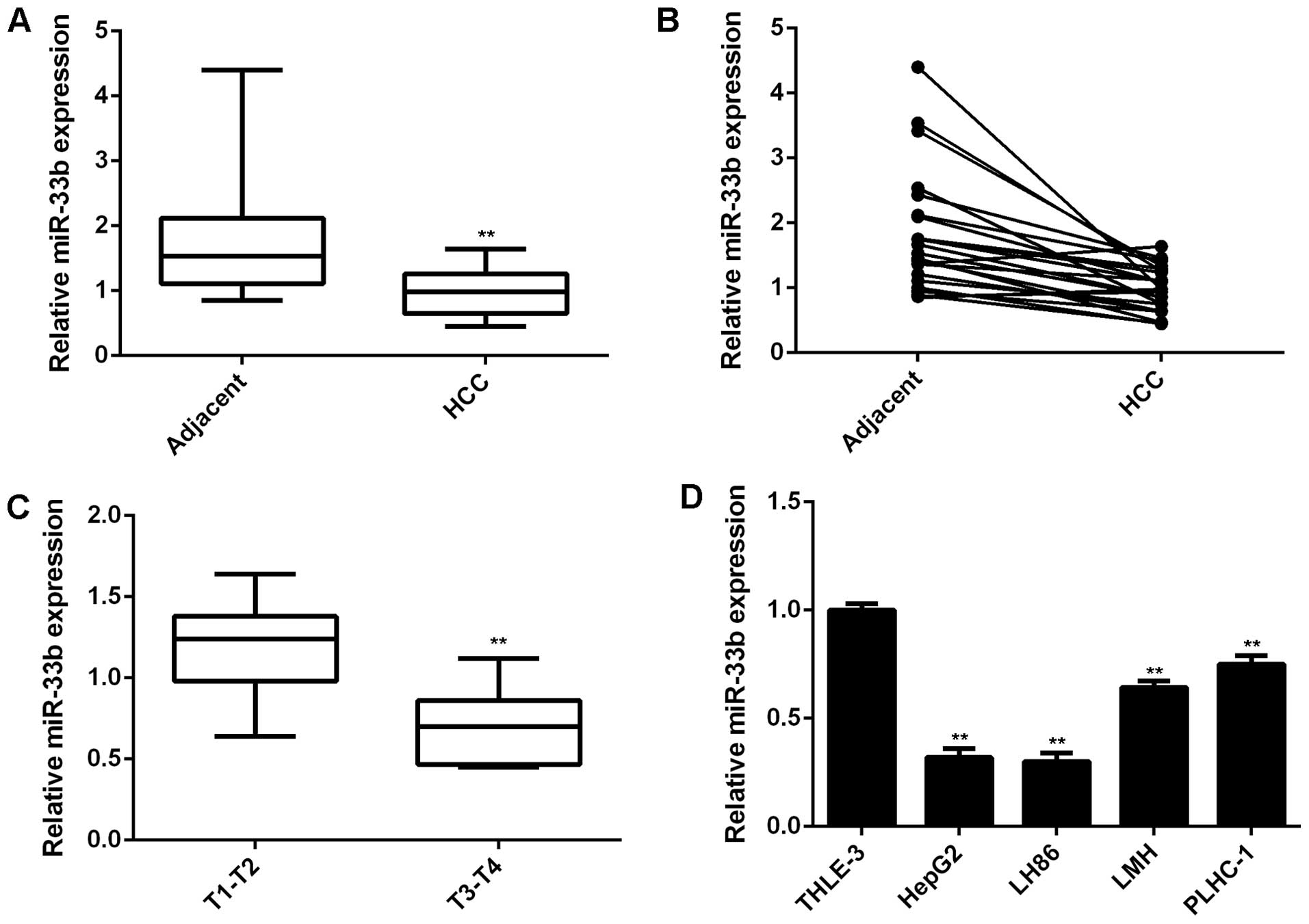
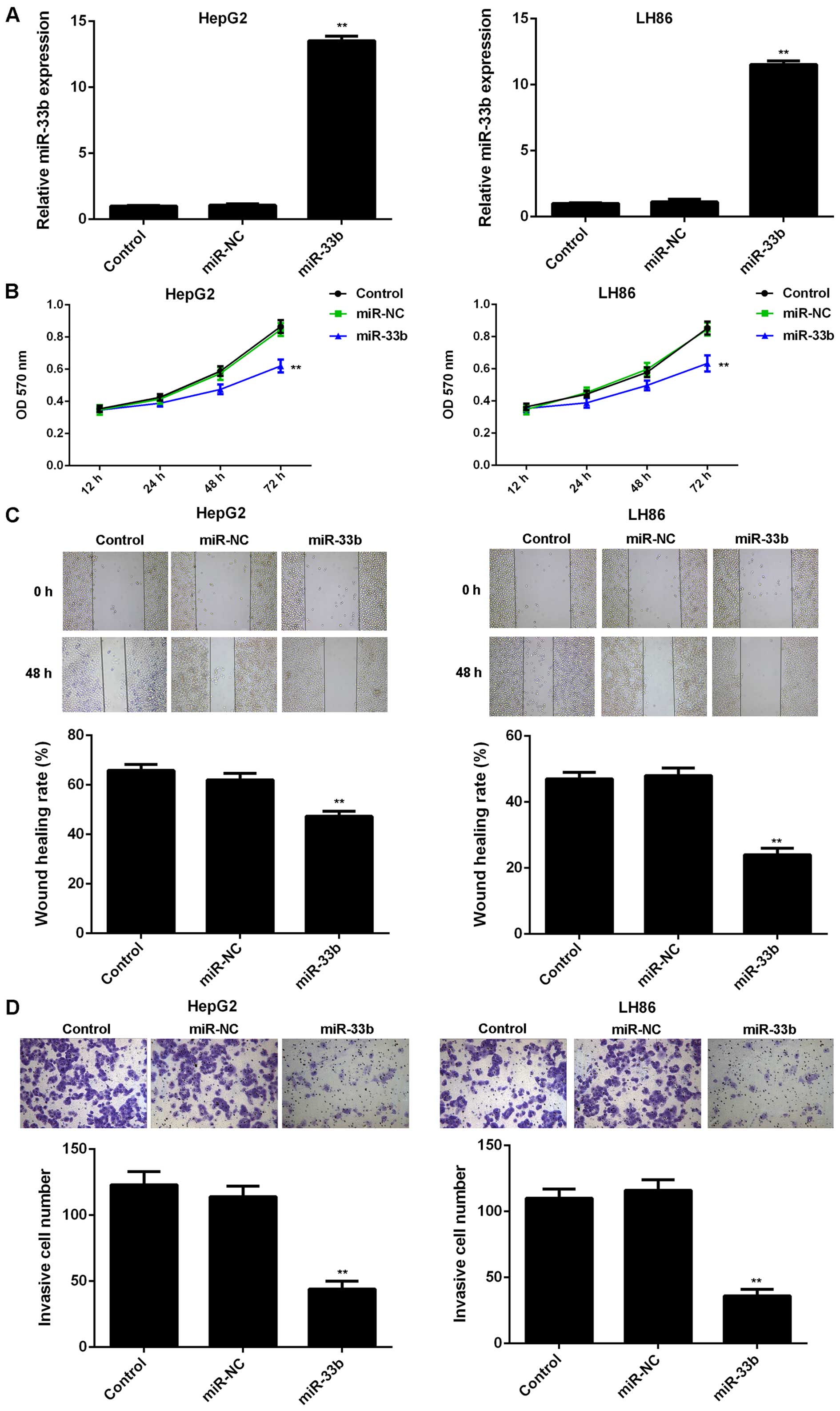
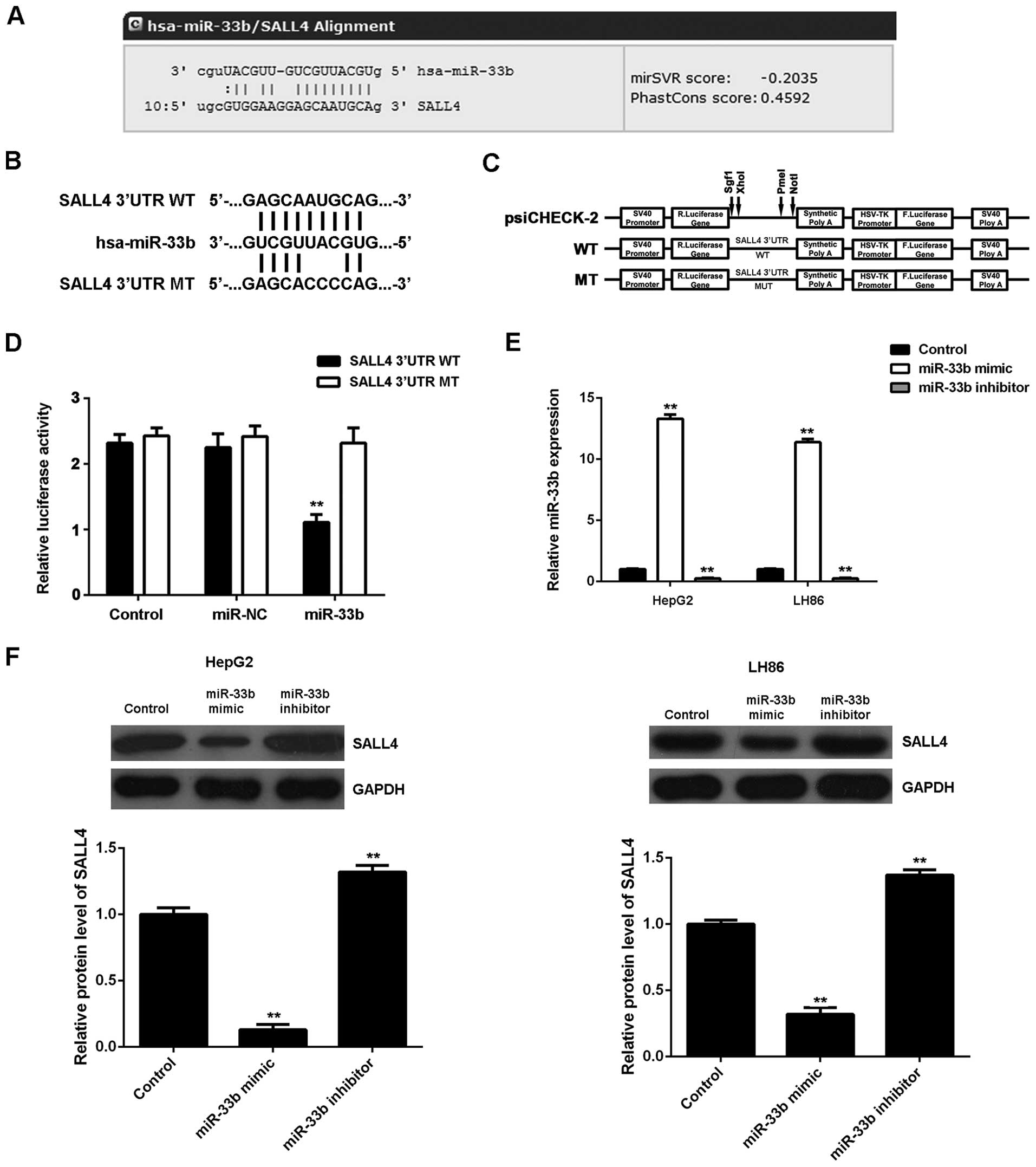
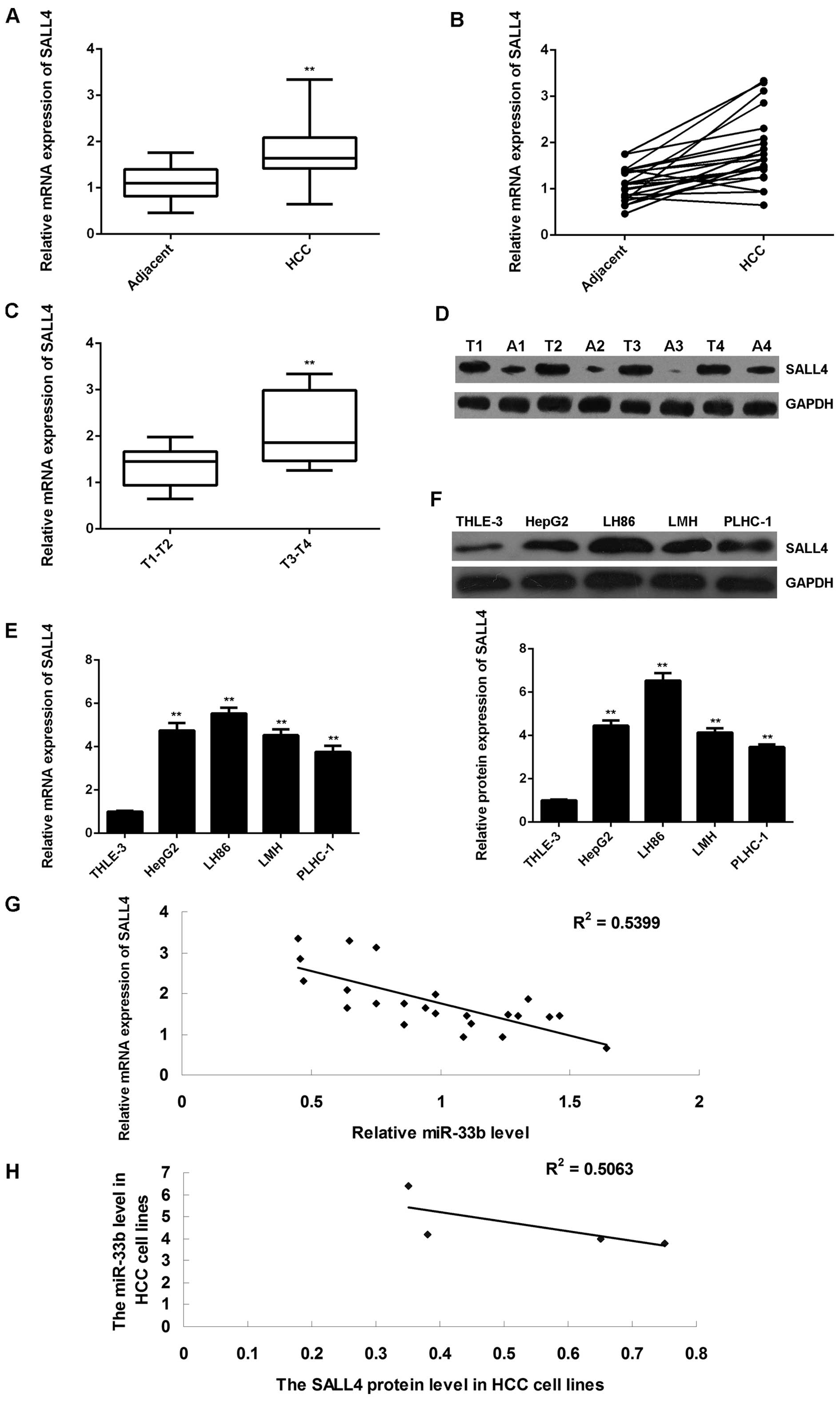
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu AX: Molecularly targeted therapy for
advanced hepatocellular carcinoma in 2012: Current status and
future perspectives. Semin Oncol. 39:493–502. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dongiovanni P, Romeo S and Valenti L:
Hepatocellular carcinoma in nonalcoholic fatty liver: Role of
environmental and genetic factors. World J Gastroenterol.
20:12945–12955. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Su CH, Lin Y and Cai L: Genetic factors,
viral infection, other factors and liver cancer: An update on
current progress. Asian Pac J Cancer Prev. 14:4953–4960. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen C and Wang G: Mechanisms of
hepatocellular carcinoma and challenges and opportunities for
molecular targeted therapy. World J Hepatol. 7:1964–1970. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen X, Vega VB and Ng HH: Transcriptional
regulatory networks in embryonic stem cells. Cold Spring Harb Symp
Quant Biol. 73:203–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Yuan X, Zhu W, Qian H and Xu W:
SALL4: An emerging cancer biomarker and target. Cancer Lett.
357:55–62. 2015. View Article : Google Scholar
|
|
8
|
Oishi N, Yamashita T and Kaneko S:
Molecular biology of liver cancer stem cells. Liver Cancer.
3:71–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oikawa T, Kamiya A, Zeniya M, Chikada H,
Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW, et
al: Sal-like protein 4 (SALL4), a stem cell biomarker in liver
cancers. Hepatology. 57:1469–1483. 2013. View Article : Google Scholar
|
|
10
|
Shikauchi Y, Saiura A, Kubo T, Niwa Y,
Yamamoto J, Murase Y and Yoshikawa H: SALL3 interacts with DNMT3A
and shows the ability to inhibit CpG island methylation in
hepatocellular carcinoma. Mol Cell Biol. 29:1944–1958. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeng SS, Yamashita T, Kondo M, Nio K,
Hayashi T, Hara Y, Nomura Y, Yoshida M, Hayashi T, Oishi N, et al:
The transcription factor SALL4 regulates stemness of EpCAM-positive
hepatocellular carcinoma. J Hepatol. 60:127–134. 2014. View Article : Google Scholar
|
|
12
|
Han SX, Wang JL, Guo XJ, He CC, Ying X, Ma
JL, Zhang YY, Zhao Q and Zhu Q: Serum SALL4 is a novel prognosis
biomarker with tumor recurrence and poor survival of patients in
hepatocellular carcinoma. J Immunol Res. 2014:2623852014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tessitore A, Cicciarelli G, Del Vecchio F,
Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D,
Zazzeroni F and Alesse E: MicroRNAs in the DNA Damage/Repair
Network and Cancer. Int J Genomics. 2014:8202482014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coulouarn C, Factor VM, Andersen JB,
Durkin ME and Thorgeirsson SS: Loss of miR-122 expression in liver
cancer correlates with suppression of the hepatic phenotype and
gain of metastatic properties. Oncogene. 28:3526–3536. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar
|
|
18
|
Wang W, Zhao LJ, Tan YX, Ren H and Qi ZT:
MiR-138 induces cell cycle arrest by targeting cyclin D3 in
hepatocellular carcinoma. Carcinogenesis. 33:1113–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yin W, Zhao Y, Ji YJ, Tong LP, Liu Y, He
SX and Wang AQ: Serum/plasma microRNAs as biomarkers for
HBV-related hepatocellular carcinoma in China. BioMed Res Int.
2015:9651852015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang Y, Feng Y, Wu T, Srinivas S, Yang W,
Fan J, Yang C and Wang S: Aflatoxin B1 negatively regulates
Wnt/β-catenin signaling pathway through activating miR-33a. PLoS
One. 8:e730042013. View Article : Google Scholar
|
|
21
|
Antaramian A, González-Gallardo A,
García-Ugalde C, Portillo W and Paredes RG: Steroid Receptors and
Aromatase Gene Expression in Different Brain Areas of Copulating
and Sexually Sluggish Male Rats. J Sex Med. 12:2267–2275. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lv SQ, Kim YH, Giulio F, Shalaby T,
Nobusawa S, Yang H, Zhou Z, Grotzer M and Ohgaki H: Genetic
alterations in microRNAs in medulloblastomas. Brain Pathol.
22:230–239. 2012. View Article : Google Scholar
|
|
24
|
Miyachi K, Sawada Y, Shida Y, Sugawara A
and Hisatomi H: Lipogenic gene expression profile in patients with
gastric cancer. Mol Clin Oncol. 1:825–827. 2013.
|
|
25
|
Hao M, Zang M, Wendlandt E, Xu Y, An G,
Gong D, Li F, Qi F, Zhang, Yang Y, et al: Low serum miR-19a
expression as a novel poor prognostic indicator in multiple
myeloma. Int J Cancer. 136:1835–1844. 2015. View Article : Google Scholar :
|
|
26
|
Li F, Hao M, Feng X, Zang M, Qin Y, Yi S,
Li Z, Xu Y, Zhou L, Sui W, et al: Downregulated miR-33b is a novel
predictor associated with disease progression and poor prognosis in
multiple myeloma. Leuk Res. 39:793–799. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshitaka T, Kawai A, Miyaki S, Numoto K,
Kikuta K, Ozaki T, Lotz M and Asahara H: Analysis of microRNAs
expressions in chondrosarcoma. J Orthop Res. 31:1992–1998. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yong KJ, Gao C, Lim JS, Yan B, Yang H,
Dimitrov T, Kawasaki A, Ong CW, Wong KF, Lee S, et al: Oncofetal
gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med.
368:2266–2276. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park H, Lee H, Seo AN, Cho JY, Choi YR,
Yoon YS, Han HS, Park YN and Kim H: SALL4 Expression in
Hepatocellular Carcinomas Is Associated with EpCAM-Positivity and a
Poor Prognosis. J Pathol Transl Med. 49:373–381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shibahara J, Ando S, Hayashi A, Sakamoto
Y, Hesegawa K, Kokudo N and Fukayama M: Clinicopathologic
characteristics of SALL4-immunopositive hepatocellular carcinoma.
Springerplus. 3:7212014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin Y, Liu AY, Fan C, Zheng H, Li Y, Zhang
C, Wu S, Yu D, Huang Z, Liu F, et al: MicroRNA-33b Inhibits Breast
Cancer Metastasis by Targeting HMGA2, SALL4 and Twist1. Sci Rep.
5:99952015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang P, Huang C, Fu C, Tian Y, Hu Y, Wang
B, Strasner A, Song Y and Song E: Cordycepin (3′-deoxyadenosine)
suppressed HMGA2, Twist1 and ZEB1-dependent melanoma invasion and
metastasis by targeting miR-33b. Oncotarget. 6:9834–9853. 2015.
View Article : Google Scholar :
|
|
33
|
Lee SA, Ho C, Roy R, Kosinski C, Patil MA,
Tward AD, Fridlyand J and Chen X: Integration of genomic analysis
and in vivo transfection to identify sprouty 2 as a candidate tumor
suppressor in liver cancer. Hepatology. 47:1200–1210. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang C, Delogu S, Ho C, Lee SA, Gui B,
Jiang L, Ladu S, Cigliano A, Dombrowski F, Evert M, et al:
Inactivation of Spry2 accelerates AKT-driven hepatocarcinogenesis
via activation of MAPK and PKM2 pathways. J Hepatol. 57:577–583.
2012. View Article : Google Scholar : PubMed/NCBI
|