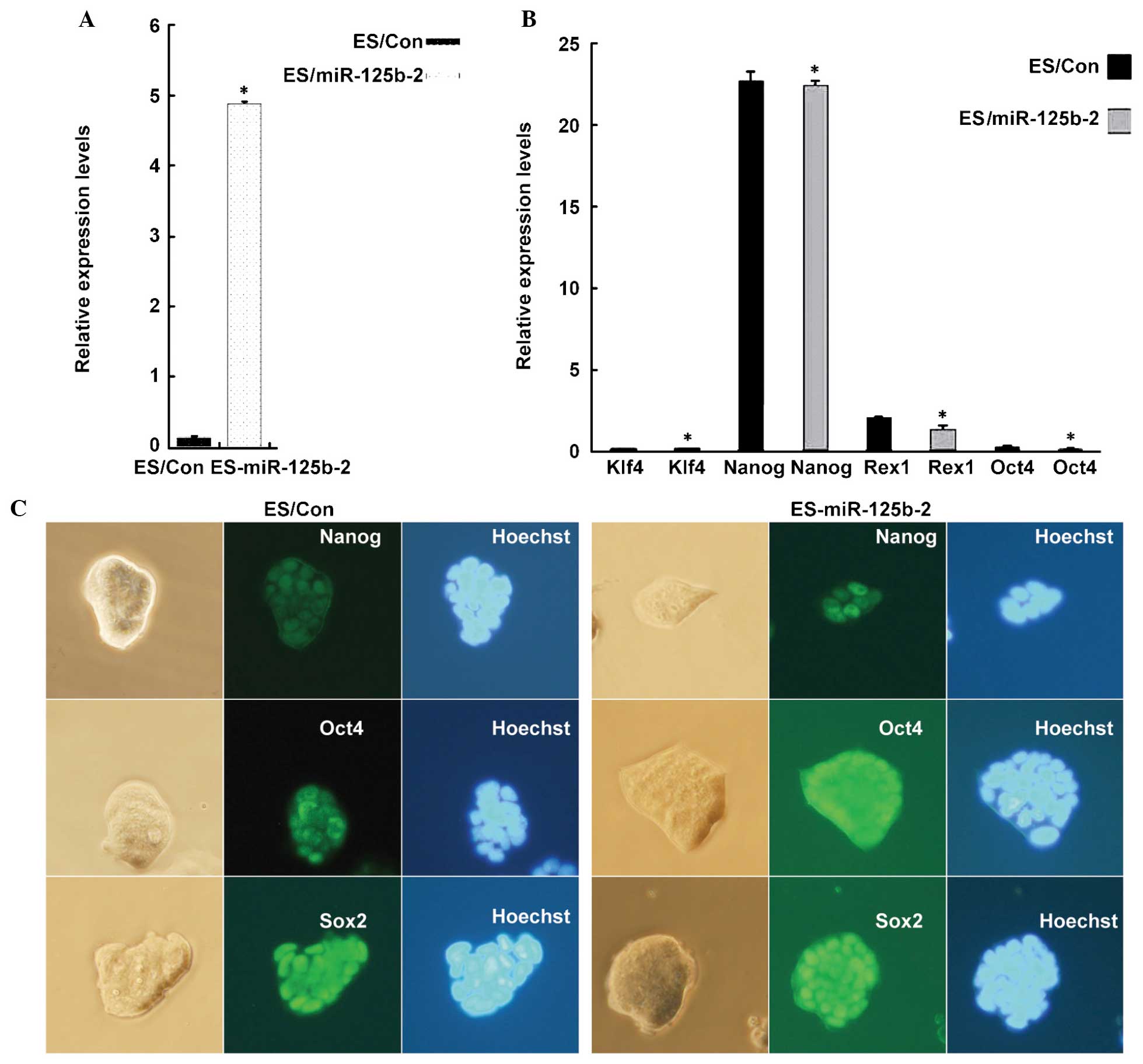 | Figure 1miR-125b-2 does not affect the
pluripotency of ESCs. (A) RT-qPCR analysis of miR-125b-2 expression
in ESCs transfected with empty vector and miR-125b-2. U6 was used
as loading control. (B) RT-qPCR analysis of self-renewal marker
gene expression. GAPDH was used as loading control. Groups: ES/Con,
empty vector-transfected ESCs; ES/miR-125b-2,
miR-125b-2-transfected ESCs. Values are expressed as the mean ±
standard error of data from one representative of three
experiments, performed in triplicate. *P<0.05
compared with the ES/Con group. (C) Phase contrast microscopy
images of miR-125b-2-transfected (right) and control (left) cells.
Immunofluorescence staining for the stem cell pluripotency markers
Nanog, Oct4 and Sox2. Cell nuclei were stained by Hoechst. RT-qPCR,
reverse transcription quantitative polymerase chain reaction; ESCs,
embryonic stem cells; miR, microRNA; Sox2, sex-determining region
Y-box 2; Oct4, octamer-binding transcription factor 4; KLF4,
kruppel-like factor 4. |
miR-125b-2 overexpression does not
promote ESC proliferation
miRNAs have important roles in living organisms and
regulate stem cell proliferation (24). To investigate the biological
effects of miR-125b-2 on ESC proliferation, CCK-8 was added at
various time-points (24, 48, 72, 96 and 120 h) after transfection
(25,26). Overexpression of miR-125b-2 did
not significantly stimulate the growth of mESCs as compared with
that in the controls (P<0.05) (Fig. 2), which implied that
miR-125b-2-overexpression had no distinct effect on ESC
proliferation.
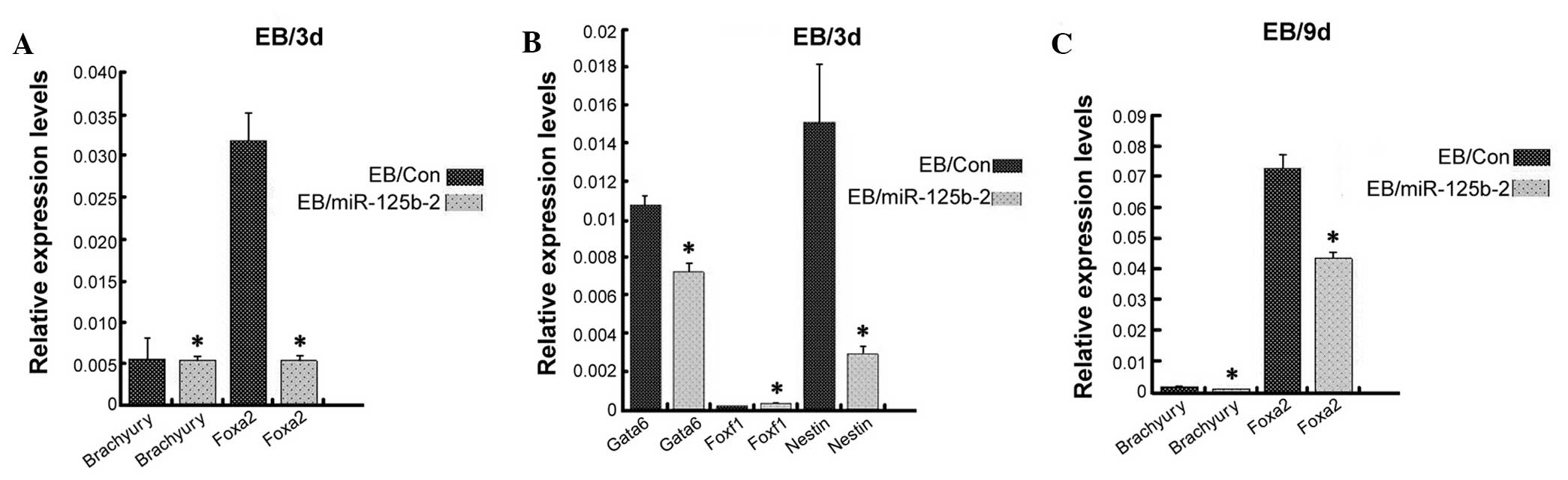
miR-125b-2 overexpression inhibits the
differentiation of mESCs into endoderm and ectoderm, but not
mesoderm
The self-renewal capacity and differentiation
potential are hallmarks of stem cells (24). In the past few years, miR-125b was
shown to be an important factor involved in stem cell development
by regulating the differentiation of stem cells (19,20,27). To evaluate the effect of
miR-125b-2 on the direction of ESC differentiation, transfected and
control ESCs were cultured in suspension for four days to form EBs
(28). RT-qPCR analysis was
performed to detect markers of endoderm, ectoderm and mesoderm on
day-3 and day-9 EB cells. The levels of Foxa2 and Gata6 (29,30), which are expressed by all
extra-embryonic endodermal cells, were significantly decreased in
miR-125b-2-overexpressing EBs compared with those in the control
EBs (P<0.05) (Fig. 3). The
ectoderm marker Nestin (30,31) was also significantly decreased in
miR-125b-2 transfectants compared with that in the control EBs
(P<0.05) (Fig. 3B). However,
there were no differences in the expression levels of the mesoderm
markers Brachyury and Foxf1 (32)
(Fig. 3). These results suggested
that miR-125b-2 overexpression suppressed the differentiation of
mESCs into endoderm and ectoderm, while there was no obvious
influence on the mesodermal differentiation of ESCs.
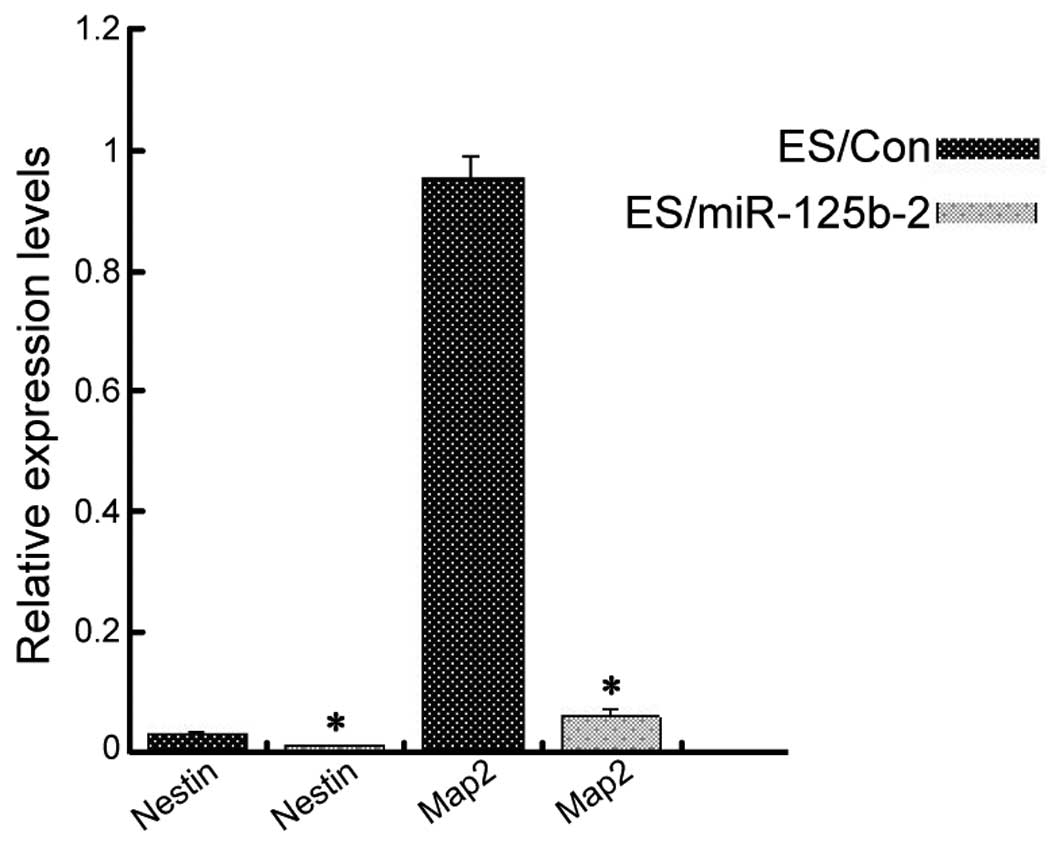
miR-125b-2 overexpression reduces neural
progenitor differentiation
Ectoderm is one of the three classic germ layers in
the early mouse embryo, with the capacity to develop into the
central nervous system (33). In
order to determine the impact of miR-125b-2 overexpression on the
nervous system, RA was used to induce EB differentiation into
neuronal cells (34). ESCs were
first cultured in suspension for four days to form EBs. RA was then
added to the medium and the cells were incubated for another four
days. The cells were then adherently cultured for 3–6 days. The
neuron-specific markers Nestin and Map2 were analyzed using RT-qPCR
(35). There was a significant
decrease in the expression of the two neuronal markers in the
ES/miR-125b-2 group as compared with those in the control group
(Fig. 4). These results suggested
that downregulation of miR-125b-2 may be required to induce the
differentiation of ESCs.
Discussion
The present study showed that miR-125b-2 has an
important role in mouse embryonic stem cells (mESCs) by inhibiting
the differentiation of mESCs into endoderm and ectoderm without
affecting their proliferation, mesodermal differentiation and
self-renewal. Functional genetic studies on EBs treated with RA
further indicated that miR-125b-2 overexpression impaired neuron
development. The present study demonstrated that it is necessary to
further investigate the regulatory mechanisms of the effects of
miR-125b on the self-renewal and differentiation of ESCs.
A stably miR-125b-2-overexpressing mESC line E14Tg2A
was established by transfection with an miR-125b-2 expression
lentivirus. RT-qPCR analysis confirmed that miR-125b-2 was
expressed in undifferentiated mESCs (Fig. 1A); however, it has remained
impossible to identify the expression levels without the threshold.
For example, Tarantino et al (36) reported that miR-125b is
undetectable in undifferentiated cells and is induced upon
differentiation in the two mESC lines E14Tg2A and MPI; however, it
was not detectable in R1 mESCs by microRNA array. By contrast, Wang
et al (19) reported that
miR-125b expression was detected in R1 mESCs using microRNA array
screening. They further showed that miR-125b is highly enriched in
undifferentiated mESCs as compared with other expressed miRNAs,
while it is markedly downregulated during early ESC differentiation
(19). Solozobova and Blattner
(37) have shown that expression
of miRNA-125b-2 during the process of EB formation is significantly
lower in all mESC lines (R1, D3 and CGR8) than that in
differentiated cells. The expression of miR-125b-2 in brains of
children with DS was found to be 1.5 times higher than that in
normal brains (14). In agreement
with these data, miR-125b-2 was shown to be highly expressed in
numerous adult mouse tissue types. The discrepancies among the
abovementioned previous studies may be due to the different ESC
lines and differentiation protocols used. The present study used RA
to induce differentiation, as its effects are similar to natural
early embryonic development (38). The differentiation protocol used
in the present study induces slow and more physiological
differentiation which mainly affected neurons (14).
The results of the present study demonstrated that
miR-125b is essential for the proper differentiation of ESCs, which
is consistent with the results recently observed in mESCs (19,20). In the present study, the
expression of four ESC self-renewal markers was found to be similar
among miR-125b-2-overexpressing and control cells, and no major
change in morphology was observed among them. Of note, a proportion
of miR-125b-overexpressing cells were resistant to differentiation
into endoderm and ectoderm. This regulatory role of miR-125b was
confirmed by the observations of previous studies, which reported
that the downregulation of miR-125b is required for the initiation
of ESC differentiation (19,20). In addition, it is known that
miRNAs have important roles in cell cycle regulation of ESCs
(39). Compared with somatic
cells, ESCs are characterized by a cell cycle with a shortened G1
phase as an adaptation to the rapid growth during early embryonic
development. Furthermore, the present study tested the
proliferation of miR-125b-2-overexpressing mESCs using the CCK-8
assay. miR-125b-2 was found to have no significant effect on the
proliferation of mESCs. A previous screening-based study, which
examined the effect of 461 individually re-introduced miRNAs on the
proliferation of DGCR8-null cells showed that the defective
proliferation was rescued by 14 different miRNAs, including
miR-290, miR-302 and the miR-17-92 cluster (40 and refs therein).
These findings showed that miR-125b may not be an ESC-specific cell
cycle-regulating miRNA. Therefore, these observations, together
with the findings of the present study, suggested a distinctive
role of miR-125b in the lineage commitment of ESCs as well as
tissue/organ generation.
The results of the present study further showed that
miR-125b is acts as a regulator of ESC-specific germ layer
commitment. Wang et al (19) reported that ectopically expressed
miR-125b-2 can impair the expression of endoderm marker genes,
which is consistent with these results. It has been demonstrated
that the endoderm forms the respiratory and digestive tracts, which
has implications for diseases of the endoderm, including cystic
fibrosis and cancer (41). In
contrast to the marked inhibitory role of miR-125b on endodermal
and ectodermal differentiation, the present study has shown that
overexpression of miR-125b-2 did not affect the expression of
mesoderm-associated markers and therefore the mesodermal
differentiation of ESCs. Furthermore, the ectoderm forms the
central nervous system (41);
therefore the decreased RA-induced differentiation of ESCs into
neurons following overexpression of miR-125b-2, as indicated by
reduced levels of neuroectodermal markers, was in line with the
decreases in ectodermal differentiation. However, other studies
have shown that miR-125b-2 promote neuronal differentiation of
SH-SY5Y, GCP and P19 cells as well as hippocampal neurons (16–18). Boissart et al (42) showed that miR-125b-2 potentiated
early neuronal specification of human embryonic stem cells (hESCs).
hESC neurons were induced by N2B27 medium supplemented with
fibroblast growth factor 2. Wu and Belasco (43) produced similar results using a
specific non-mESC line, mouse P19 embryonal carcinoma cells.
Differences in experimental protocols may in part explain the
differences between the results of the present study and those of
previous studies. The discrepant conclusions may also be explained
by the limitations of the methods of the present study regarding
mESC differentiation in vitro. Thus, it cannot be excluded
that miR-125b also regulates ectoderm formation and neural
differentiation, which therefore requires further study.
Of note, miR-125b was shown to target Lin28 in
cardiac differentiation, whereas it targets Dies1 in neuronal
differentiation (20). A
bioinformatics study and a luciferase reporter assay have been
performed in our group and will be published shortly. The
preliminary results showed that miR-125b inhibited the expression
of at least five genes, among which at least two genes were
associated with nervous system development (data not shown). In
addition, miR-125a was shown to be essential for the proper
differentiation of ESCs (44),
which suggested that at least two miRNAs work cooperatively to
inhibit Dies1. It is well known that redundancy characterizes miRNA
functions: In most cases, one single miRNA targets a number of
mRNAs and, on the other hand, one mRNA is often targeted by a
number of miRNAs (45). Whether
miR-125b coordinately targets Dies1 and other genes requires
further investigation.
In conclusion, the results of the present study
indirectly demonstrated that miR-125b is required for the
initiation of ESC differentiation. It negatively regulates
endodermal and ectodermal differentiation and terminal
differentiation of neurons, while not affecting mesodermal
differentiation. Therefore, the ongoing identification of novel
targets of miR-125b will further elucidate the molecular mechanisms
of ESC differentiation and may provide tools to direct ESC
differentiation toward specific lineages.
Acknowledgments
The authors would like to thank Professor Ping Li
(Key Laboratory of Molecular Medicine, Fudan University, Shanghai,
China) for providing the E14Tg2a cells. This study was supported by
grants from the National Natural Science Foundation of China (no.
81371269), and the Shanghai Scientific and Technology Committee,
China (nos. 14140902600 and 14DJ1400103).
References
|
1
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ketting RF, Fischer SE, Bernstein E, Sijen
T, Hannon GJ and Plasterk RH: Dicer functions in RNA interference
and in synthesis of small RNA involved in developmental timing in
C. elegans. Genes Dev. 15:2654–2659. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vo N, Klein ME, Varlamova O, Keller DM,
Yamamoto T, Goodman RH and Impey S: A cAMP-response element binding
protein-induced microRNA regulates neuronal morphogenesis. Proc
Natl Acad Sci USA. 102:16426–16431. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wayman GA, Davare M, Ando H, Fortin D,
Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman
RH, et al: An activity-regulated microRNA controls dendritic
plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA.
105:9093–9098. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schratt GM, Tuebing F, Nigh EA, Kane CG,
Sabatini ME, Kiebler M and Greenberg ME: A brain-specific microRNA
regulates dendritic spine development. Nature. 439:283–289. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel G, Obernosterer G, Fiore R, Oehmen
M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch
CJ, Kane C, et al: A functional screen implicates
microRNA-138-dependent regulation of the depalmitoylation enzyme
APT1 in dendritic spine morphogenesis. Nat Cell Biol. 11:705–716.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim J, Krichevsky A, Grad Y, Hayes GD,
Kosik KS, Church GM and Ruvkun G: Identification of many microRNAs
that copurify with polyribosomes in mammalian neurons. Proc Natl
Acad Sci USA. 101:360–365. 2004. View Article : Google Scholar :
|
|
10
|
Kosik KS: The neuronal microRNA system.
Nat Rev Neurosci. 7:911–920. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Krichevsky AM, King KS, Donahue CP,
Khrapko K and Kosik KS: A microRNA array reveals extensive
regulation of microRNAs during brain development. RNA. 9:1274–1281.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Elton TS, Sansom SE and Martin MM:
Trisomy-21 gene dosage over-expression of miRNAs results in the
haploinsufficiency of specific target proteins. RNA Biol.
7:540–547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Y, Li W, Liu X, Chen H, Tan K, Chen Y,
Tu Z and Dai Y: Identification of dysregulated microRNAs in
lymphocytes from children with Down syndrome. Gene. 530:278–286.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klusmann JH, Li Z, Böhmer K, Maroz A, Koch
ML, Emmrich S, Godinho FJ, Orkin SH and Reinhardt D: miR-125b-2 is
a potential oncomiR on human chromosome 21 in megakaryoblastic
leukemia. Genes Dev. 24:478–490. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bian S and Sun T: Functions of noncoding
RNAs in neural development and neurological diseases. Mol
Neurobiol. 44:359–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um
M, Udolph G, Yang H, Lim B and Lodish HF: MicroRNA-125b promotes
neuronal differentiation in human cells by repressing multiple
targets. Mol Cell Biol. 29:5290–5305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edbauer D, Neilson JR, Foster KA, Wang CF,
Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA and Sheng M:
Regulation of synaptic structure and function by FMRP-associated
microRNAs miR-125b and miR-132. Neuron. 65:373–384. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ferretti E, De Smaele E, Miele E, Laneve
P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E,
Screpanti I, et al: Concerted microRNA control of Hedgehog
signalling in cerebellar neuronal progenitor and tumour cells. EMBO
J. 27:2616–2627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Cao N, Yuan M, Cui H, Tang Y, Qin
L, Huang X, Shen N and Yang HT: MicroRNA-125b/Lin28 pathway
contributes to the mesendodermal fate decision of embryonic stem
cells. Stem Cells Dev. 21:1524–1537. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Battista M, Musto A, Navarra A, Minopoli
G, Russo T and Parisi S: miR-125b Regulates the Early Steps of ESC
Differentiation through Dies1 in a TGF-Independent Manner. Int J
Mol Sci. 14:13482–13496. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alexopoulou AN, Couchman JR and Whiteford
JR: The CMV early enhancer/chicken beta actin (CAG) promoter can be
used to drive transgene expression during the differentiation of
murine embryonic stem cells into vascular progenitors. BMC Cell
Biol. 9:22008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen CM, Krohn J, Bhattacharya S and
Davies B: A comparison of exogenous promoter activity at the ROSA26
locus using a ΦiC31 integrase mediated cassette exchange approach
in mouse ES cells. PLoS One. 6:e233762011. View Article : Google Scholar
|
|
23
|
Liew CG, Draper JS, Walsh J, Moore H and
Andrews PW: Transient and stable transgene expression in human
embryonic stem cells. Stem Cells. 25:1521–1528. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mathieu J and Ruohola-Baker H: Regulation
of stem cell populations by microRNAs. Adv Exp Med Biol.
786:329–351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sasaki N, Okishio K, Ui-Tei K, Saigo K,
Kinoshita-Toyoda A, Toyoda H, Nishimura T, Suda Y, Hayasaka M,
Hanaoka K, et al: Heparan sulfate regulates self-renewal and
pluripotency of embryonic stem cells. J Biol Chem. 283:3594–3606.
2008. View Article : Google Scholar
|
|
26
|
Li J, Bei Y, Liu Q, Lv D, Xu T, He Y, Chen
P and Xiao J: MicroRNA-221 is required for proliferation of mouse
embryonic stem cells via P57 targeting. Stem Cell Rev. 11:39–49.
2015. View Article : Google Scholar
|
|
27
|
Wan Y, Sun G, Wang Z, Guo J and Shi L:
miR-125b promotes cell proliferation by directly targeting Lin28 in
glioblastoma stem cells with low expression levels of miR-125b.
Neuroreport. 25:289–296. 2014.
|
|
28
|
Wobus AM, Guan K, Yang HT and Boheler KR:
Embryonic stem cells as a model to study cardiac, skeletal muscle,
and vascular smooth muscle cell differentiation. Methods Mol Biol.
185:127–156. 2002.PubMed/NCBI
|
|
29
|
Hwang JT and Kelly GM: GATA6 and FOXA2
regulate Wnt6 expression during extraembryonic endoderm formation.
Stem Cells Dev. 21:3220–3232. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roche E, Sepulcre P, Reig JA, Santana A
and Soria B: Ectodermal commitment of insulin-producing cells
derived from mouse embryonic stem cells. FASEB J. 19:1341–1343.
2005.PubMed/NCBI
|
|
31
|
Lobo MV, Arenas MI, Alonso FJ, Gomez G,
Bazán E, Paíno CL, Fernández E, Fraile B, Paniagua R, Moyano A, et
al: Nestin, a neuroectodermal stem cell marker molecule, is
expressed in Leydig cells of the human testis and in some specific
cell types from human testicular tumours. Cell Tissue Res.
316:369–376. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rosa A, Spagnoli FM and Brivanlou AH: The
miR-430/427/302 family controls mesendodermal fate specification
via species-specific target selection. Dev Cell. 16:517–527. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li L, Liu C, Biechele S, Zhu Q, Song L,
Lanner F, Jing N and Rossant J: Location of transient ectodermal
progenitor potential in mouse development. Development.
140:4533–4543. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Addae C, Yi X, Gernapudi R, Cheng H, Musto
A and Martinez-Ceballos E: All-trans-retinoid acid induces the
differentiation of encapsulated mouse embryonic stem cells into
GABAergic neurons. Differentiation. 83:233–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu J, Wang H, Liang T, Cai X, Rao X, Huang
Z and Sheng G: Retinoic acid promotes neural conversion of mouse
embryonic stem cells in adherent monoculture. Mol Biol Rep.
39:789–795. 2012. View Article : Google Scholar
|
|
36
|
Tarantino C, Paolella G, Cozzuto L,
Minopoli G, Pastore L, Parisi S and Russo T: miRNA 34a, 100, and
137 modulate differentiation of mouse embryonic stem cells. FASEB
J. 24:3255–3263. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Solozobova V and Blattner C: Regulation of
p53 in embryonic stem cells. Exp Cell Res. 316:2434–2446. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gaulden J and Reiter JF: Neur-ons and
neur-offs: regulators of neural induction in vertebrate embryos and
embryonic stem cells. Hum Mol Genet. 17:R60–R66. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Berardi E, Pues M, Thorrez L and
Sampaolesi M: miRNAs in ESC differentiation. Am J Physiol Heart
Circ Physiol. 303:H931–H939. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tiscornia G and Izpisúa Belmonte JC:
MicroRNAs in embryonic stem cell function and fate. Genes Dev.
24:2732–2741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wells JM and Melton DA: Vertebrate
endoderm development. Annu Rev Cell Dev Biol. 15:393–410. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Boissart C, Nissan X, Giraud-Triboult K,
Peschanski M and Benchoua A: miR-125 potentiates early neural
specification of human embryonic stem cells. Development.
139:1247–1257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu L and Belasco JG: Micro-RNA regulation
of the mammalian lin-28 gene during neuronal differentiation of
embryonal carcinoma cells. Mol Cell Biol. 25:9198–9208. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Parisi S, Battista M, Musto A, Navarra A,
Tarantino C and Russo T: A regulatory loop involving Dies1 and
miR-125a controls BMP4 signaling in mouse embryonic stem cells.
FASEB J. 26:3957–3968. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun YM, Lin KY and Chen YQ: Diverse
functions of miR-125 family in different cell contexts. J Hematol
Oncol. 6:62013. View Article : Google Scholar : PubMed/NCBI
|