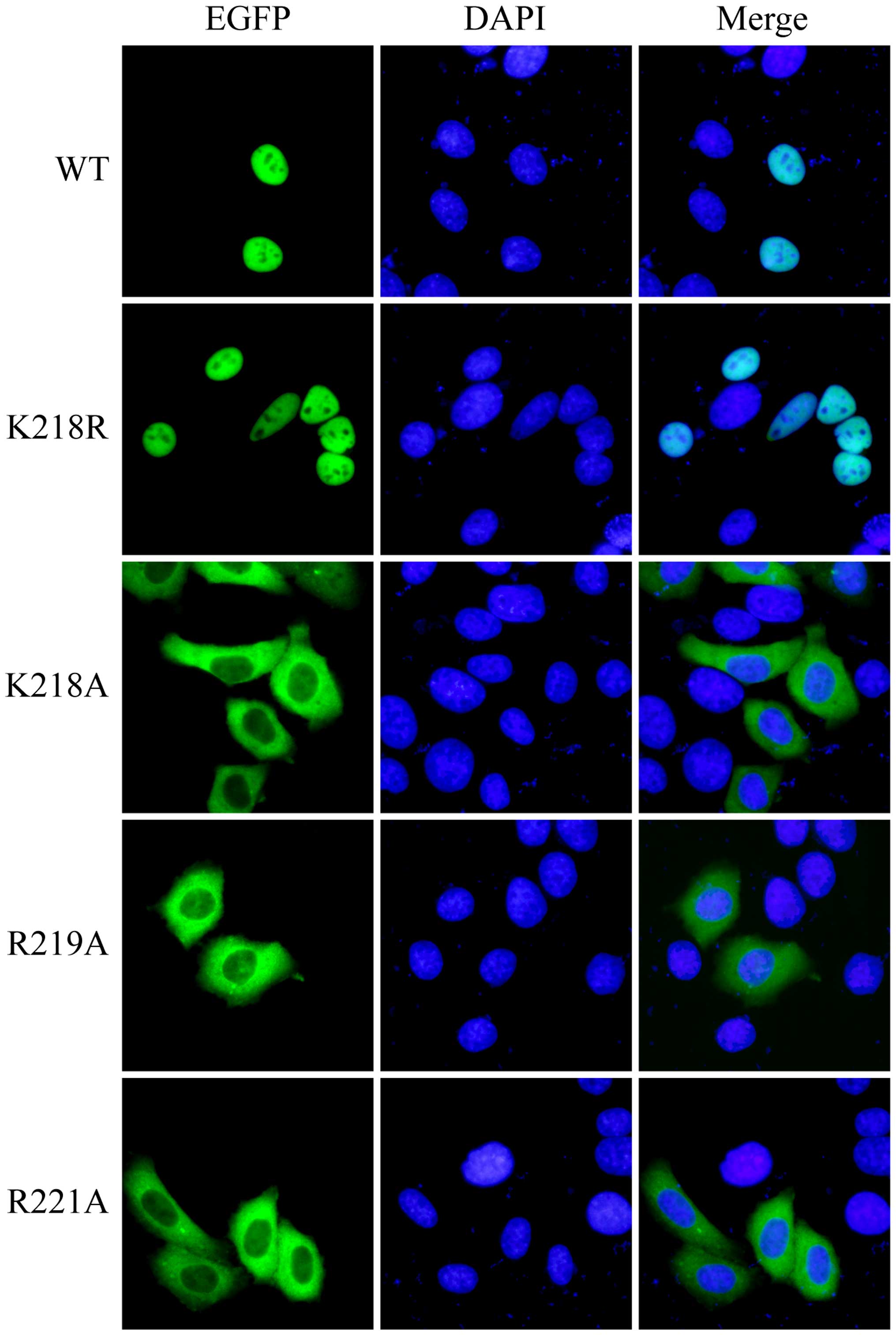
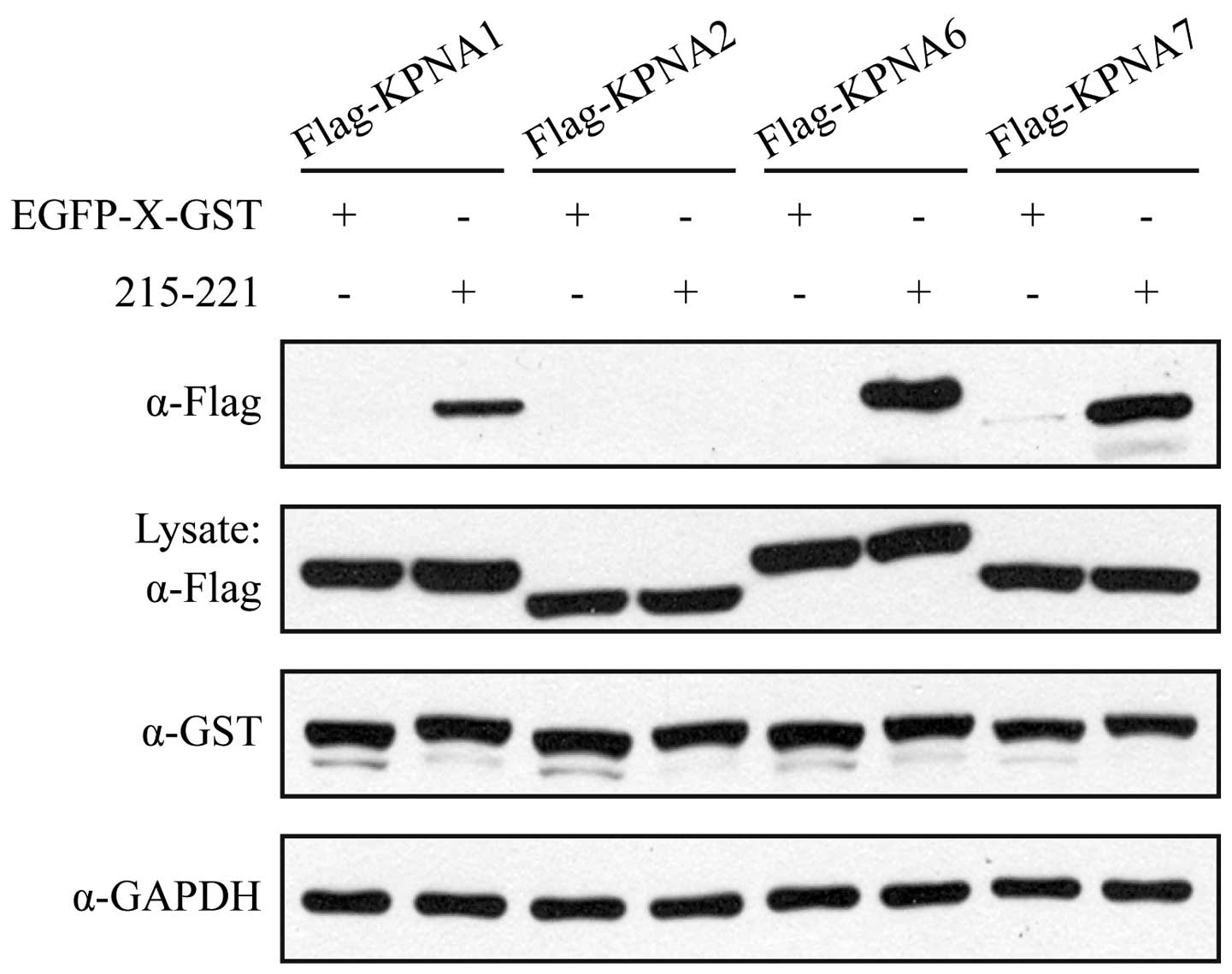
|
1
|
Broussard SR, Comuzzie AG, Leighton KL,
Leland MM, Whitehead EM and Allan JS: Characterization of new
simian foamy viruses from African nonhuman primates. Virology.
237:349–359. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hatama S, Otake K, Omoto S, Murase Y,
Ikemoto A, Mochizuki M, Takahashi E, Okuyama H and Fujii Y:
Isolation and sequencing of infectious clones of feline foamy virus
and a human/feline foamy virus Env chimera. J Gen Virol.
82:2999–3004. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herchenröder O, Renne R, Loncar D, Cobb
EK, Murthy KK, Schneider J, Mergia A and Luciw PA: Isolation,
cloning, and sequencing of simian foamy viruses from chimpanzees
(SFVcpz): high homology to human foamy virus (HFV). Virology.
201:187–199. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Materniak M, Bicka L and Kuźmak J:
Isolation and partial characterization of bovine foamy virus from
Polish cattle. Pol J Vet Sci. 9:207–211. 2006.
|
|
5
|
Tobaly-Tapiero J, Bittoun P, Neves M,
Guillemin MC, Lecellier CH, Puvion-Dutilleul F, Gicquel B, Zientara
S, Giron ML, de Thé H, et al: Isolation and characterization of an
equine foamy virus. J Virol. 74:4064–4073. 2000. View Article : Google Scholar
|
|
6
|
Löchelt M, Zentgraf H and Flügel RM:
Construction of an infectious DNA clone of the full-length human
spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology.
184:43–54. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He F, Blair WS, Fukushima J and Cullen BR:
The human foamy virus Bel-1 transcription factor is a
sequence-specific DNA binding protein. J Virol. 70:3902–3908.
1996.PubMed/NCBI
|
|
8
|
Kang Y, Blair WS and Cullen BR:
Identification and functional characterization of a high-affinity
Bel-1 DNA binding site located in the human foamy virus internal
promoter. J Virol. 72:504–511. 1998.PubMed/NCBI
|
|
9
|
Löchelt M, Flügel RM and Aboud M: The
human foamy virus internal promoter directs the expression of the
functional Bel 1 transactivator and Bet protein early after
infection. J Virol. 68:638–645. 1994.PubMed/NCBI
|
|
10
|
Chang J, Lee KJ, Jang KL, Lee EK, Baek GH
and Sung YC: Human foamy virus Bel1 transactivator contains a
bipartite nuclear localization determinant which is sensitive to
protein context and triple multimerization domains. J Virol.
69:801–808. 1995.PubMed/NCBI
|
|
11
|
Venkatesh LK, Theodorakis PA and
Chinnadurai G: Distinct cis-acting regions in U3 regulate
trans-activation of the human spumaretrovirus long terminal repeat
by the viral bel1 gene product. Nucleic Acids Res. 19:3661–3666.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Flügel RM: Spumaviruses: a group of
complex retroviruses. J Acquir Immune Defic Syndr. 4:739–750.
1991.PubMed/NCBI
|
|
13
|
He F, Sun JD, Garrett ED and Cullen BR:
Functional organization of the Bel-1 trans activator of human foamy
virus. J Virol. 67:1896–1904. 1993.PubMed/NCBI
|
|
14
|
Venkatesh LK and Chinnadurai G: The
carboxy-terminal transcription enhancement region of the human
spumaretrovirus transactivator contains discrete determinants of
the activator function. J Virol. 67:3868–3876. 1993.PubMed/NCBI
|
|
15
|
Venkatesh LK, Yang C, Theodorakis PA and
Chinnadurai G: Functional dissection of the human spumaretrovirus
transactivator identifies distinct classes of dominant-negative
mutants. J Virol. 67:161–169. 1993.PubMed/NCBI
|
|
16
|
Lee CW, Chang J, Lee KJ and Sung YC: The
Bel1 protein of human foamy virus contains one positive and two
negative control regions which regulate a distinct activation
domain of 30 amino acids. J Virol. 68:2708–2719. 1994.PubMed/NCBI
|
|
17
|
Ma Q, Tan J, Cui X, Luo D, Yu M, Liang C
and Qiao W: Residues R(199)H(200) of prototype foamy virus
transactivator Bel1 contribute to its binding with LTR and IP
promoters but not its nuclear localization. Virology. 449:215–223.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Görlich D, Prehn S, Laskey RA and Hartmann
E: Isolation of a protein that is essential for the first step of
nuclear protein import. Cell. 79:767–778. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van der Watt PJ, Stowell CL and Leaner VD:
The nuclear import receptor Kpnβ1 and its potential as an
anticancer therapeutic target. Crit Rev Eukaryot Gene Expr.
23:1–10. 2013. View Article : Google Scholar
|
|
20
|
Flores K and Seger R: Stimulated nuclear
import by β-like importins. F1000Prime Rep. 5:412013. View Article : Google Scholar
|
|
21
|
Zehorai E and Seger R: Beta-like importins
mediate the nuclear translocation of mitogen-activated protein
kinases. Mol Cell Biol. 34:259–270. 2014. View Article : Google Scholar :
|
|
22
|
Goldfarb DS, Corbett AH, Mason DA,
Harreman MT and Adam SA: Importin alpha: a multipurpose
nuclear-transport receptor. Trends Cell Biol. 14:505–514. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cimica V, Chen HC, Iyer JK and Reich NC:
Dynamics of the STAT3 transcription factor: nuclear import
dependent on Ran and importin-β1. PLoS One. 6:e201882011.
View Article : Google Scholar
|
|
24
|
Pumroy RA and Cingolani G: Diversification
of importin-α isoforms in cellular trafficking and disease states.
Biochem J. 466:13–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mattaj IW and Englmeier L:
Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem.
67:265–306. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kelley JB, Talley AM, Spencer A, Gioeli D
and Paschal BM: Karyopherin alpha7 (KPNA7), a divergent member of
the importin alpha family of nuclear import receptors. BMC Cell
Biol. 11:632010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Köhler M, Speck C, Christiansen M,
Bischoff FR, Prehn S, Haller H, Görlich D and Hartmann E: Evidence
for distinct substrate specificities of importin alpha family
members in nuclear protein import. Mol Cell Biol. 19:7782–7791.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Life RB, Lee EG, Eastman SW and Linial ML:
Mutations in the amino terminus of foamy virus Gag disrupt
morphology and infectivity but do not target assembly. J Virol.
82:6109–6119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cassany A and Gerace L: Reconstitution of
nuclear import in permeabilized cells. Methods Mol Biol.
464:181–205. 2009. View Article : Google Scholar
|
|
30
|
Adam SA, Marr RS and Gerace L: Nuclear
protein import in permeabilized mammalian cells requires soluble
cytoplasmic factors. J Cell Biol. 111:807–816. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McLane LM and Corbett AH: Nuclear
localization signals and human disease. IUBMB Life. 61:697–706.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Marfori M, Lonhienne TG, Forwood JK and
Kobe B: Structural basis of high-affinity nuclear localization
signal interactions with importin-α. Traffic. 13:532–548. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jans DA, Xiao CY and Lam MH: Nuclear
targeting signal recognition: a key control point in nuclear
transport? BioEssays. 22:532–544. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stewart M: Molecular mechanism of the
nuclear protein import cycle. Nat Rev Mol Cell Biol. 8:195–208.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Marfori M, Mynott A, Ellis JJ, Mehdi AM,
Saunders NF, Curmi PM, Forwood JK, Bodén M and Kobe B: Molecular
basis for specificity of nuclear import and prediction of nuclear
localization. Biochim Biophys Acta. 1813:1562–1577. 2011.
View Article : Google Scholar
|
|
36
|
Fontes MR, Teh T and Kobe B: Structural
basis of recognition of monopartite and bipartite nuclear
localization sequences by mammalian importin-alpha. J Mol Biol.
297:1183–1194. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Keller A, Partin KM, Löchelt M, Bannert H,
Flügel RM and Cullen BR: Characterization of the transcriptional
trans activator of human foamy retrovirus. J Virol. 65:2589–2594.
1991.PubMed/NCBI
|
|
38
|
Meertens L, Chevalier S, Weil R, Gessain A
and Mahieux R: A 10-amino acid domain within human T-cell leukemia
virus type 1 and type 2 tax protein sequences is responsible for
their divergent subcellular distribution. J Biol Chem.
279:43307–43320. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gu L, Tsuji T, Jarboui MA, Yeo GP, Sheehy
N, Hall WW and Gautier VW: Intermolecular masking of the HIV-1 Rev
NLS by the cellular protein HIC: novel insights into the regulation
of Rev nuclear import. Retrovirology. 8:172011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Korlimarla A, Bhandary L, Prabhu JS,
Shankar H, Sankaranarayanan H, Kumar P, Remacle J, Natarajan D and
Sridhar TS: Identification of a non-canonical nuclear localization
signal (NLS) in BRCA1 that could mediate nuclear localization of
splice variants lacking the classical NLS. Cell Mol Biol Lett.
18:284–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Soniat M and Chook YM: Nuclear
localization signals for four distinct karyopherin-β nuclear import
systems. Biochem J. 468:353–362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lange A, Mills RE, Lange CJ, Stewart M,
Devine SE and Corbett AH: Classical nuclear localization signals:
Definition, function, and interaction with importin alpha. J Biol
Chem. 282:5101–5105. 2007. View Article : Google Scholar
|
|
43
|
Larkin MA, Blackshields G, Brown NP,
Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm
A, Lopez R, et al: Clustal W and Clustal X version 2.0.
Bioinformatics. 23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Henikoff S and Henikoff JG: Amino acid
substitution matrices from protein blocks. Proc Natl Acad Sci USA.
89:10915–10919. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Köhler M, Ansieau S, Prehn S, Leutz A,
Haller H and Hartmann E: Cloning of two novel human importin-alpha
subunits and analysis of the expression pattern of the
importin-alpha protein family. FEBS Lett. 417:104–108. 1997.
View Article : Google Scholar : PubMed/NCBI
|