Introduction
Hyperthermia (HT) therapy in combination with either
chemotherapy, radiotherapy or both are used for patients with
cancer in various organs. The anticancer effects of these
combination therapies have been verified in many clinical trials
(1–4). However, the acquisition of
thermotolerance in cancer cells, which is at least partly due to an
increase in the levels of heat shock proteins (HSPs), attenuates
the therapeutic effects of HT (5,6).
HSPs function as molecular chaperones, and their epxression is
induced by various stresses, particularly heat. Moreover, it has
been recognized that these proteins exert potent cytoprotective
effects, which prevent cell death (7,8).
HSPs consist of several family members, including DnaJ (Hsp40
homolog (DNAJ), heat shock 70 kDa protein (HSPA), heat shock 27 kDa
protein (HSPB), heat shock 60 kDa protein (HSPD) and heat shock 105
kDa/110 kDa protein (HSPH), and among these, HSPA1A plays a major
role as a molecular chaperone (9,10).
BCL2-associated athanogene (BAG) family proteins, an
ubiquitous family of chaperone regulators, have been found to be
associated with the anti-apoptotic protein, BCL2, and also to
interact with HSPA proteins, such as HSPA1A and HSPA8 (11,12). Among the BAG proteins, the
expression of BAG3 has been reported to be regulated, at least in
part, by the activation of heat shock transcription factor 1 as in
the cases of HSPs (13,14). Under normal conditions, the
expression level of BAG3 is relatively low, whereas a significant
elevation in its protein level is observed in cells exposed to
stressors, such as heavy metals (15), heat (16–18), oxidative stress (19) and ultrasound (20). It has also been indicated that
BAG3 is abundantly expressed in a variety of cancers, and is
involved in cellular processes such as cell growth and cell death
(11,12,16,21–24). Liu et al (25) previously reported that silencing
the BAG3 gene sensitizes leukemic cells to compound-induced cell
injury. Recently, we clearly demonstrated that the inhibition of
BAG3 improves cell death sensitivity to HT in cancer cells
(17,18). However, the detailed molecular
mechanisms underling the enhancement of HT sensitivity by BAG3
knockdown (KD) in cancer cells have not yet been elucidated.
In the present study, we examined gene expression
patterns in human oral squamous cell carcinoma (OSCC) HSC-3 cells
exposed to HT and transfected with small interfering RNA (siRNA)
against BAG3 using a global-scale microarray system. In addition,
gene network analysis of differentially expressed genes was
performed using computational gene expression analysis tools.
Materials and methods
Cell culture and exposure to HT
Human OSCC HSC-3 cells were obtained from the Human
Science Research Resources Bank, Japan Health Sciences Foundation
(Tokyo, Japan). The HSC-3 cells were cultured in E-MEM (Wako Pure
Chemical Industries, Ltd., Osaka, Japan) supplemented with 10%
fetal bovine serum (FBS) at 37°C in humidified air with 5%
CO2 and 95% air. Exposure to HT was were performed by
immersing plastic culture vessels containing the attached cells in
a water bath at 44°C for 90 min. Following exposure to HT, the
cells were incubated for 6–24 h at 37°C, as previously described
(26).
siRNA transfection
A siRNA (siBAG; GGUGGAUUCUAAA CCUGUU) targeting BAG3
for BAG3-KD was designed by Nippon EGT Co., Ltd. (Toyama, Japan).
Luciferase siRNA (siLuc; CGUACGCGGAAUACUUCGA) was used as a
negative control siRNA. The cells were incubated in
Opti-MEM® I Reduced Serum Medium containing 20 nM siRNA
and Lipofectamine™ RNAiMAX (both from Life Technologies Japan,
Ltd., Tokyo, Japan) at 37°C. Six hours following transfection, the
medium was exchanged for E-MEM supplemented with 10% FBS, and the
cells were then maintained at 37°C for 42 h, as previously
described (18).
Measurements of cell growth and cell
death
The number of cells was counted using a
hematocytometer. When the cell death was evaluated, the cells were
treated with 0.2% trypan blue solution (NanoEnTek Inc., Seoul,
Korea) at room temperature for 5 min. The number of dead cells
(stained) and viable cells (unstained) was counted using an EVE™
automatic cell counter (NanoEnTek Inc.).
Sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and western blot analysis
The cells were dissolved in lysis buffer (150 mM
NaCl, 1% Nonidet P-40 and 50 mM Tris-HCl, pH 8.0) containing a
protease inhibitor cocktail (Nacalai Tesque Inc., Kyoto, Japan).
SDS-PAGE and western blot analysis were carried out as previously
described (27,28). The primary antibodies used were as
follows: a rabbit monoclonal anti-BAG3 antibody (GTX62327; GeneTex
Inc., Irvine, CA, USA) and a mouse monoclonal anti-glyceraldehyde
3-phosphate dehydrogenase (GAPDH) antibody (MAB374; Millipore Co.,
Temecula, CA, USA). Immunoreactive proteins were visualized using a
luminescent image analyzer (LAS-4000 mini; GE Healthcare, Tokyo,
Japan) using an enhanced chemiluminescence detection system. GAPDH
served as a loading control.
RNA isolation
Total RNA was extracted from cells using a
NucleoSpin® RNA isolation kit (Macherey-Nagel GmbH &
Co., Düren, Germany) along with on-column DNase I treatment. The
RNA quality was analyzed using a Bioanalyzer 2100 (Agilent
Technologies, Inc., Santa Clara, CA, USA). RNA samples with RNA
integrity number (RIN) values >9.5 were considered
acceptable.
Quantitative (real-time) polymerase chain
reaction (qPCR)
qPCR was performed on a Real-Time PCR system Mx3005P
(Agilent Technologies, Inc.) using SYBR® Premix Ex Taq™
II (Takara Bio, Inc., Shiga, Japan) according to the manufacturer's
instructions. Reverse transcriptase reaction was carried out with
total RNA using a random 6 mers and an oligo dT primer (PrimeScript
RT reagent kit; Takara Bio, Inc.). The reaction was carried out
using the specific primers: human BAG3 forward and reverse,
CGACCAGGCTACATTCCCAT and TCTGGCT GAGTGGTTTCTGG, respectively; human
GAPDH forward and reverse, AAGGCTGGGGCTCATTTGCA and ATGACC
TTGCCCACAGCCTT, respectively. The temperature cycling conditions
for each primer consisted of 10 min at 95°C followed by 40 cycles
for 10 sec at 95°C and 40 sec at 60°C. The mRNA expression level of
BAG3 was normalized with respect to the mRNA expression level of
GAPDH, as described in a previous study of ours (18).
Microarray gene expression analysis
Microarray gene expression analysis was performed
using a GeneChip® system with a Human Genome U133-plus
2.0 array, which was spotted with 54,675 probe sets (Affymetrix,
Inc., Santa Clara, CA, USA) according to the manufacturer's
instructions. In brief, 500 ng of total RNA was used to synthesize
cRNA with a GeneChip® 3′ IVT Express kit (Affymetrix,
Inc.). Fragmentated biotin-labeled cRNA was hybridized to the array
at 45°C for 16 h. After the staining with
streptavidin-phycoerythrin, the array was scanned using a probe
array scanner. The obtained hybridization intensity data were
analyzed using GeneSpring® GX software (Agilent
Technologies, Inc.) to extract the significant genes. To examine
gene ontology, including biological processes, cellular components,
molecular functions and gene networks, the obtained data were
analyzed using Ingenuity® pathway analysis tools
(Ingenuity Systems, Inc., Mountain View, CA, USA), as previously
described (29,30).
Statistical analysis
Data are shown as the means ± SD. The Student's
t-test was used for statistical analysis and P-values <0.05 were
considered to indicate statistically significant differences.
Results
Effects of BAG3-KD on the growth and
death of HSC-3 cells exposed to HT
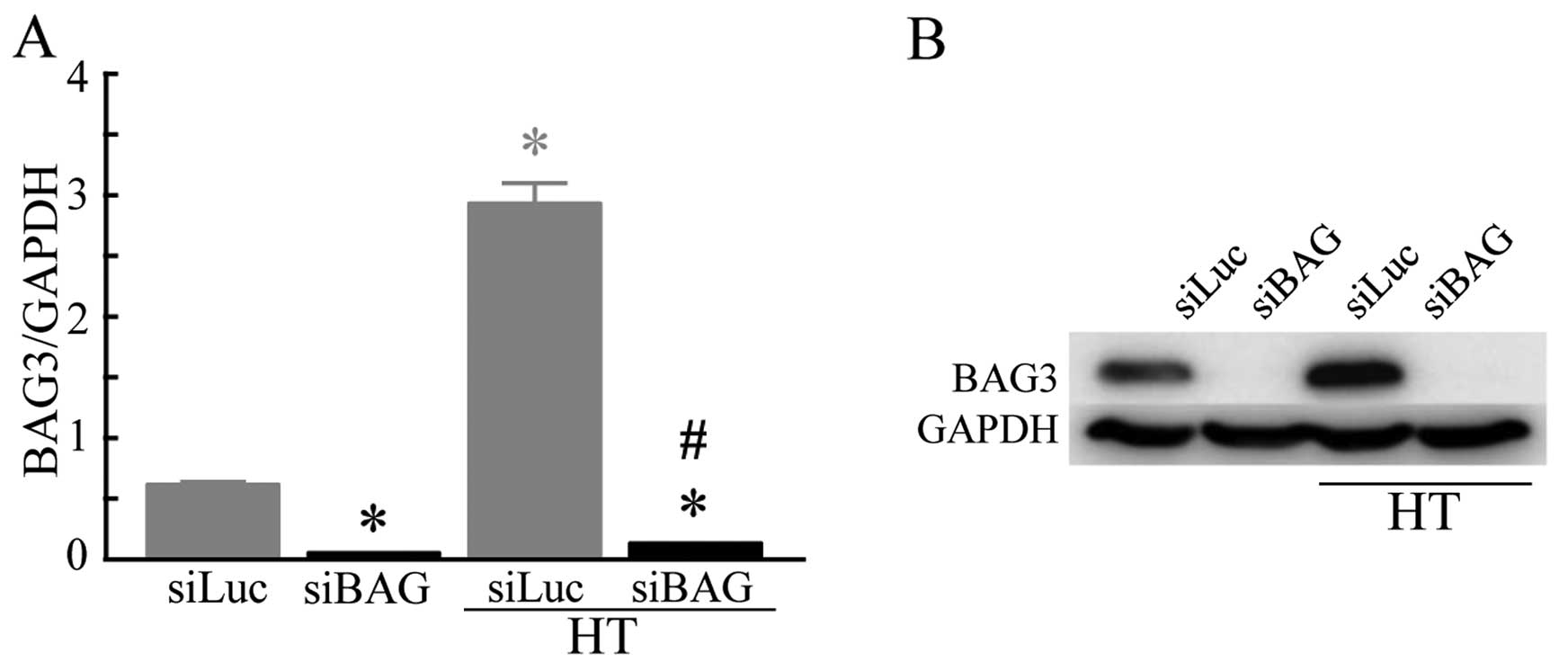
Although the mRNA expression level of BAG3 was
relatively low in the HSC-3 cells transfected with siLuc (control),
in the cells subjected to both siLuc transfection and HT exposure
at 44°C (HT control), a significantly increased expression level of
BAG3 was observed. A significant decrease in the mRNA expression
level of BAG3 was detected in the cells transfected with siBAG
under both the control (siLuc) and HT conditions (Fig. 1A). The results of western blot
analysis clearly demonstrated that the protein expression level of
BAG3 was significantly increased in the cells exposed to HT.
Transfection of the cells with siBAG almost completely inhibited
the protein expression level of BAG3 under either condition
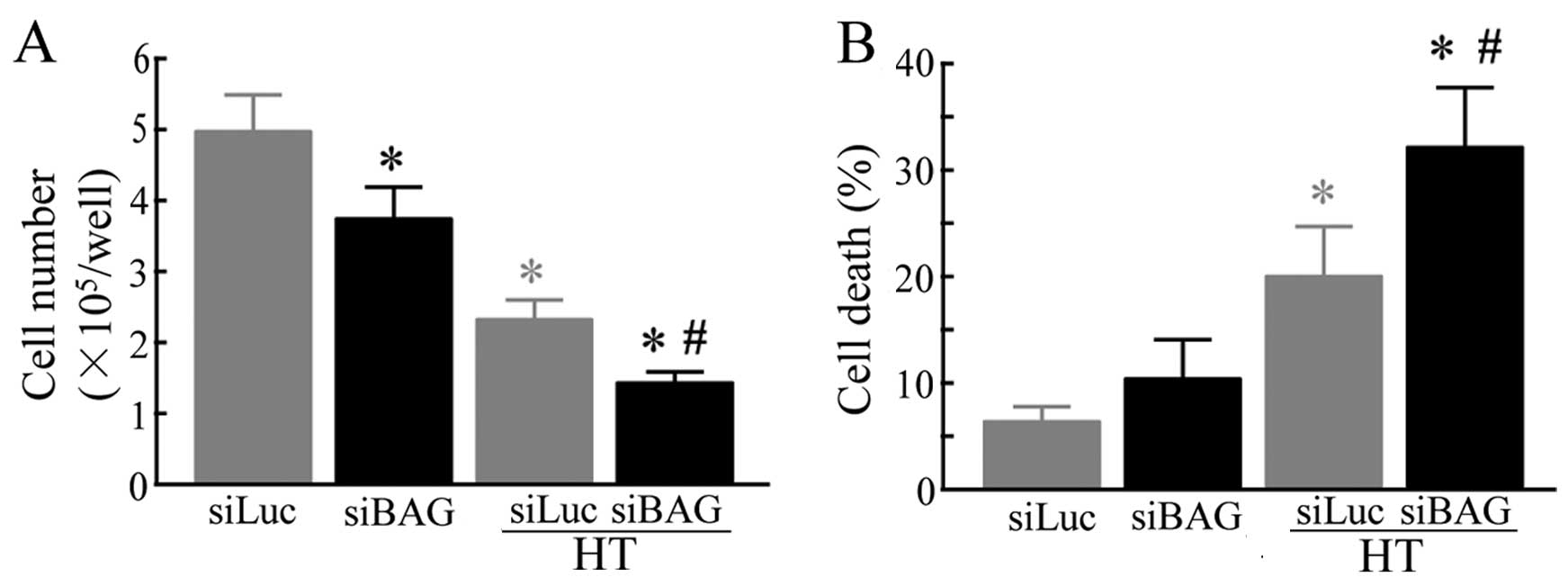
(Fig. 1B). We then evaluated
whether BAG3-KD affected the growth and death of HSC-3 cells
exposed to HT. At the normal temperature, transfection of the cells
with siBAG significantly suppressed the cell number compared to the
control group. HT markedly decreased cell growth, and a further
decrease in the number of cells was observed in the cells subjected
to both siBAG transfection and exposure to HT to those exposed to
HT alone (Fig. 2A). HT
significantly enhanced cell death. Moreover, a significant increase
in cell death was observed in the cells subjected to both siBAG
transfection and exposure to HT compared to those exposed to HT
alone. These results indicate that the silencing of BAG3 enhances
the sensitivity of human OSCC HSC-3 cells to HT (Fig. 2B).
Global gene expression analysis
To identify genes involved in the enhancement of HT
sensitivity by BAG3-KD, global-scale gene expression analysis was
carried out using a GeneChip® system with a Human Genome
U133-plus 2.0 array, which was spotted with 54,675 probe sets.
Complete lists of probe sets from all samples are available on the
Gene Expression Omnibus, a public database (accession number,
GSE75127). GeneSpring software was used to analyze gene expression
in the HSC-3 cells subjected to both HT exposure and siLuc (HT
control) or siBAG transfection (HT + BAG3-KD), and revealed that
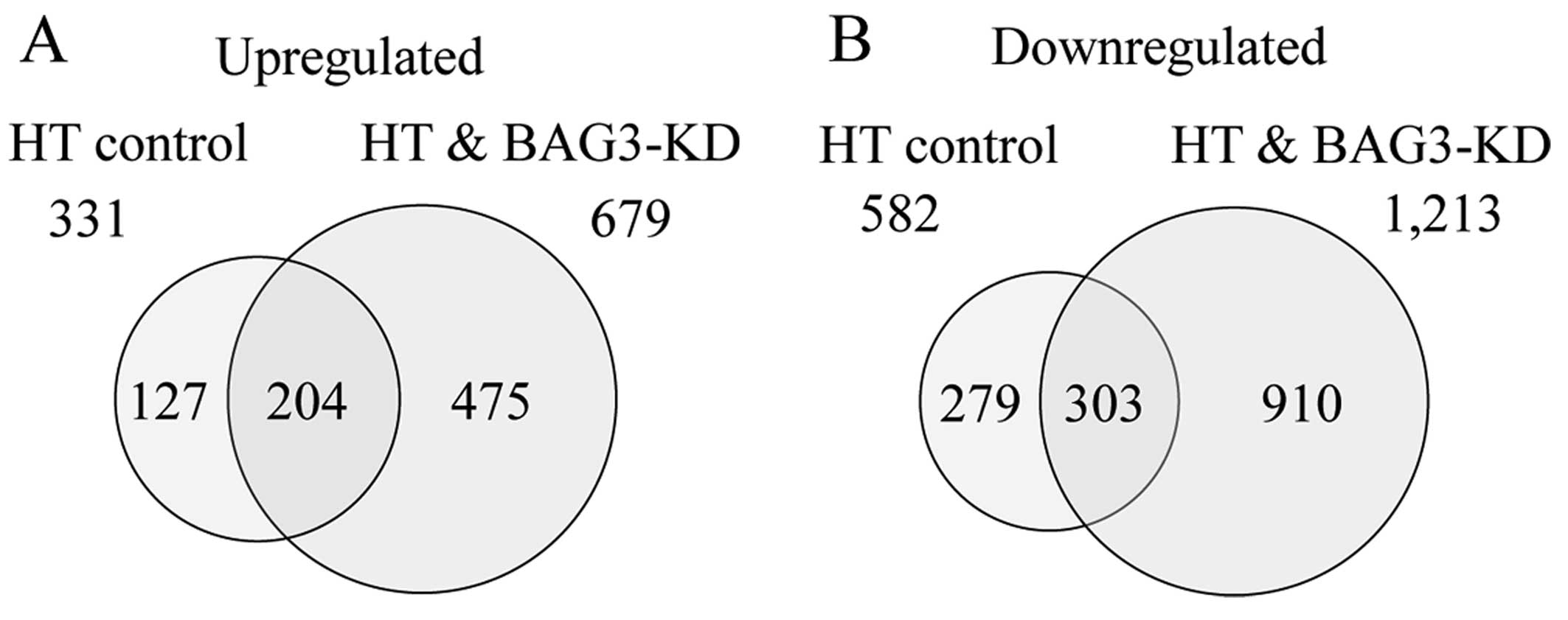
many genes were differentially regulated by a factor of ≥2.0. The
Venn diagram in Fig. 3 summarizes
the numbers of specifically and commonly expressed genes in each
group. The total numbers of genes that were found to be
differentially expressed were 913 (331 up- and 582 downregulated
genes) and 1,892 (679 up- and 1,213 downregulated genes) in the HT
control and HT + BAG3-KD groups, respectively. The numbers of
commonly up- and downregulated genes were 204 and 303, respectively
(Fig. 3A and B).
Identification of biological functions
and gene networks
In order to identify the biological functions and
gene networks in differentially expressed genes involved in the
enhancement of HT sensitivity by BAG3-KD, functional category and
gene network analyses were conducted by use of the Ingenuity
Pathways Knowledge Base. We identified many functionally annotated
genes, and the top 3 biological functions in each group are
summarized in Table I. In the
upregulated genes, biological functions including cell death and
survival, and/or cell growth and proliferation were observed in all
3 groups: i) the HT control only; ii) the HT + BAG3-KD only; and
iii) the commonly regulated groups. On the other hand, these 2
biological functions were observed only in the downregulated genes
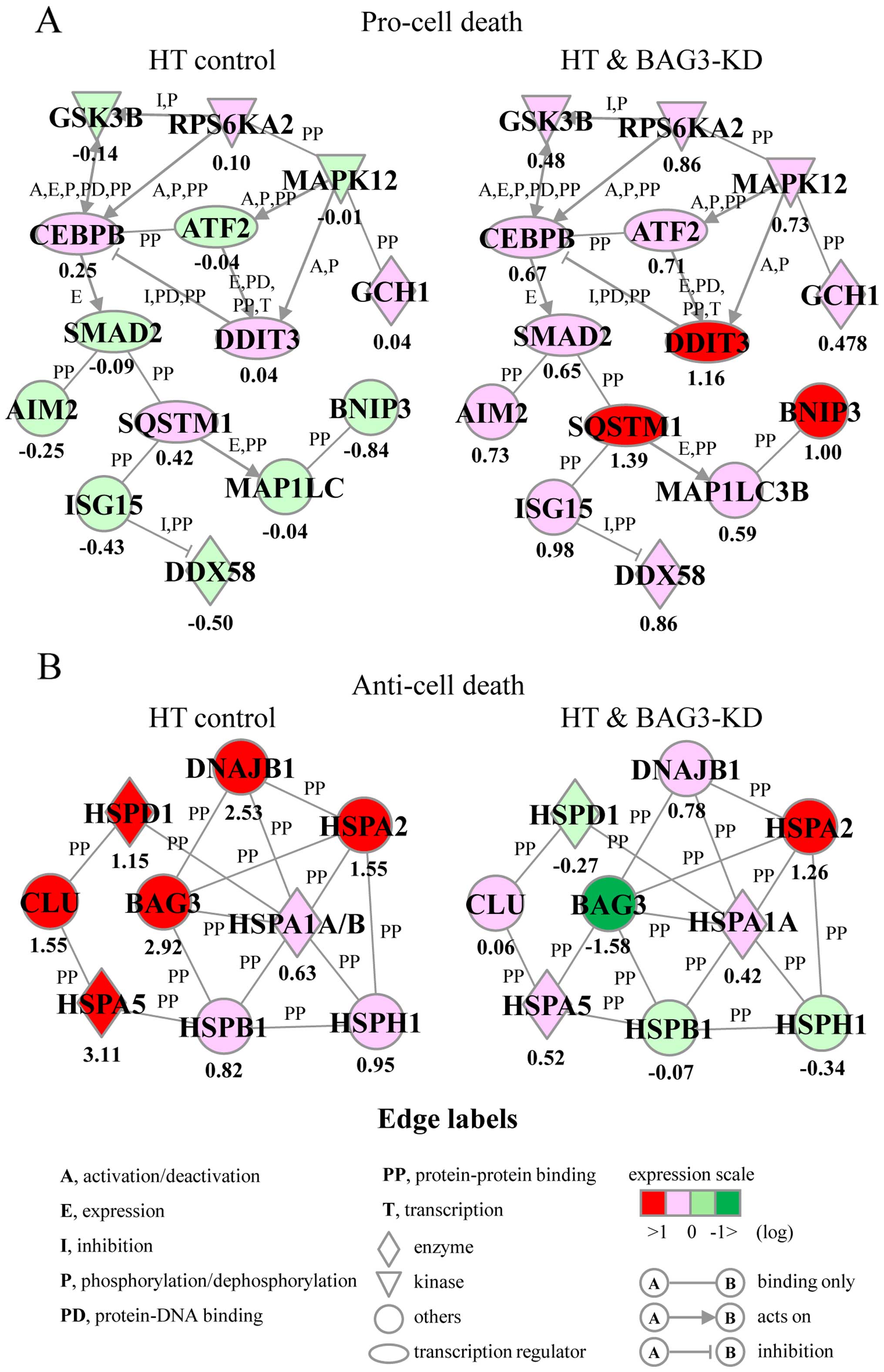
of the HT + BAG3-KD only group. In addition, we identified 2 unique
gene networks, and these are designated as Pro-cell death and
Anti-cell death, that were obtained from the upregulated genes
(Fig. 4). The Pro-cell death gene
network included several transcription factors, such as activating
transcription factor 2 (ATF2), CCAAT/enhancer binding protein β
(CEBPB), DNA damage inducible transcript 3 (DDIT3), SMAD family
member 2 (SMAD2) and sequestosome 1 (SQSTM1), as well as
BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), and was
associated with the biological function of the induction of cell
death (Fig. 4A). The Anti-cell
death gene network contained several HSPs, such as DNAJB1, HSPA1A,
HSPA5, HSPB1, HSPD1, and HSPH1, as well as BAG3 and clusterin
(CLU), and was associated with the biological function of the
prevention of cell death (Fig.
4B). The expression levels of genes in the Pro-cell death and
Anti-cell death gene networks were significantly elevated and
reduced in the HT + BAG3-KD group compared to those in the HT
control group, respectively (Fig. 4A
and B). As expected, the mRNA expression level of BAG3 was
markedly decreased in the HT + BAG3-KD group as detected by the
microarray system (Fig. 4B).
 | Table ITop three biological functions in
differentially expressed genes. |
Table I
Top three biological functions in
differentially expressed genes.
| Name | P-value | Number of
molecules |
|---|
| Upregulated | | |
| HT control only
(75)a | | |
| Cell growth and
proliferation |
5.02E-05–4.19E-02 | 36 |
|
Post-translational modification |
1.20E-04–3.51E-02 | 6 |
| Protein
folding |
1.20E-04–2.12E-02 | 4 |
| HT + BAG3-KD only
(263)a | | |
| Cell growth and
proliferation |
1.47E-05–2.73E-02 | 123 |
| Cell death and
survival |
3.35E-05–2.73E-02 | 121 |
| Cellular
development |
1.93E-04–2.73E-02 | 92 |
| Commonly
regulated (133)a | | |
| Cell death and
survival |
2.03E-17–2.70E-03 | 82 |
| Cell growth and
proliferation |
4.39E-14–2.70E-03 | 84 |
| Cell cycle |
1.26E-12–2.70E-03 | 39 |
| Downregulated | | |
| HT control only
(62)a | | |
| Cell cycle |
7.27E-04–4.99E-02 | 17 |
| Gene
expression |
3.52E-03–4.63E-02 | 6 |
| Protein
synthesis |
4.88E-04–1.57E-02 | 3 |
| HT + BAG3-KD only
(432)a | | |
| Cellular
development |
2.20E-06–2.64E-02 | 123 |
| Cell growth and
proliferation |
2.20E-06–2.70E-02 | 121 |
| Cell death and
survival |
1.28E-05–2.70E-02 | 92 |
| Commonly
regulated (108)a | | |
| RNA
post-transcriptional modification |
4.19E-05–3.58E-02 | 9 |
| Cell cycle |
2.90E-04–4.74E-02 | 32 |
| Cell
morphology |
2.90E-04–3.58E-02 | 26 |
Discussion
BAG3, a co-chaperone of the HSPA family of proteins,
is well known as a cytoprotective protein that acts against various
stresses, including heat stress (11,12,16,21–25). In the present study, the almost
complete silencing of BAG3 significantly enhanced sensitivity of
human OSCC HSC-3 cells to HT. This finding is compatible with those
of our previous studies (17,18). In addition, using global-scale
microarray and bioinformatics analyses, we herein identified genes
and gene networks involved in the enhancement of HT sensitivity in
BAG3-KD OSCC cells.
Our functional category analysis demonstrated that
biological functions including cell death and survival, and cell
growth and proliferation were observed in the upregulated genes in
the cells from the HT + BAG3-KD group (Table I). Of note, we also successfully
identified 2 unique gene networks, designated as Pro-cell death and
Anti-cell death (Fig. 4). The
Pro-cell death gene network consisted of 14 genes and was
principally associated with the biological function of the
induction of cell death. A marked induction of genes in this
network was observed in the HT + BAG3-KD group compared to the HT
control group (Fig. 4A). This
network included 3 basic-region leucine zipper (bZIP) transcription
factors, ATF2 (31), CEBPB
(32) and DDIT3 (33), which have been reported to induce
cell death. Homo- or hetero-dimeric protein complexes of the bZIP
protein function as repressors and activators of transcription
(34); associations have been
identified between DDIT3 and both ATF2 and CEBPB (34–36). The activation of these bZIP
transcription factors has also been reported to be regulated by
kinases, such as glycogen synthase kinase 3β (GSK3β) (37), ribosomal protein S6 kinase, 90
kDa, polypeptide 2 (RPS6KA2) (38) and mitogen-activated protein kinase
12 (MAPK12) (39). Moreover,
absent in melanoma 2 (AIM2) (40), BNIP3 (41), DEAD box polypeptide 58 (DDX58)
(42), GTP cyclohydrolase 1
(GCH1) (43), ISG15
ubiquitin-like modifier (ISG15) (44), microtubule-associated protein 1
light chain 3 beta (MAP1LC3B) (45), SMAD2 (46), and SQSTM1 (47) have been reported to exert
cell-damaging effects.
On the other hand, the expression levels of genes in
the Anti-cell death gene network were significantly decreased in
the HT + BAG3-KD group compared to those in the HT control group
(Fig. 4B). This gene network
consisted of 9 chaperone genes, 7 HSPs, CLU and BAG3. It is well
known that HSPs protect cells both by protein chaperoning and
refolding and by directly interfering with the cell death pathway
(7,8). HSPs such as DNAJB1 (48), HSPA1A (48,49), HSPA2 (50), HSPA5 (51), HSPB1 (52), HSPD1 (49) and HSPH1 (53) were found to be associated with the
prevention of cell death. Of note, BAG3 silencing markedly
decreased the expression levels of CLU, DNAJB1, HSPA5, HSPB1, HSPD1
and HSPH1 in HSC-3 cells induced by HT exposure (Fig. 4B). CLU is a secreted or cytosolic
chaperone that is expressed under certain stress conditions such as
heat shock (54), and secretory
human CLU has been reported to decrease the rate of cell death of
human breast cancer cells (55).
In addition, protein-protein interactions have been reported
between BAG3 and DNAJB1 (12),
HSPA5 (56) and HSPB1 (12) under in vitro experimental
conditions.
Taken together, our results suggest that an increase
in gene expression in the Pro-cell death gene network, and the
decrease in gene expression in the Anti-cell death gene network may
be closely associated with the enhancement of HT-induced cell death
by BAG3-KD in OSCC cells. However, the interaction between gene
expression and the enhancement of the HT effects remains a subject
for further study. In clinical fields, HT combined with
radiotherapy and/or chemotherapy has been used as a possible
treatment modality for various types of cancer (1–4).
However, the thermotolerance resulting from the elevation of HSP
expression and other cytoprotective proteins in some cancer cells
remains a disadvantage, diminishing the effects of HT (5,6).
The functional silencing of BAG3, a co-chaperone of the HSPA family
of proteins, may effectively enhance the sensitivity of cancer
cells to HT. Therefore, the targeting of BAG3 in combination with
HT may become a promising therapeutic approach for the treatment of
cancer (17,18).
Abbreviations:
|
AIM2
|
absent in melanoma 2
|
|
ATF2
|
activating transcription factor 2
|
|
BAG
|
BCL2-associated athanogene
|
|
BNIP3
|
BCL2/adenovirus E1B 19 kDa interacting
protein 3
|
|
bZIP
|
basic-region leucine zipper
|
|
CEBPB
|
CCAAT/enhancer binding protein β
|
|
CLU
|
clusterin
|
|
DDIT3
|
DNA damage inducible transcript 3
|
|
DDX58
|
DEAD box polypeptide 58
|
|
DNAJ
|
DnaJ (Hsp40) homolog
|
|
FBS
|
fetal bovine serum
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
GCH1
|
GTP cyclohydrolase 1
|
|
GSK3B
|
glycogen synthase kinase 3β
|
|
HSPA
|
heat shock 70 kDa protein
|
|
HSPB
|
heat shock 27 kDa protein
|
|
HSPD
|
heat shock 60 kDa protein
|
|
HSPH
|
heat shock 105 kDa/110 kDa protein
|
|
HSPs
|
heat shock proteins
|
|
HT
|
hyperthermia
|
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
KD
|
knockdown
|
|
MAP1LC3B
|
microtubule-associated protein 1 light
chain 3 beta
|
|
MAPK12
|
mitogen-activated protein kinase
12
|
|
OSCC
|
oral squamous cell carcinoma
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90 kDa,
polypeptide 2
|
|
SDS-PAGE
|
sodium dodecyl sulfate-polyacrylamide
gel electrophoresis
|
|
siRNA
|
small interfering RNA
|
|
SMAD2
|
SMAD family member 2
|
|
SQSTM1
|
sequestosome 1
|
Acknowledgments
The present study was supported in part by a
Grant-in-Aid for Challenging Exploratory Research (23650303) and a
Grant-in-Aid for Scientific Research B (24310046) from Japan
Society for the Promotion of Science, and by research grants from
the University of Toyama.
References
|
1
|
van der Zee J, González González D, van
Rhoon GC, van Dijk JD, van Putten WL and Hart AA: Comparison of
radiotherapy alone with radiotherapy plus hyperthermia in locally
advanced pelvic tumours: a prospective, randomised, multicentre
trial. Dutch Deep Hyperthermia Group. Lancet. 355:1119–1125. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harima Y, Nagata K, Harima K, Ostapenko
VV, Tanaka Y and Sawada S: A randomized clinical trial of radiation
therapy versus thermoradiotherapy in stage IIIB cervical carcinoma.
Int J Hyperthermia. 17:97–105. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Westermann A, Mella O, Van Der Zee J,
Jones EL, Van Der Steen-Banasik E, Koper P, Uitterhoeve AL, De Wit
R, Van Der Velden J, Burger C, et al: Long-term survival data of
triple modality treatment of stage IIB-III-IVA cervical cancer with
the combination of radiotherapy, chemotherapy and hyperthermia - an
update. Int J Hyperthermia. 28:549–553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cihoric N, Tsikkinis A, van Rhoon G,
Crezee H, Aebersold DM, Bodis S, Beck M, Nadobny J, Budach V, Wust
P, et al: Hyperthermia-related clinical trials on cancer treatment
within the http://ClinicalTrials.govurisimpleClinicalTrials.gov
registry. Int J Hyperthermia. 31:609–614. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li GC, Mivechi NF and Weitzel G: Heat
shock proteins, thermotolerance, and their relevance to clinical
hyperthermia. Int J Hyperthermia. 11:459–488. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nollen EA, Brunsting JF, Roelofsen H,
Weber LA and Kampinga HH: In vivo chaperone activity of heat shock
protein 70 and thermotolerance. Mol Cell Biol. 19:2069–2079. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Beere HM: 'The stress of dying': the role
of heat shock proteins in the regulation of apoptosis. J Cell Sci.
117:2641–2651. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lanneau D, Wettstein G, Bonniaud P and
Garrido C: Heat shock proteins: cell protection through protein
triage. Scientific World Journal. 10:1543–1552. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ohtsuka K and Hata M: Molecular chaperone
function of mammalian Hsp70 and Hsp40 - a review. Int J
Hyperthermia. 16:231–245. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vos MJ, Hageman J, Carra S and Kampinga
HH: Structural and functional diversities between members of the
human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry.
47:7001–7011. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kabbage M and Dickman MB: The BAG
proteins: a ubiquitous family of chaperone regulators. Cell Mol
Life Sci. 65:1390–1402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Taipale M, Tucker G, Peng J, Krykbaeva I,
Lin ZY, Larsen B, Choi H, Berger B, Gingras AC and Lindquist S: A
quantitative chaperone interaction network reveals the architecture
of cellular protein homeostasis pathways. Cell. 158:434–448. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Franceschelli S, Rosati A, Lerose R, De
Nicola S, Turco MC and Pascale M: Bag3 gene expression is regulated
by heat shock factor 1. J Cell Physiol. 215:575–577. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du ZX, Zhang HY, Meng X, Gao YY, Zou RL,
Liu BQ, Guan Y and Wang HQ: Proteasome inhibitor MG132 induces BAG3
expression through activation of heat shock factor 1. J Cell
Physiol. 218:631–637. 2009. View Article : Google Scholar
|
|
15
|
Pagliuca MG, Lerose R, Cigliano S and
Leone A: Regulation by heavy metals and temperature of the human
BAG-3 gene, a modulator of Hsp70 activity. FEBS Lett. 541:11–15.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liao Q, Ozawa F, Friess H, Zimmermann A,
Takayama S, Reed JC, Kleeff J and Büchler MW: The anti-apoptotic
protein BAG-3 is overexpressed in pancreatic cancer and induced by
heat stress in pancreatic cancer cell lines. FEBS Lett.
503:151–157. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yunoki T, Kariya A, Kondo T, Hayashi A and
Tabuchi Y: The combination of silencing BAG3 and inhibition of the
JNK pathway enhances hyperthermia sensitivity in human oral
squamous cell carcinoma cells. Cancer Lett. 335:52–57. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yunoki T, Tabuchi Y, Hayashi A and Kondo
T: BAG3 protects against hyperthermic stress by modulating NF-κB
and ERK activities in human retinoblastoma cells. Graefes Arch Clin
Exp Ophthalmol. 253:399–407. 2015. View Article : Google Scholar
|
|
19
|
Bonelli P, Petrella A, Rosati A, Romano
MF, Lerose R, Pagliuca MG, Amelio T, Festa M, Martire G, Venuta S,
et al: BAG3 protein regulates stress-induced apoptosis in normal
and neoplastic leukocytes. Leukemia. 18:358–360. 2004. View Article : Google Scholar
|
|
20
|
Tabuchi Y, Ando H, Takasaki I, Feril LB
Jr, Zhao QL, Ogawa R, Kudo N, Tachibana K and Kondo T:
Identification of genes responsive to low intensity pulsed
ultrasound in a human leukemia cell line Molt-4. Cancer Lett.
246:149–156. 2007. View Article : Google Scholar
|
|
21
|
Chiappetta G, Ammirante M, Basile A,
Rosati A, Festa M, Monaco M, Vuttariello E, Pasquinelli R, Arra C,
Zerilli M, et al: The antiapoptotic protein BAG3 is expressed in
thyroid carcinomas and modulates apoptosis mediated by tumor
necrosis factor-related apoptosis-inducing ligand. J Clin
Endocrinol Metab. 92:1159–1163. 2007. View Article : Google Scholar
|
|
22
|
Festa M, Del Valle L, Khalili K, Franco R,
Scognamiglio G, Graziano V, De Laurenzi V, Turco MC and Rosati A:
BAG3 protein is overexpressed in human glioblastoma and is a
potential target for therapy. Am J Pathol. 178:2504–2512. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rosati A, Graziano V, De Laurenzi V,
Pascale M and Turco MC: BAG3: a multifaceted protein that regulates
major cell pathways. Cell Death Dis. 2:e1412011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nymoen DA, Hetland Falkenthal TE, Holth A,
Ow GS, Ivshina AV, Tropé CG, Kuznetsov VA, Staff AC and Davidson B:
Expression and clinical role of chemoresponse-associated genes in
ovarian serous carcinoma. Gynecol Oncol. 139:30–39. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu P, Xu B, Li J and Lu H: BAG3 gene
silencing sensitizes leukemic cells to bortezomib-induced
apoptosis. FEBS Lett. 583:401–406. 2009. View Article : Google Scholar
|
|
26
|
Kariya A, Furusawa Y, Yunoki T, Kondo T
and Tabuchi Y: A microRNA-27a mimic sensitizes human oral squamous
cell carcinoma HSC-4 cells to hyperthermia through downregulation
of Hsp110 and Hsp90. Int J Mol Med. 34:334–340. 2014.PubMed/NCBI
|
|
27
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: procedure and some applications. Proc Natl
Acad Sci USA. 76:4350–4354. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tabuchi Y, Takasaki I, Doi T, Ishii Y,
Sakai H and Kondo T: Genetic networks responsive to sodium butyrate
in colonic epithelial cells. FEBS Lett. 580:3035–3041. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tabuchi Y, Yunoki T, Hoshi N, Suzuki N and
Kondo T: Genes and gene networks involved in sodium
fluoride-elicited cell death accompanying endoplasmic reticulum
stress in oral epithelial cells. Into J Mol Sci. 15:8959–8978.
2014. View Article : Google Scholar
|
|
31
|
Baan B, van Dam H, van der Zon GC, Maassen
JA and Ouwens DM: The role of c-Jun N-terminal kinase, p38, and
extracellular signal-regulated kinase in insulin-induced Thr69 and
Thr71 phosphorylation of activating transcription factor 2. Mol
Endocrinol. 20:1786–1795. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan HC, Yang CN, Hung YW, Lee WJ, Tien HR,
Shen CC, Sheehan J, Chou CT and Sheu ML: Reciprocal modulation of
C/EBP-α and C/EBP-β by IL-13 in activated microglia prevents
neuronal death. Eur J Immunol. 43:2854–2865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zinszner H, Kuroda M, Wang X, Batchvarova
N, Lightfoot RT, Remotti H, Stevens JL and Ron D: CHOP is
implicated in programmed cell death in response to impaired
function of the endoplasmic reticulum. Genes Dev. 12:982–995. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Newman JR and Keating AE: Comprehensive
identification of human bZIP interactions with coiled-coil arrays.
Science. 300:2097–2101. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Reinke AW, Baek J, Ashenberg O and Keating
AE: Networks of bZIP protein-protein interactions diversified over
a billion years of evolution. Science. 340:730–734. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Behrends C, Sowa ME, Gygi SP and Harper
JW: Network organization of the human autophagy system. Nature.
466:68–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang QQ, Grønborg M, Huang H, Kim JW, Otto
TC, Pandey A and Lane MD: Sequential phosphorylation of CCAAT
enhancer-binding protein beta by MAPK and glycogen synthase kinase
3beta is required for adipogenesis. Proc Natl Acad Sci USA.
102:9766–9771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee S, Shuman JD, Guszczynski T,
Sakchaisri K, Sebastian T, Copeland TD, Miller M, Cohen MS, Taunton
J, Smart RC, et al: RSK-mediated phosphorylation in the C/EBPβ
leucine zipper regulates DNA binding, dimerization, and growth
arrest activity. Mol Cell Biol. 30:2621–2635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tibbles LA and Woodgett JR: The
stress-activated protein kinase pathways. Cell Mol Life Sci.
55:1230–1254. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Beamer WG, Shultz KL, Coombs HF III,
DeMambro VE, Reinholdt LG, Ackert-Bicknell CL, Canalis E, Rosen CJ
and Donahue LR: BMD regulation on mouse distal chromosome 1,
candidate genes, and response to ovariectomy or dietary fat. J Bone
Miner Res. 26:88–99. 2011. View Article : Google Scholar
|
|
41
|
Wang EY, Gang H, Aviv Y, Dhingra R,
Margulets V and Kirshenbaum LA: p53 mediates autophagy and cell
death by a mechanism contingent on Bnip3. Hypertension. 62:70–77.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hiscott J, Lin R, Nakhaei P and Paz S:
MasterCARD: a priceless link to innate immunity. Trends Mol Med.
12:53–56. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pickert G, Lim HY, Weigert A, Häussler A,
Myrczek T, Waldner M, Labocha S, Ferreirós N, Geisslinger G, Lötsch
J, et al: Inhibition of GTP cyclohydrolase attenuates tumor growth
by reducing angiogenesis and M2-like polarization of tumor
associated macrophages. Int J Cancer. 132:591–604. 2013. View Article : Google Scholar
|
|
44
|
Yángüez E, García-Culebras A, Frau A,
Llompart C, Knobeloch KP, Gutierrez-Erlandsson S, García-Sastre A,
Esteban M, Nieto A and Guerra S: ISG15 regulates peritoneal
macrophages functionality against viral infection. PLoS Pathog.
9:e10036322013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu L, Wan F, Dutta S, Welsh S, Liu Z,
Freundt E, Baehrecke EH and Lenardo M: Autophagic programmed cell
death by selective catalase degradation. Proc Natl Acad Sci USA.
103:4952–4957. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lin Y, Zhang B, Liang H, Lu Y, Ai X, Zhang
B and Chen X: JNK inhibitor SP600125 enhances TGF-β-induced
apoptosis of RBE human cholangiocarcinoma cells in a Smad-dependent
manner. Mol Med Rep. 8:1623–1629. 2013.PubMed/NCBI
|
|
47
|
Huang S, Yang ZJ, Yu C and Sinicrope FA:
Inhibition of mTOR kinase by AZD8055 can antagonize
chemotherapy-induced cell death through autophagy induction and
down-regulation of p62/sequestosome 1. J Biol Chem.
286:40002–40012. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Evert BO, Wüllner U and Klockgether T:
Cell death in polyglutamine diseases. Cell Tissue Res. 301:189–204.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Veereshwarayya V, Kumar P, Rosen KM,
Mestril R and Querfurth HW: Differential effects of mitochondrial
heat shock protein 60 and related molecular chaperones to prevent
intracellular beta-amyloid-induced inhibition of complex IV and
limit apoptosis. J Biol Chem. 281:29468–29478. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dix DJ, Allen JW, Collins BW,
Poorman-Allen P, Mori C, Blizard DR, Brown PR, Goulding EH, Strong
BD and Eddy EM: HSP70-2 is required for desynapsis of synaptonemal
complexes during meiotic prophase in juvenile and adult mouse
spermatocytes. Development. 124:4595–4603. 1997.PubMed/NCBI
|
|
51
|
Zhou H, Zhang Y, Fu Y, Chan L and Lee AS:
Novel mechanism of anti-apoptotic function of 78-kDa
glucose-regulated protein (GRP78): endocrine resistance factor in
breast cancer, through release of B-cell lymphoma 2 (BCL-2) from
BCL-2-interacting killer (BIK). J Biol Chem. 286:25687–25696. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tchivilev I, Madamanchi NR, Vendrov AE,
Niu XL and Runge MS: Identification of a protective role for
protein phosphatase 1cgamma1 against oxidative stress-induced
vascular smooth muscle cell apoptosis. J Biol Chem.
283:22193–22205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Saito Y, Yamagishi N, Ishihara K and
Hatayama T: Identification of alpha-tubulin as an
hsp105alpha-binding protein by the yeast two-hybrid system. Exp
Cell Res. 286:233–240. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Viard I, Wehrli P, Jornot L, Bullani R,
Vechietti JL, Schifferli JA, Tschopp J and French LE: Clusterin
gene expression mediates resistance to apoptotic cell death induced
by heat shock and oxidative stress. J Invest Dermatol. 112:290–296.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Flanagan L, Whyte L, Chatterjee N and
Tenniswood M: Effects of clusterin over-expression on metastatic
progression and therapy in breast cancer. BMC Cancer. 10:1072010.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kong DH, Zhang Q, Meng X, Zong ZH, Li C,
Liu BQ, Guan Y and Wang HQ: BAG3 sensitizes cancer cells exposed to
DNA damaging agents via direct interaction with GRP78. Biochim
Biophys Acta. 1833:3245–3253. 2013. View Article : Google Scholar : PubMed/NCBI
|