Introduction
Osteosarcoma is the most common non-hematological
primary bone tumor in children and adolescents, and for
osteocarcoma patients the prognosis remains poor, particularly when
metastasis is present at the time of diagnosis (1–4).
Current strategies focus on the primary tumor and have limited
efficacy for metastatic osteosarcoma (5). Treating metastatic osteosarcoma
remains a challenge in oncology. Therefore, broadening our
understanding of the pathogenesis and biology of metastatic
osteosarcoma tumors is key to improving treatment efficacy
(6), and there is an urgent need
for new, targeted therapies based on biomarkers (7).
MicroRNAs (miRNAs or miRs) are small, non-coding,
single-stranded RNAs of ~24 nt in length that regulate gene output
at the post-transcriptional level either by mRNA degradation or
translation repression (8,9).
Their target genes include numerous regulators of cell
proliferation, differentiation, apoptosis, development, metabolism
and immunity (10,11). The deregulation of miRNA
expression may contribute to tumorigenesis and other
pathophysiologies associated with cancer (12). miRNAs have been proven to cause
the widespread disruption of various function in many types of
cancer (13). Emerging evidence
has indicated that miRNAs participate in human carcinogenesis as
oncogenic miRNAs (increasing tumor progression) and tumor
suppressor miRNAs (inhibiting tumor progression) (11,14–16).
miR-221, which is highly conserved in vertebrates,
is encoded on the X chromosome in humans (17). A number of studies, to date, have
examined the role of miR-221 in cancer development either as an
oncogenic miRNA or a tumor suppressor miRNA. The overexpression of
miR-221 has been observed in a several types of cancer, including
glioblastoma, hepatocellular carcinoma, and gastric, breast,
prostate and pancreatic cancer (17–21). Fornari et al (19) suggested that miR-221 had an
oncogenic function in hepatocarcinogenesis. Garofalo et al
(21) reported that miR-221 was
overexpressed in aggressive hepatocarcinoma and non-small cell lung
cancer cells, as compared with less invasive and/or normal liver
and lung cells. The high levels of miR-221 in breast cancer results
in a poorly differentiated, mesenchymal-like phenotype (22). Galardi et al (23) demonstrated that the overexpression
of miR-221 was one of the factors contributing to the oncogenesis
and progression of prostate carcinoma. He et al (24) demonstrated that miR-221 silencing
inhibited the malignant properties of liver cancer in vitro
and in vivo. The antagonism of miR-221 has been shown to
reduce the growth of colon tumors in mice (25). Gan et al (26) demonstrated that the
down-regulation of miR-221 enhanced the sensitivity of breast
cancer cells to tamoxifen through the upregulation of tissue
inhibitor of metalloproteinases 3 (TIMP3). In view of the above, we
hypothesized that miR-221 may serve as an oncogenic miRNA in
multiple types of cancer. However, Felli et al (27) indicated that miR-221 inhibited
normal erythropoiesis and erythroleukemic cell growth at least in
part via the down-modulation of the kit receptor. In the present
study, we provide evidence that miR-221 plays the role of a tumor
suppressor in erythrocytes, which indicates that miRNA function is
exclusively dependent on the cellular context and tumor type.
Nonetheless, the question of whether miR-221 is
deregulated in osteosarcoma and its role in osteosarcoma
carcinogenesis and progression remain to be elucidated. Thus, the
aim of the present study was to examine miR-221 expression in
osteosarcoma tissues and cells by RT-qPCR and to observe the
changes in thee proliferation, invasion and migration ability of
MG-63 cells following the increase and reduction in the expression
of miR-221. Our data may provide important scientific information
for prognosis prediction and targeted therapy for osteosarcoma.
Materials and methods
Reagents
All cell culture reagents were purchased from
Gibco-BRL (Grand Island, NY, USA). The mirVana miRNA Isolation kit,
SuperTaq polymerase and the mirVana qRT-PCR miRNA detection kit
were purchased from Ambion (Austin, TX, USA). Osteosarcoma cell
lines (MG-63 and U2OS) and a human osteoblast cell line (hFOB1.19)
were obtained from the American Type Culture Collection (Rockville,
MD, USA). miR-221 mimic and inhibitor were chemically synthesized
by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Transfection
reagents were purchased from Guangzhou RiboBio Co., Ltd. Protein
extraction and quantification kits, propidium iodide (PI), dimethyl
sulfoxide (DMSO) and RNase A were purchased from the Beyotime
Institute of Biotechnology (Haimen, China). The ECL
chemiluminescence kit was purchased from Pierce (Rockford, IL,
USA). Matrigel was purchased from Collaborative Biomedical
(Bedford, MA, USA). The Transwell invasion chamber was purchased
from Costar (Cambridge, MA, USA). The rabbit anti-phosphatase and
tensin homolog (PTEN; Cat. no. 251264) and rabbit anti-β-actin
(Cat. no. 250920) polyclonal antibodies were purchased from
Abbiotec LLC (San Diego, CA, USA). The horseradish
peroxidase-conjugated goat anti-rabbit IgG (H+L; Cat. no. 626100)
was purchased from Life Technologies (Carlsbad, CA, USA).
Specimens
Fresh osteosarcoma tissues were collected from 16
patients who had been subjected to resection surgery in our
hospital (the Affiliated Hospital of Nantong University, Nantong,
China) between January 2009 and July 2014. None of the patients had
recevied any pre-operative treatment, and all cases were
pathologically diagnosed as osteosarcoma post-operatively. Twelve
fresh normal bone tissues were also obtained at this time. All
tissue samples were immediately frozen in liquid nitrogen for use
in subsequent experiments. The study protocol was approved by the
Ethics Committee of The Affiliated Hospital of Nantong University,
Nantong, China. All patients provided written informed consent
prior to the isolation of the samples.
Cell culture and transfection
The osteosarcoma cell lines (MG-63 and U2OS) were
maintained in RPMI-1640 (Gibco-BRL) supplemented with 10% fetal
bovine serum (FBS) and 1% penicillin-streptomycin (Gibco-BRL) at
37°C. Human osteoblast hFOB1.19 cells were maintained in DMEM:Ham's
F-12 containing 10% FBS and geneticin (400 µg/ml; Gibco-BRL)
at 34°C in an incubator with 5% CO2.
To enforce miR-221 expression or suppress miR-221
expression in MG-63 cells, the cells were transfected with miR-221
mimic or inhibitor, and these cells then served as the
overexpression group and knockdown group, respectively. MG-63 cells
transfected with control miRNA were used as the control group. On
the day of transfection, MG-63 cells at approximately 70–80%
confluence were moved to antibiotic-free medium. The following day,
the cells were transfected with 50 nM miR-221 mimic and 100 nM
miR-221 inhibitor using Lipofectamine™ 2000 reagent (Thermo Fisher
Scientific, Waltham, MA, USA), according to the manufacturer's
instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the osteosarcoma tissues
and cells using the mirVana miRNA Isolation kit according to the
manufacturer's instructions. Reverse transcription was performed
using the TaqMan MicroRNA Reverse Transcription kit. qPCR was
performed using TaqMan Universal PCR Master Mix, and TaqMan
MicroRNA assay primers for human miR-221. TaqMan microRNA assays
(Applied Biosystems, Foster City, CA, USA) were performed using the
7500 Fast Real-Time PCR system. The levels of miRNA were normalized
to U6 controls. The cycle threshold (Ct) values, corresponding to
the PCR cycle number at which fluorescence emission reaches a
threshold above baseline emission, were determined and the relative
miRNA expression was calculated using the 2−ΔΔCt method,
as previously described (28).
Western blot analysis
Total proteins were extracted from the osteosarcoma
tissues and cells using RIPA buffer (Beyotime Institute of
Biotechnology) and quantified with a BCA assay kit (Beyotime
Institute of Biotechnology). Proteins were then transferred to
polyvinylidene fluoride (PVDF; Amresco, Inc., Solon, OH, USA)
membranes following SDS-PAGE. The PVDF membranes were blocked for 1
h at an ambient temperature in blocking buffer. They were then
incubated with primary antibody at 4°C overnight in blocking
buffer. The PVDF membranes were then washed in TBST for 3×5 min,
and probed with corresponding secondary antibody for 2 h at ambient
temperature. After being washed with TBST, autoradiography was
conducted with ECL chemiluminescence reagents (Pierce, Rockford,
IL, USA). The relative expression of the target protein was
evaluated, with the gray value ratio of the target protein content
being compared to the to β-actin content (target
protein/β-actin).
Cell viability and proliferation
When the population of the MG-63 cells cultured in
96-well plates reached optimal densities, we replaced the culture
medium with 100 µl fresh culture medium. We then added 10
µl of the 12 mM MTT stock solution to each well and
incubated it for 4 h at 37°C. Subsequently, we added 100 µl
SDS-HCl solution (10 ml 0.01 M HCl was mixed with 1 mg) to each
well, mixed thoroughly and incubated the plates at 37°C for 4 h in
a humidified chamber. Finally, we mixed each sample again using a
pipette. The absorbance at 570 nm was measured using a plate
reader, and a measurement of 690 nm was used as a reference. By
using this procedure, only viable cells with functioning
mitochondria oxidized MTT to a violet-red reaction product.
The osteosarcoma MG-63 cells were cultured in
serum-free medium for 24 h to synchronize and then cultured in
complete medium for 24 h. The MG-63 cells were then harvested and
fixed in 0.5 ml 70% precooled ethanol. The fixed MG-63 cells were
stored on ice for 1 day. The following day, the fixed MG-63 cells
were washed and then pre-treated with RNAse (DNAse-free) (10
µg/ml) for 30 min at 37°C. PI was then added to the solution
at a final concentration of 100 µg/ml and incubated with the
MG-63 cells for 30 min at room temperature in the dark. The cell
cycle was evaluated by flow cytometry (BD FACSCalibur™; BD
Biosciences, San Jose, CA, USA). For cell cycle analysis, data were
expressed as fractions of cells in different cycle phases, and each
experiment was repeated 3 times.
Wound healing assay
Cell mobility was assessed using a wound healing
assay. Equal numbers of MG-63 cells from each group were seeded
near confluence as a monolayer in 6-well plates. Gently and slowly,
we scratched the monolayer with a sterile plastic pipette tip,
across the center of the well. After scratching, we gently washed
the well twice with serum-free culture medium to remove the
detached cells. We then replenished the well with fresh medium.
Twenty-four hours later, MG-63 cells that had migrated into the
wounded area or cells with extended protrusion from the border of
the wound were visualized and photographed under an inverted
microscope (XDS-1B; Wuzhou New Found Instrument Co., Ltd, Guangxi,
China). The distance that MG-63 cells migrated into the wounded
area was measured by subtracting the distance at 24 h after wound
healing from the initial distance. The results were based on 3
independent experiments.
Transwell invasion assay
The mechanisms involved in tumor cell invasion are
very important. The most commonly used in vitro invasion
assay is a modified Boyden chamber assay. Transwell inserts with
8-µm porous membranes were used for the assay. Matrigel (20
µl) (1 mg/ml) was added in order to evenly cover the surface
of the Transwell insert and create the Matrigel membrane after
washing the chamber with serum-free medium, which divided the
chamber into upper and lower chambers. The MG-63 cells were washed
3 times with serum-free medium and added to the top chamber in
serum-free medium. The bottom chamber was filled with medium
containing 10% FBS, which acted as chemoattractant. The MG-63 cells
were incubated for 48 h at 37°C in a 5% CO2 humidified
atmosphere. Following incubation for 48 h, the MG-63 cells on the
top surface of the chamber were removed with cotton swabs and the
MG-63 cells which had invaded into the Matrigel membrane were fixed
with 4% formaldehyde for 15 min, stained with a crystal violet
solution for 10 min, visualized under a phase-contrast microscope
(Leica DMIL 090–135.001; Leica Company, Wetzlar, Germany), and
photographed. The results are presented as the means± SD, and the
experiment was repeated 3 times for each group.
Statistical analysis
SPSS 17.0 software was used to analyze the data in
the present study. One-way ANOVA was used to compare 3 or more
groups or variables for statistical significance. A P-value
<0.05 was considered to indicate a statistically significant
difference.
Results
miR-221 is overexpressed in
osteosarcoma
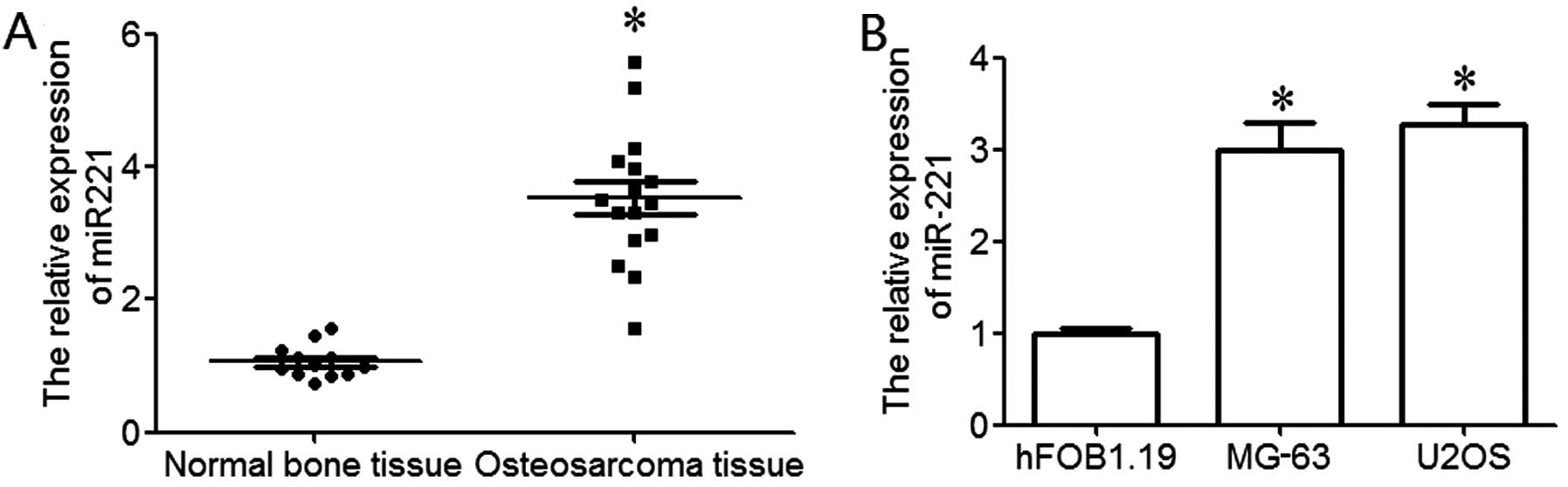
RT-qPCR was used to examine the expression level of
miR-221 in osteosarcoma. The results revealed that miR-221 was
significantly upregulated in osteosarcoma tissues compared to
normal bone tissues (P<0.05; Fig.
1A). In order to further explore the expression of miR-221 in
osteosarcoma cell lines, 2 osteosarcoma cell lines (MG-63 and U2OS)
and a human osteoblastic cell line (hFOB1.192) were used in the
present study. The results revealed that miR-221 expression in
osteosarcoma cell lines was significantly upregulated compared to
the normal human osteoblastic cell line, hFOB1.19 (P<0.05;
Fig. 1B). These data indicate
that the upregulation of miR-221 plays an important role in the
development and progression of osteosarcoma.
miR-221 increases MG-63 cell
viability
MG-63 cells were used to further examine the effects
of miR-221. To enforce miR-221 expression or inhibit miR-221
expression in the MG-63 cells, the cells were transfected with
miR-221 mimic or inhibitor, and these cells then served as the
overexpression group and knockdown group, respectively. The MG-63
cells transfected with scrambled oligonucleotides was used as the
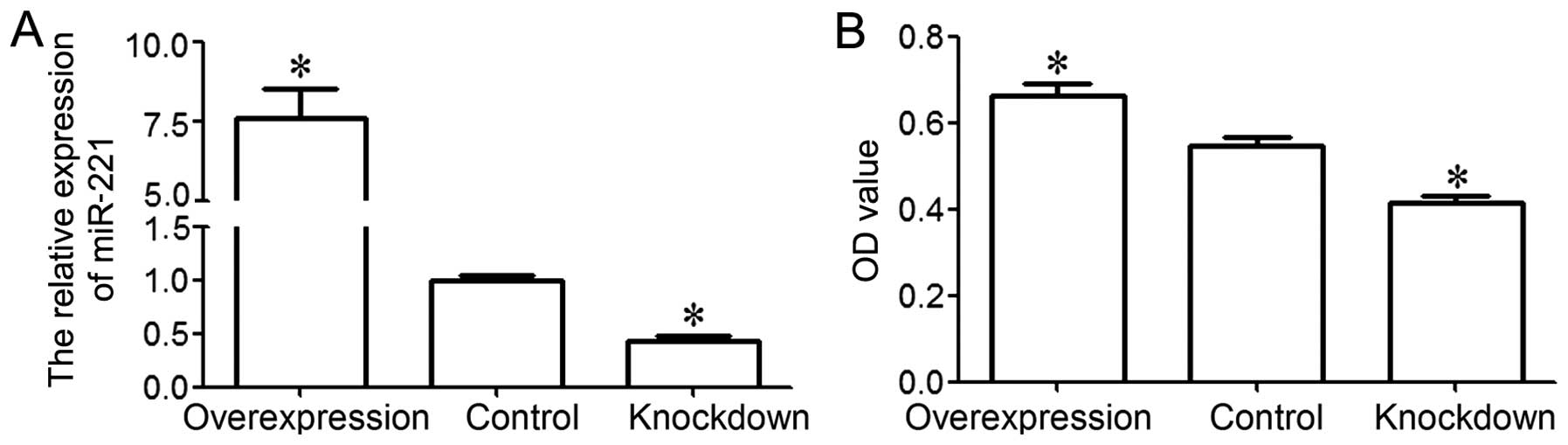
control group. The results from RT-qPCR revealed that miR-221 was
significantly upregulated in the overexpression group and
significantly downregulated in the knockdown group, as compared to
the control group (P<0.05; Fig.
2A). These data demonstrated that we effectively enforced or
inhibited miR-221 expression in the MG-63 cells.
An MTT assay was performed to examine the viability
of the MG-63 cells. The results indicated that MG-63 cell viability
in the overexpression group was significantly higher than that in
the control group, and that MG-63 cell viability in the knockdown
group was significantly lower than that in the control group
(P<0.05; Fig. 2B). These
results indicate that miR-221 is associated with the increase in
MG-63 cell viability.
miR-221 accelerates MG-63 cell cycle
progression
A flow cytometric assay was performed in order to
examine the cell cycle of MG-63 cells. The results demonstrated
that the percentage of MG-63 cells in the S and G2M
phase in the overexpression group was higher than that in the
control group (P<0.05), and the percentage of MG-63 cells in the
S and G2M phase in the knockdown group was lower than
that in the control group (P<0.05; Fig. 3). Based on the above results, we
conclude that miR-221 enhances the proliferation of MG-63
cells.
miR-221 increases MG-63 cell invasion and
migration
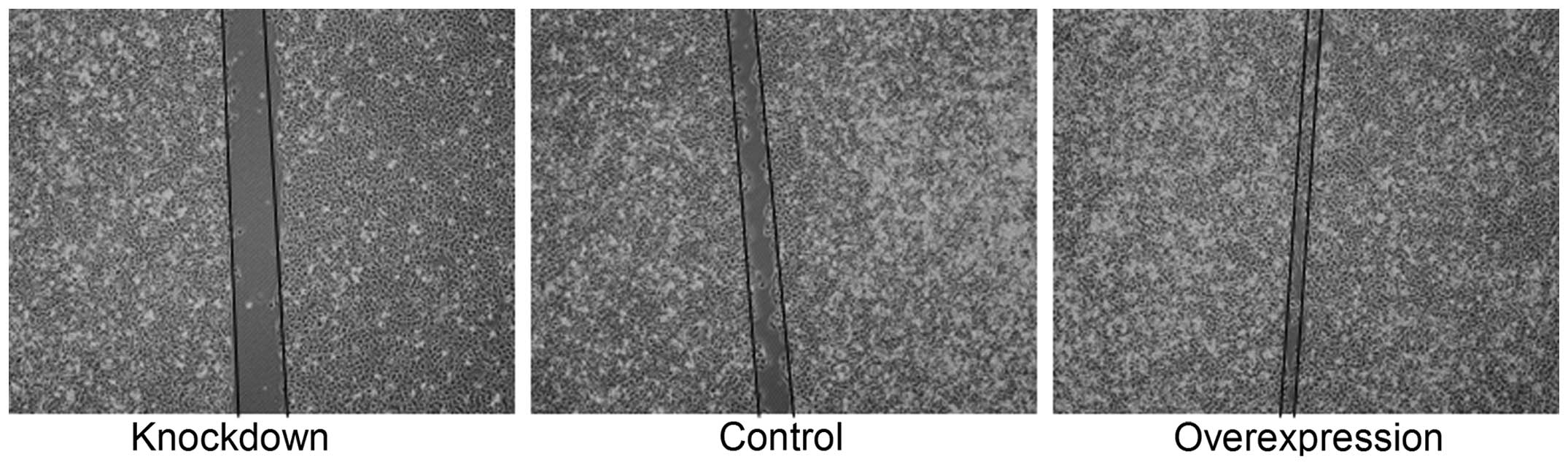
To determine the influence of miR-221 on the
migratory potential of MG-63 cells, we examined the effects of
miR-221 on cell migration using a classical scratch-wound healing
assay. The MG-63 cells in the overexpression group exhibited a
higher migration rate as compared to the MG-63 cells in the control
group, and the MG-63 cells in the knockdown group exhibited a lower
migration rate than the MG-63 cells in the control group (Fig. 4).
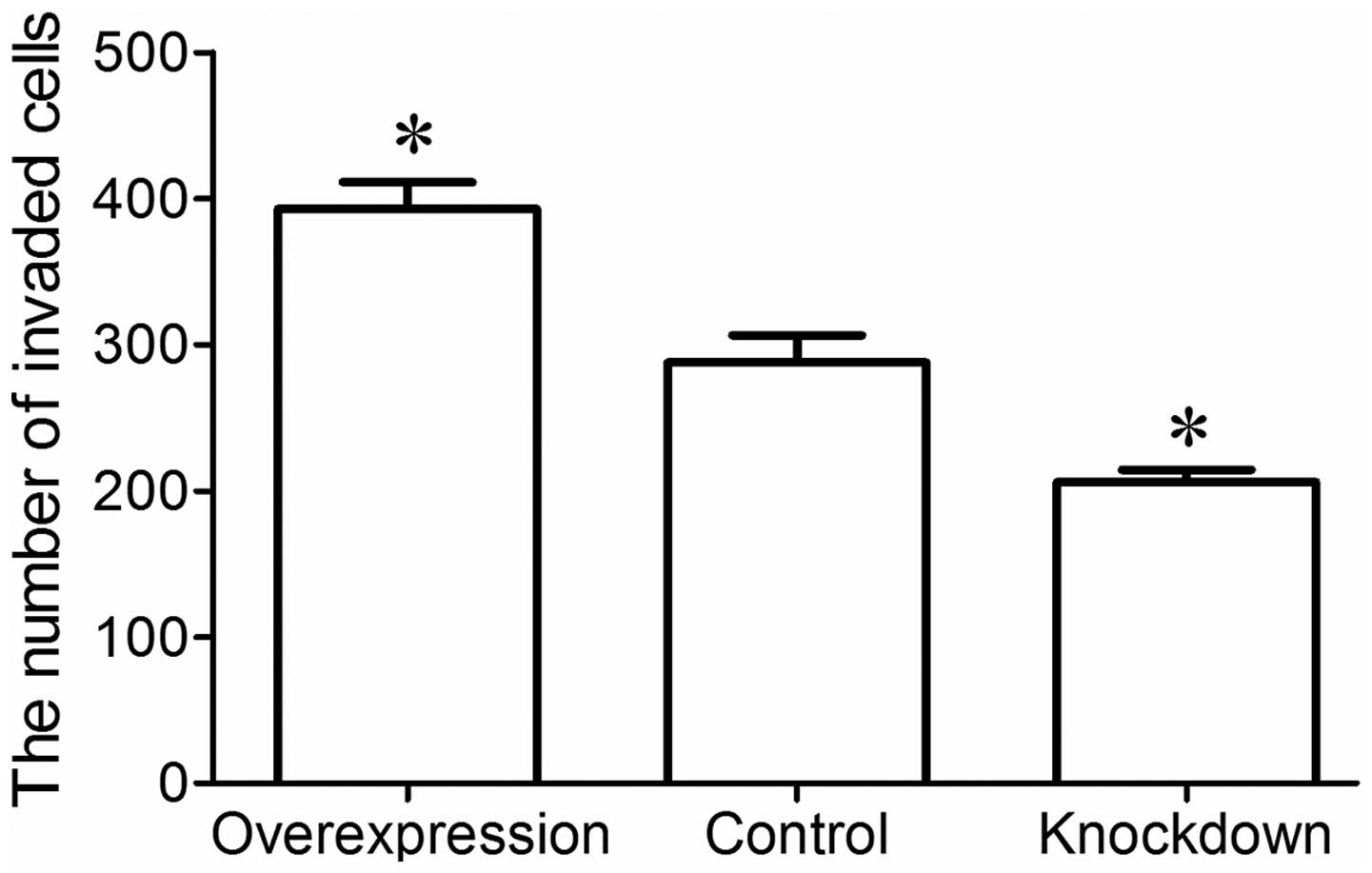
Invasion assay was performed to examine the invasion
ability of the MG-63 cells using a Transwell invasion chamber. The
results revealed that a greater number of MG-63 cells in the
overexpression group had invaded the Matrigel membrane, whereas
fewer MG-63 cells in the knockdown group invaded the Matrigel
membrane compared to the control group (P<0.05; Fig. 5). These data indicate that miR-221
enhances the invasion and metastatic ability of MG-63 osteosarcoma
cells.
Effects of miR-221 on the expression of
PTEN in MG-63 cells
A previous study demonstrated that miR-221 increases
the invasive ability of breast cancer cells by suppressing the
expression of PTEN (29). In the
present study, western blot analysis revealed that PTEN was
downregulated in osteosarcoma tissues and cell lines compared to
the normal bone tissue or the human osteoblastic cell line,
hFOB1.19 (P<0.05; Fig. 6).
These data indicate the inverse correlation between miR-221
expression and PTEN expression that exists in osteosarcoma.
Additionally, PTEN was downregulated in the overexpression group
and upregulated in the knockdown group compared to the control
group (P<0.05; Fig. 6). These
results indicate that miR-221 suppresses the expression of PTEN in
osteosarcoma.
Discussion
miRNAs are small non-coding RNAs that control the
expression of many target mRNAs involved in a great number of
normal cell functions, including differentiation, proliferation and
apoptosis (30). Emerging
evidence indicates that the altered expression of miRNAs is
involved in cancer development and progression (31). In recent years, the dysregulation
of miR-221 has been associated with a variety of human cancers,
including liver and breast cancer, glioblastoma, and gastric,
colon, prostate and pancreatic cancer (17–21,24,26,32). It has been shown that miR-221 is
significantly upregulated in liver cancer, and that it plays the
role of a critical modulator in the liver cancer signaling pathway,
and that miR-221 knockdown inhibits liver cancer cell
proliferation, clonogenicity, migration and invasion in
vitro and in vivo (24). A recent study suggested that
miR-221 is responsible for resistance to tamoxifen in breast cancer
(33). The downregulation of
miRNA-221 increases the sensitivity of estrogen-receptor
(ER)-positive MCF-7 breast cancer cells to tamoxifen (26). However, whether or not miR-221 is
deregulated in osteosarcoma, and its role in osteosarcoma
carcinogenesis and progression remains to be elucidated.
In the present study, we demonstrated that miR-221
was overexpressed in osteosarcoma tissues and cell lines compared
to normal bone tissues and the non-cancerous osteoblastic cell
line, hFOB1.19. The expression model of miR-221 in osteosarcoma in
this study was consistent with that in other types of cancer, such
as liver, breast and colon cancer (24,26,33). Yang et al (34) demonstrated that miR-221 was highly
expressed in PC-3 cells, and miR-221 inhibition led to reduced cell
proliferation and migration in prostate cancer cells. miR-221 is
frequently overexpressed in breast cancer and is associated with
increased malignancy (35). It
has been previously demonstrated that miR-221 knockdown induces
cell apoptosis, inhibits cell cycle progression, cell
proliferation, cell migration and cell invasion in breast cancer
(36). Therefore, we hypothesized
that miR-221 plays a positive regulatory role in tumor cell
proliferation, migration and invasion. In order to determine the
effects of miR-221 on osteosarcoma cells, we examined whether
miR-221 was overexpressed or suppressed in MG-63 cells.
MTT assay demonstrated that the viability of MG-63
cells was significantly enhanced in the overexpression group and
significantly inhibited in the knockdown group compared to the
control group. Transfection with miR-221 mimic increased cell
viability and miR-221 inhibitor significantly inhibited cell
growth, as has also been previously demonstrated (37). These data indicate that miR-221
plays a key role in the viability of MG-63 cells. A cell cycle
assay was also performed, and the results indicated that miR-221
increased MG-63 cell proliferation. Yang et al (34) found that miR-221 inhibition led to
reduced cell proliferation in prostate cancer cells. It has also
been shown that miR-221 targets SUN2 directly, increasing cell
proliferation and tumor malignancy in medulloblastoma in
vitro and in vivo (38). Sarkar et al (39) found that miR-221 inhibition
upregulated the expression of PTEN and p27kip1, and led
to the inhibition of the proliferation of MiaPaCa-2 and Panc-1
cells. The above results suggest that miR-221 promotes the growth
and proliferation of osteosarcoma cells.
Increasing evidence has demonstrated that miR-221
plays an important role in the process of tumor metastasis and cell
invasion (29,40,41). In the present study, we
investigated the effects of miR-221 on the invasion and metastasic
ability of osteosarcoma cells in vitro. The scratch-wound
healing assay demonstrated that miR-221 contributed to the
migration of MG-63 cells, and that miR-221 knockdown inhibited the
migration of MG-63 cells. The invasion assay demonstrated that
miR-221 enhanced the invasive potential of MG-63 cells, and that
miR-221 knockdown suppressed the invasive potential of MG-63 cells.
These data indicate that miR-221 increases MG-63 cell invasiveness
and metastasis, which was consistent with findings in relation to
other types of cancer. miR-221 has been shown to be significantly
upregulated in metastatic minimally invasive follicular thyroid
carcinoma, and some cases show a poor prognosis due to severe
distant metastasis (42). Ye
et al (29) demonstrated
that miR-221 promoted the metastasis of human epidermal growth
factor receptor 2 (HER2)-positive breast cancers by targeting PTEN.
miR-221 has been proven to enhance cell survival and to induce
TNF-related apoptosis-inducing ligand resistance in non-small cell
lung cancer (NSCLC) cell lines through the downregulation of PTEN
(43). Previous studies have
demonstrated that miR-221 enhances cellular migration by
suppressing PTEN (21,44). Therefore, we speculated that
miR-221 plays an important role in osteosarcoma, possibly by
suppressing PTEN. Thus, the expression of PTEN was examined in this
study. Our results indicated that PTEN was downregulated in
osteosarcoma tissues and cell lines, which indicated that there was
an inverse correlation between miR-221 expression and PTEN
expression in osteosarcoma. The expression of PTEN was inhibited by
miR-221 in the overexpression group and was improved in the miR-221
knockdown group, which indicated that miR-221 suppresses the
expression of PTEN in osteosarcoma. Based on these data, we suggest
that miR-221 contributes to osteosarcoma cell proliferation,
invasion and migration partly through the downregulation of
PTEN.
Our data demonstrated that miR-221 was significantly
upregulated in osteosarcoma tissues and cell lines, suggesting that
increased miR-221 expresion plays an important role in osteosarcoma
oncogenesis and progression. The suppression or overexpression of
miR-221 expression in MG-63 osteosarcoma cells was achieved through
transfection with miR-221 inhibitor or mimic, respectively. miR-221
overexpression increased the proliferative ability of MG-63 cells,
as well as invasion and migration, and miR-221 knockdown suppressed
the proliferative ability of MG-63 cells, as well as invasion and
migration. The downregulation of miR-221 increased the expression
of PTEN, and the upregulation of miR-221 decreased the expression
of PTEN. Taken together, our results suggest that miR-221
facilitates the proliferation, invasion and migration of
osteosarcoma cells, at least, partly by suppressing PTEN.
Acknowledgments
The present study was supported by the Natural
Science Foundation of Jiangsu Province (grant no. BK20131199) and
by the fifty-fifth batch of the China Postdoctoral Science
Foundation (no. 2014M551640).
References
|
1
|
Endo-Munoz L, Evdokiou A and Saunders NA:
The role of osteoclasts and tumour-associated macrophages in
osteosarcoma metastasis. Biochim Biophys Acta. 1826.434–442.
2012.
|
|
2
|
Lamora A, Talbot J, Bougras G, Amiaud J,
Leduc M, Chesneau J, Taurelle J, Stresing V, Le Deley MC, Heymann
MF, et al: Overexpression of smad7 blocks primary tumor growth and
lung metastasis development in osteosarcoma. Clin Cancer Res.
20:5097–5112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harting MT and Blakely ML: Management of
osteosarcoma pulmonary metastases. Semin Pediatr Surg. 15:25–29.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jia S and Li B: Osteosarcoma of the jaws:
case report on synchronous multicentric osteosarcomas. J Clin Diagn
Res. 8:ZD01–ZD03. 2014.PubMed/NCBI
|
|
5
|
He JP, Hao Y, Wang XL, Yang XJ, Shao JF,
Guo FJ and Feng JX: Review of the molecular pathogenesis of
osteosarcoma. Asian Pac J Cancer Prev. 15:5967–5976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Poletajew S, Fus L and Wasiutyński A:
Current concepts on pathogenesis and biology of metastatic
osteosarcoma tumors. Ortop Traumatol Rehabil. 13:537–545. 2011.
View Article : Google Scholar
|
|
7
|
Egas-Bejar D, Anderson PM, Agarwal R,
Corrales-Medina F, Devarajan E, Huh WW, Brown RE and Subbiah V:
Theranostic profiling for actionable aberrations in advanced high
risk osteosarcoma with aggressive biology breveals high molecular
diversity: the human fingerprint hypothesis. Oncoscience.
1:167–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu CW, Cheng YW, Hsu NY, Yeh KT, Tsai YY,
Chiang CC, Wang WR and Tung JN: MiRNA-221 negatively regulated
downstream p27Kip1 gene expression involvement in pterygium
pathogenesis. Mol Vis. 20:1048–1056. 2014.PubMed/NCBI
|
|
9
|
Wolter JM, Kotagama K, Pierre-Bez AC,
Firago M and Mangone M: 3′LIFE: a functional assay to detect miRNA
targets in high-throughput. Nucleic Acids Res. 42:e1322014.
View Article : Google Scholar
|
|
10
|
Gaál Z and Oláh E: MicroRNA-s and their
role in malignant hematologic diseases. Orv Hetil. 153:2051–2059.
2012.In Hungarian. View Article : Google Scholar
|
|
11
|
Lin S, Pan L, Guo S, Wu J, Jin L, Wang JC
and Wang S: Prognostic role of microRNA-181a/b in hematological
malignancies: a meta-analysis. PLoS One. 8:e595322013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Sarkar N, Roy R, Mitra JK, Ghose S,
Chakraborty A, Paul RR, Mukhopadhyay I and Roy B: a quest for miRNA
bio-marker: a track back approach from gingivo buccal cancer to two
different types of precancers. PLoS One. 9:e1048392014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu C, Ping Y and Li X, Zhao H, Wang L, Fan
H, Xiao Y and Li X: Prioritizing candidate disease miRNAs by
integrating phenotype associations of multiple diseases with
matched miRNA and mRNA expression profiles. Mol Biosyst.
10:2800–2809. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye S, Yang L, Zhao X, Song W, Wang W and
Zheng S: Bioinformatics method to predict two regulation mechanism:
TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell
Biochem Biophys. 70:1849–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kushwaha D, Ramakrishnan V, Ng K, Steed T,
Nguyen T, Futalan D, Akers JC, Sarkaria J, Jiang T, Chowdhury D, et
al: A genome-wide miRNA screen revealed miR-603 as a
MGMT-regulating miRNA in glioblastomas. Oncotarget. 5:4026–4039.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kalinowski FC, Brown RA, Ganda C, Giles
KM, Epis MR, Horsham J and Leedman PJ: microRNA-7: a tumor
suppressor miRNA with therapeutic potential. Int J Biochem Cell
Biol. 54:312–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garofalo M, Quintavalle C, Romano G, Croce
CM and Condorelli G: miR221/222 in cancer: their role in tumor
progression and response to therapy. Curr Mol Med. 12:27–33. 2012.
View Article : Google Scholar
|
|
18
|
le Sage C, Nagel R, Egan DA, Schrier M,
Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et
al: Regulation of the p27Kip1 tumor suppressor by
miR-221 and miR-222 promotes cancer cell proliferation. EMBO J.
26:3699–3708. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fornari F, Gramantieri L, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM,
Bolondi L and Negrini M: MiR-221 controls CDKN1C/p57 and CDKN1B/p27
expression in human hepatocellular carcinoma. Oncogene.
27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting
p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Garofalo M, Di Leva G, Romano G, Nuovo G,
Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P,
et al: miR-221&222 regulate TRAIL resistance and enhance
tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell.
16:498–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Howe EN, Cochrane DR and Richer JK: The
miR-200 and miR-221/222 microRNA families: opposing effects on
epithelial identity. J Mammary Gland Biol Neoplasia. 17:65–77.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem.
282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He XX, Guo AY, Xu CR, Chang Y, Xiang GY,
Gong J, Dan ZL, Tian DA, Liao JZ and Lin JS: Bioinformatics
analysis identifies miR-221 as a core regulator in hepatocellular
carcinoma and its silencing suppresses tumor properties. Oncol Rep.
32:1200–1210. 2014.PubMed/NCBI
|
|
25
|
Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang
Y, Liu Z, Cao X, Chen P, Liu Z, et al: A microRNA miR-221- and
miR-222-mediated feedback loop, via PDLIM2, maintains constitutive
activation of nuclear factor kappaB and STAT3 in colorectal cancer
cells. Gastroenterology. 147:847–859. 2014. View Article : Google Scholar
|
|
26
|
Gan R, Yang Y, Yang X, Zhao L, Lu J and
Meng QH: Downregulation of miR-221/222 enhances sensitivity of
breast cancer cells to tamoxifen through upregulation of TIMP3.
Cancer Gene Ther. 21:290–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Felli N, Fontana L, Pelosi E, Botta R,
Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et
al: MicroRNAs 221 and 222 inhibit normal erythropoiesis and
erythroleukemic cell growth via kit receptor down-modulation. Proc
Natl Acad Sci USA. 102:18081–18086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang
L, Yang A, Zhao J and Jia L: MiR-221 promotes
trastuzumab-resistance and metastasis in HER2-positive breast
cancers by targeting PTEN. BMB Rep. 47:268–273. 2014. View Article : Google Scholar :
|
|
30
|
De Tullio G, De Fazio V, Sgherza N, Minoia
C, Serratì S, Merchionne F, Loseto G, Iacobazzi A, Rana A, Petrillo
P, et al: Challenges and opportunities of microRNAs in lymphomas.
Molecules. 19:14723–14781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu Y, Huang Z and Liu Y: Reduced
miR-125a-5p expression is associated with gastric carcinogenesis
through the targeting of E2F3. Mol Med Rep. 10:2601–2608.
2014.PubMed/NCBI
|
|
32
|
Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H
and Yu H: Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p
expression in colon cancer. Am J Transl Res. 6:391–401.
2014.PubMed/NCBI
|
|
33
|
Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y,
Li H, Zhu X, Yao L and Zhang J: Exosomal miR-221/222 enhances
tamoxifen resistance in recipient ER-positive breast cancer cells.
Breast Cancer Res Treat. 147:423–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang X, Yang Y, Gan R, Zhao L, Li W, Zhou
H, Wang X, Lu J and Meng QH: Down-regulation of mir-221 and mir-222
restrain prostate cancer cell proliferation and migration that is
partly mediated by activation of SIRT1. PLoS One. 9:e988332014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Falkenberg N, Anastasov N, Rappl K,
Braselmann H, Auer G, Walch A, Huber M, Höfig I, Schmitt M, Höfler
H, et al: MiR-221/-222 differentiate prognostic groups in advanced
breast cancers and influence cell invasion. Br J Cancer.
109:2714–2723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nassirpour R, Mehta PP, Baxi SM and Yin
MJ: miR-221 promotes tumorigenesis in human triple negative breast
cancer cells. PLoS One. 8:e621702013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen G, Dang YW and Luo DZ: Effect of
miR-221 on the viability and apoptosis of hepatocellular carcinoma
HepG2 cells. Zhonghua Gan Zang Bing Za Zhi. 19:582–587. 2011.In
Chinese. PubMed/NCBI
|
|
38
|
Hsieh TH, Chien CL, Lee YH, Lin CI, Hsieh
JY, Chao ME, Liu DJ, Chu SS, Chen W, Lin SC, et al: Downregulation
of SUN2, a novel tumor suppressor, mediates miR-221/222-induced
malignancy in central nervous system embryonal tumors.
Carcinogenesis. 35:2164–2174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27kip1, p57kip2, and
PUMA. Am J Cancer Res. 3:465–477. 2013.
|
|
40
|
Hwang MS, Yu N, Stinson SY, Yue P, Newman
RJ, Allan BB and Dornan D: miR-221/222 targets adiponectin receptor
1 to promote the epithelial-to-mesenchymal transition in breast
cancer. PLoS One. 8:e665022013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun T, Wang X, He HH, Sweeney CJ, Liu SX,
Brown M, Balk S, Lee GS and Kantoff PW: MiR-221 promotes the
development of androgen independence in prostate cancer cells via
downregulation of HECTD2 and RAB1A. Oncogene. 33:2790–2800. 2014.
View Article : Google Scholar :
|
|
42
|
Jikuzono T, Kawamoto M, Yoshitake H,
Kikuchi K, Akasu H, Ishikawa H, Hirokawa M, Miyauchi A, Tsuchiya S,
Shimizu K and Takizawa T: The miR-221/222 cluster, miR-10b and
miR-92a are highly upregulated in metastatic minimally invasive
follicular thyroid carcinoma. Int J Oncol. 42:1858–1868.
2013.PubMed/NCBI
|
|
43
|
Acunzo M, Visone R, Romano G, Veronese A,
Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli
G, et al: miR-130a targets MET and induces TRAIL-sensitivity in
NSCLC by downregulating miR-221 and 222. Oncogene. 31:634–642.
2012.
|
|
44
|
Wang H, Xu C, Kong X, Li X, Kong X, Wang
Y, Ding X and Yang Q: Trail resistance induces
epithelial-mesenchymal transition and enhances invasiveness by
suppressing PTEN via miR-221 in breast cancer. PLoS One.
9:e990672014. View Article : Google Scholar : PubMed/NCBI
|