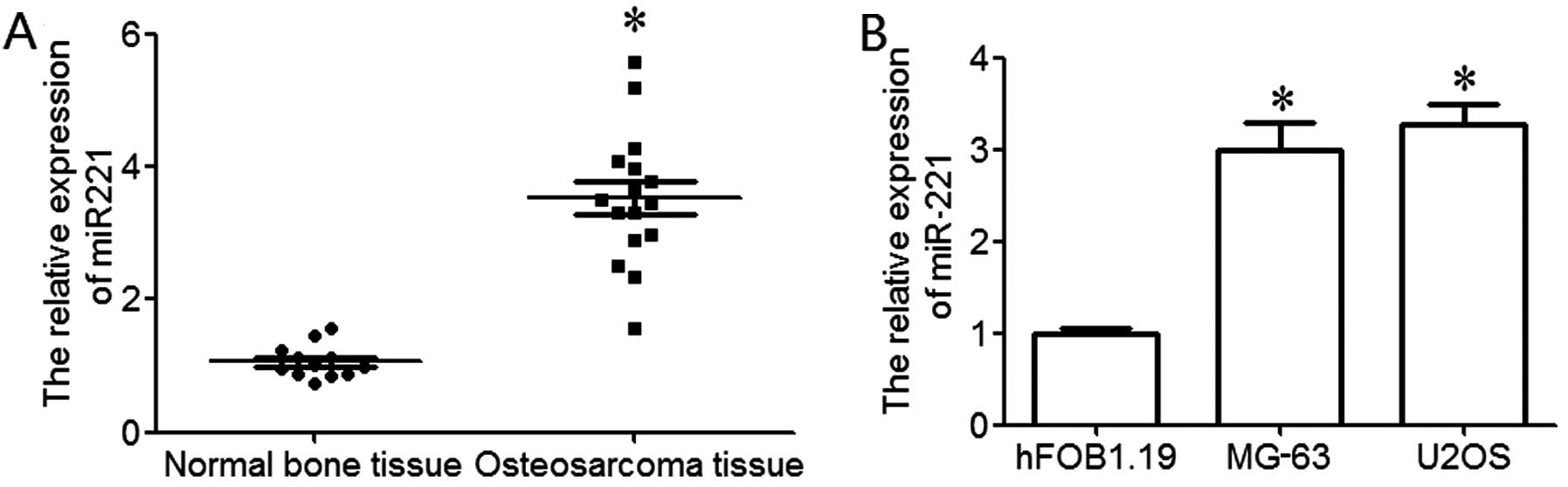
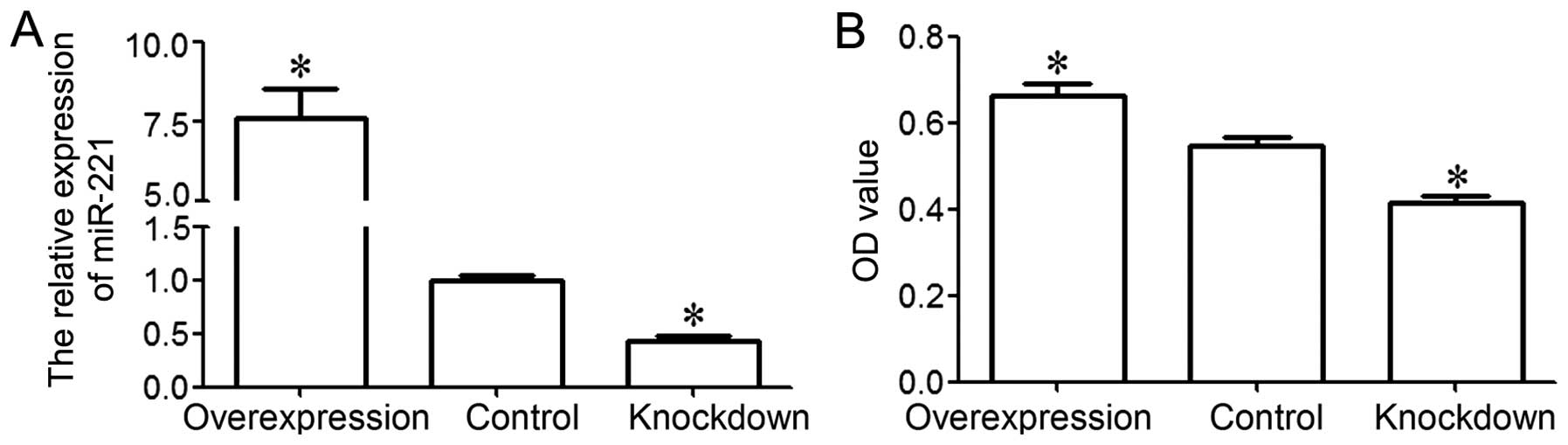
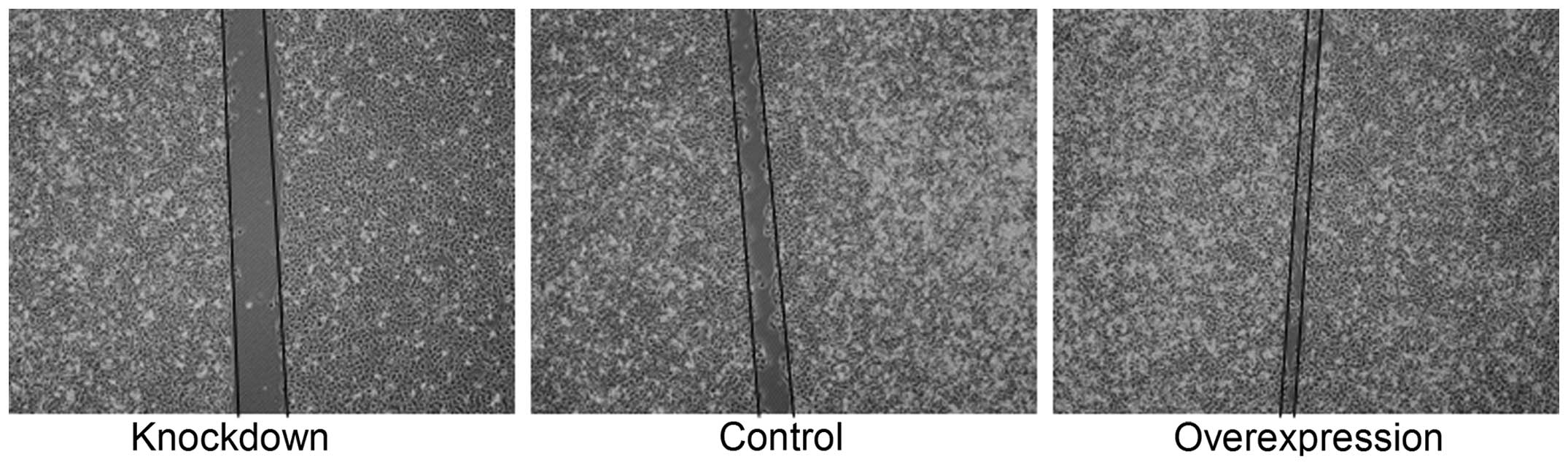
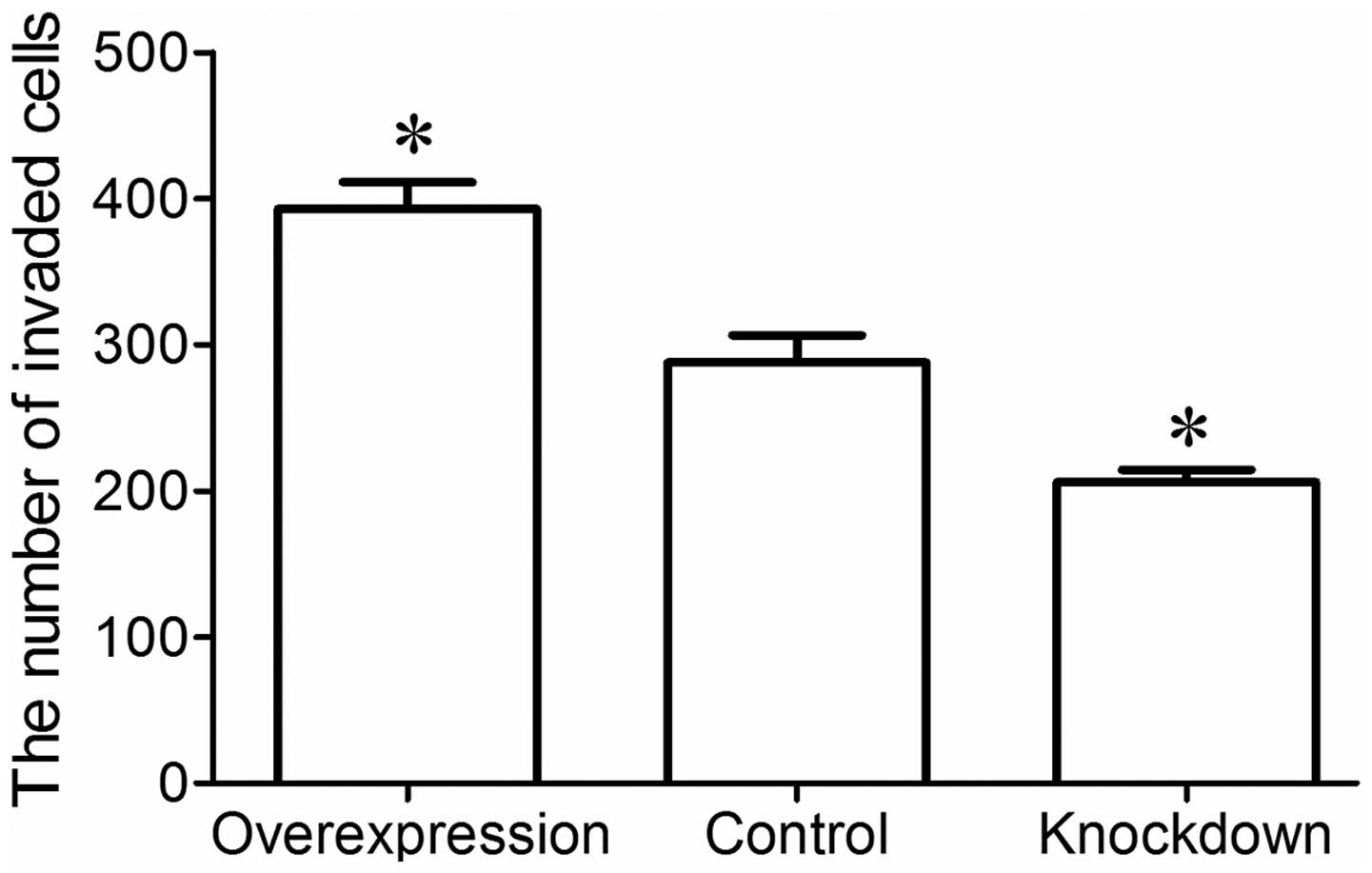
|
1
|
Endo-Munoz L, Evdokiou A and Saunders NA:
The role of osteoclasts and tumour-associated macrophages in
osteosarcoma metastasis. Biochim Biophys Acta. 1826.434–442.
2012.
|
|
2
|
Lamora A, Talbot J, Bougras G, Amiaud J,
Leduc M, Chesneau J, Taurelle J, Stresing V, Le Deley MC, Heymann
MF, et al: Overexpression of smad7 blocks primary tumor growth and
lung metastasis development in osteosarcoma. Clin Cancer Res.
20:5097–5112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harting MT and Blakely ML: Management of
osteosarcoma pulmonary metastases. Semin Pediatr Surg. 15:25–29.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jia S and Li B: Osteosarcoma of the jaws:
case report on synchronous multicentric osteosarcomas. J Clin Diagn
Res. 8:ZD01–ZD03. 2014.PubMed/NCBI
|
|
5
|
He JP, Hao Y, Wang XL, Yang XJ, Shao JF,
Guo FJ and Feng JX: Review of the molecular pathogenesis of
osteosarcoma. Asian Pac J Cancer Prev. 15:5967–5976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Poletajew S, Fus L and Wasiutyński A:
Current concepts on pathogenesis and biology of metastatic
osteosarcoma tumors. Ortop Traumatol Rehabil. 13:537–545. 2011.
View Article : Google Scholar
|
|
7
|
Egas-Bejar D, Anderson PM, Agarwal R,
Corrales-Medina F, Devarajan E, Huh WW, Brown RE and Subbiah V:
Theranostic profiling for actionable aberrations in advanced high
risk osteosarcoma with aggressive biology breveals high molecular
diversity: the human fingerprint hypothesis. Oncoscience.
1:167–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu CW, Cheng YW, Hsu NY, Yeh KT, Tsai YY,
Chiang CC, Wang WR and Tung JN: MiRNA-221 negatively regulated
downstream p27Kip1 gene expression involvement in pterygium
pathogenesis. Mol Vis. 20:1048–1056. 2014.PubMed/NCBI
|
|
9
|
Wolter JM, Kotagama K, Pierre-Bez AC,
Firago M and Mangone M: 3′LIFE: a functional assay to detect miRNA
targets in high-throughput. Nucleic Acids Res. 42:e1322014.
View Article : Google Scholar
|
|
10
|
Gaál Z and Oláh E: MicroRNA-s and their
role in malignant hematologic diseases. Orv Hetil. 153:2051–2059.
2012.In Hungarian. View Article : Google Scholar
|
|
11
|
Lin S, Pan L, Guo S, Wu J, Jin L, Wang JC
and Wang S: Prognostic role of microRNA-181a/b in hematological
malignancies: a meta-analysis. PLoS One. 8:e595322013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Sarkar N, Roy R, Mitra JK, Ghose S,
Chakraborty A, Paul RR, Mukhopadhyay I and Roy B: a quest for miRNA
bio-marker: a track back approach from gingivo buccal cancer to two
different types of precancers. PLoS One. 9:e1048392014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu C, Ping Y and Li X, Zhao H, Wang L, Fan
H, Xiao Y and Li X: Prioritizing candidate disease miRNAs by
integrating phenotype associations of multiple diseases with
matched miRNA and mRNA expression profiles. Mol Biosyst.
10:2800–2809. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye S, Yang L, Zhao X, Song W, Wang W and
Zheng S: Bioinformatics method to predict two regulation mechanism:
TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell
Biochem Biophys. 70:1849–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kushwaha D, Ramakrishnan V, Ng K, Steed T,
Nguyen T, Futalan D, Akers JC, Sarkaria J, Jiang T, Chowdhury D, et
al: A genome-wide miRNA screen revealed miR-603 as a
MGMT-regulating miRNA in glioblastomas. Oncotarget. 5:4026–4039.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kalinowski FC, Brown RA, Ganda C, Giles
KM, Epis MR, Horsham J and Leedman PJ: microRNA-7: a tumor
suppressor miRNA with therapeutic potential. Int J Biochem Cell
Biol. 54:312–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garofalo M, Quintavalle C, Romano G, Croce
CM and Condorelli G: miR221/222 in cancer: their role in tumor
progression and response to therapy. Curr Mol Med. 12:27–33. 2012.
View Article : Google Scholar
|
|
18
|
le Sage C, Nagel R, Egan DA, Schrier M,
Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et
al: Regulation of the p27Kip1 tumor suppressor by
miR-221 and miR-222 promotes cancer cell proliferation. EMBO J.
26:3699–3708. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fornari F, Gramantieri L, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM,
Bolondi L and Negrini M: MiR-221 controls CDKN1C/p57 and CDKN1B/p27
expression in human hepatocellular carcinoma. Oncogene.
27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting
p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Garofalo M, Di Leva G, Romano G, Nuovo G,
Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P,
et al: miR-221&222 regulate TRAIL resistance and enhance
tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell.
16:498–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Howe EN, Cochrane DR and Richer JK: The
miR-200 and miR-221/222 microRNA families: opposing effects on
epithelial identity. J Mammary Gland Biol Neoplasia. 17:65–77.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem.
282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He XX, Guo AY, Xu CR, Chang Y, Xiang GY,
Gong J, Dan ZL, Tian DA, Liao JZ and Lin JS: Bioinformatics
analysis identifies miR-221 as a core regulator in hepatocellular
carcinoma and its silencing suppresses tumor properties. Oncol Rep.
32:1200–1210. 2014.PubMed/NCBI
|
|
25
|
Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang
Y, Liu Z, Cao X, Chen P, Liu Z, et al: A microRNA miR-221- and
miR-222-mediated feedback loop, via PDLIM2, maintains constitutive
activation of nuclear factor kappaB and STAT3 in colorectal cancer
cells. Gastroenterology. 147:847–859. 2014. View Article : Google Scholar
|
|
26
|
Gan R, Yang Y, Yang X, Zhao L, Lu J and
Meng QH: Downregulation of miR-221/222 enhances sensitivity of
breast cancer cells to tamoxifen through upregulation of TIMP3.
Cancer Gene Ther. 21:290–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Felli N, Fontana L, Pelosi E, Botta R,
Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et
al: MicroRNAs 221 and 222 inhibit normal erythropoiesis and
erythroleukemic cell growth via kit receptor down-modulation. Proc
Natl Acad Sci USA. 102:18081–18086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang
L, Yang A, Zhao J and Jia L: MiR-221 promotes
trastuzumab-resistance and metastasis in HER2-positive breast
cancers by targeting PTEN. BMB Rep. 47:268–273. 2014. View Article : Google Scholar :
|
|
30
|
De Tullio G, De Fazio V, Sgherza N, Minoia
C, Serratì S, Merchionne F, Loseto G, Iacobazzi A, Rana A, Petrillo
P, et al: Challenges and opportunities of microRNAs in lymphomas.
Molecules. 19:14723–14781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu Y, Huang Z and Liu Y: Reduced
miR-125a-5p expression is associated with gastric carcinogenesis
through the targeting of E2F3. Mol Med Rep. 10:2601–2608.
2014.PubMed/NCBI
|
|
32
|
Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H
and Yu H: Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p
expression in colon cancer. Am J Transl Res. 6:391–401.
2014.PubMed/NCBI
|
|
33
|
Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y,
Li H, Zhu X, Yao L and Zhang J: Exosomal miR-221/222 enhances
tamoxifen resistance in recipient ER-positive breast cancer cells.
Breast Cancer Res Treat. 147:423–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang X, Yang Y, Gan R, Zhao L, Li W, Zhou
H, Wang X, Lu J and Meng QH: Down-regulation of mir-221 and mir-222
restrain prostate cancer cell proliferation and migration that is
partly mediated by activation of SIRT1. PLoS One. 9:e988332014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Falkenberg N, Anastasov N, Rappl K,
Braselmann H, Auer G, Walch A, Huber M, Höfig I, Schmitt M, Höfler
H, et al: MiR-221/-222 differentiate prognostic groups in advanced
breast cancers and influence cell invasion. Br J Cancer.
109:2714–2723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nassirpour R, Mehta PP, Baxi SM and Yin
MJ: miR-221 promotes tumorigenesis in human triple negative breast
cancer cells. PLoS One. 8:e621702013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen G, Dang YW and Luo DZ: Effect of
miR-221 on the viability and apoptosis of hepatocellular carcinoma
HepG2 cells. Zhonghua Gan Zang Bing Za Zhi. 19:582–587. 2011.In
Chinese. PubMed/NCBI
|
|
38
|
Hsieh TH, Chien CL, Lee YH, Lin CI, Hsieh
JY, Chao ME, Liu DJ, Chu SS, Chen W, Lin SC, et al: Downregulation
of SUN2, a novel tumor suppressor, mediates miR-221/222-induced
malignancy in central nervous system embryonal tumors.
Carcinogenesis. 35:2164–2174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27kip1, p57kip2, and
PUMA. Am J Cancer Res. 3:465–477. 2013.
|
|
40
|
Hwang MS, Yu N, Stinson SY, Yue P, Newman
RJ, Allan BB and Dornan D: miR-221/222 targets adiponectin receptor
1 to promote the epithelial-to-mesenchymal transition in breast
cancer. PLoS One. 8:e665022013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun T, Wang X, He HH, Sweeney CJ, Liu SX,
Brown M, Balk S, Lee GS and Kantoff PW: MiR-221 promotes the
development of androgen independence in prostate cancer cells via
downregulation of HECTD2 and RAB1A. Oncogene. 33:2790–2800. 2014.
View Article : Google Scholar :
|
|
42
|
Jikuzono T, Kawamoto M, Yoshitake H,
Kikuchi K, Akasu H, Ishikawa H, Hirokawa M, Miyauchi A, Tsuchiya S,
Shimizu K and Takizawa T: The miR-221/222 cluster, miR-10b and
miR-92a are highly upregulated in metastatic minimally invasive
follicular thyroid carcinoma. Int J Oncol. 42:1858–1868.
2013.PubMed/NCBI
|
|
43
|
Acunzo M, Visone R, Romano G, Veronese A,
Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli
G, et al: miR-130a targets MET and induces TRAIL-sensitivity in
NSCLC by downregulating miR-221 and 222. Oncogene. 31:634–642.
2012.
|
|
44
|
Wang H, Xu C, Kong X, Li X, Kong X, Wang
Y, Ding X and Yang Q: Trail resistance induces
epithelial-mesenchymal transition and enhances invasiveness by
suppressing PTEN via miR-221 in breast cancer. PLoS One.
9:e990672014. View Article : Google Scholar : PubMed/NCBI
|