Introduction
The liver plays a central role in drug metabolism
and detoxification. Acute liver failure (ALF) can lead to sudden
death, mostly due to hepatic encephalopathy (HE), intracranial
hypertension, multiple organ failure and sepsis (1). Due to the shortage of liver donors
for transplantation, non-bioartificial liver (NBAL) and
bioartificial liver (BAL) devices have been used either as a bridge
to transplantation, allowing a more efficient utilization of
available donor organs, or to 'buy time' for a patient's own liver
to recover, reducing the high demand for donor organs (2). Many new companies have been formed
in an attempt to profit from NBAL development and have recently
made promising progress (3).
However, the inability to mimic the biotransformation and metabolic
functions of liver cells in vitro is a common disadvantage
of NBAL devices. By contrast, by incorporating metabolically-active
liver cells which can efficiently biotransform toxic substances and
allow self-recovery and regeneration, BAL devices are likely to be
a better alternative for the treatment of ALF (4).
Potential BAL cell sources include primary porcine
hepatocytes, established hepatic cell-lines and primary human
hepatocytes. Unfortunately, all cell types tested and cultured
in vitro have failed to reach the same functionality
observed in primary human hepatocytes (5). Since liver-specific functions and
the proliferation of primary hepatocytes are rapidly lost during
culture (6), hepatoma or
hepatocellular carcinoma (HCC)-derived cell lines also need to be
dramatically manipulated to retain liver-specific functions and
safety-related requirements (5).
In particular, established cell lines originated from hepatic
tumors are known to lack a substantial set of liver-specific
functions. For example, the expression levels of cytochrome P450
(CYP450s) are very low or even undetectable (7). Therefore, it has been a challenge to
maintain viable and functional hepatocytes for extended periods of
time (8–10). C3A, a subclone of the
hepatoma-derived HepG2 cell line, is considered to be a suitable
cell source for the study of bioartificial liver systems, due to
its well-characterized cellular and biochemical properties and
well-preserved hepatic functions (11), and Huh7, a commercially available
human hepatoma cell line, is frequently used as an in vitro
system to study hepatotoxicity (12). Since C3A cells possess a better
differentiated hepatic phenotype, the cell line has already been
used in one of the most developed BALs currently under research
(13–15). Therefore, we primarily employed
these cell lines in order to perform our experiments in this
study.
In this study, to address the in vitro
culture challenges, we developed a novel culture method by
introducing 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid
methyl ester (ITE), a tryptophan derivative that acts as an
endogenous aryl hydrocarbon receptor (AhR) ligand (16), into the culture medium. AhR is a
transcription factor that increases xenobiotic metabolism, histone
modification and tumorigenesis (17). Due to its role in regulating drug
detoxification in a diverse group of xenobiotics, including
polychlorinated dioxins and dibenzofurans (18), AhR has been extensively studied as
a ligand-specific nuclear receptor compared to other members of the
basic helix-loop-helix/PAS protein family (19). Among other functions, the role of
AhR in regulating the expression of several isozymes of the CYP450
drug-metabolizing enzymes has been extensively studied (20). Furthermore, ITE isolated from
porcine lung tissue (21), has
been identified as a very potent endogenous agonist for AhR. In
contrast to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), another
potent but artificial ligand of AhR (20), ITE has no obvious toxicity as
previously reported.
In this study, we examined the effects of culturing
Huh7 and C3A cells with ITE on cell viability and metabolic
functions using monolayer cell cultures and microspheres. This is,
to the best of our knowledge, the first study of an in vitro
culture system that enhances the metabolic functions of Huh7 and
C3A cells without toxicity, which may improve the functions of
hepatocytes and may thus be useful in the future for the treatment
of liver diseases.
Materials and methods
Reagents and antibodies
ITE was a gift from Dr Jiasheng Song, (AhR
Pharmaceuticals, Inc., Madison, WI, USA). Anti-CYP450 isoenzyme 1A1
antibody (ab126828), anti-CYP450 isoenzyme 1B1 antibody (ab33586),
anti-CYP450 isoenzyme 1A2 antibody (ab56073), CYP3A4 antibody
(ab135813), CYP3A5 antibody (ab108624), CYP2D6 antibody (ab62204),
CYP2C9 antibody (ab150364) and CYP2E1 antibody (ab90564) were all
purchased from Abcam (Cambridge, MA, USA).
Cell culture
All cell cultures were incubated in a humidified
atmosphere at 37°C and 5% CO2. The Huh7 and C3A cells
(CRL-10741; ATCC, Manassas, VA, USA) were cultured in Dulbecco's
modified Eagle's medium (DMEM) (12430; Gibco, Auckland, New
Zealand) supplemented with 10% fetal bovine serum (FBS) (10099;
Gibco Life Technologies, Grand Island, NY, USA) as well as 1%
penicillin/streptomycin (Gibco). The cells were detached following
incubation with 0.05% trypsin-EDTA (25300; Gibco), counted, and
finally diluted to 3×106/ml with 2.0% alginate
solution.
Cell treatment
ITE was dissolved in DMSO, serial 1,000X stock
solutions in DMSO were prepared and 1:1,000 diluted with culture
medium into final concentrations of 0.2, 0.5 and 1 μM,
respectively. For the 0 μM ITE control group, only the same
volume of DMSO was added to the culture medium so that the final
concentration of DMSO was also kept at 0.1% (v/v). In most cases,
the cells were treated with serially diluted ITE for 24 and 48 h
with ITE being replenished everyday.
Alginate microsphere production
Alginate encapsulation was performed as previously
described (5) with modifications.
Briefly, the Huh7 and C3A cells were suspended in 2.0% sodium
alginate solution (154 mM NaCl, 10 mM HEPES, pH 7.4). Each mixture
was sprayed at a flow rate of 9.5 ml/min through a 300 μm
nozzle using an electrostatic microencapsulator unit (Nisco
Engineering AG, Zurich, Switzerland). The vibration frequency of
the nozzle was kept at 0.30 kHz. The alginate droplets were
collected in a calcium chloride gelation bath (154 mM NaCl, 10 mM
HEPES, 115 mM CaCl2, pH 7.4), followed by 10 min of
gelling and normal saline washes for three times. Eventually, the
Huh7 and the C3A microspheres with the designed 800 μm
diameter were produced.
MTT assay
Cell viability was determined by MTT assay
(11465007001; Roche Diagnostics, Basel, Switzerland) according to
the manufacturer's instructions. Briefly, the cells were cultured
with 100 μl of medium per well in 96-well microplates.
Following the addition of 10 μl of the MTT labeling reagent
(final concentration, 0.5 mg/ml) to each well, the plates were
incubated for 4 h in a humidified atmosphere at 37°C and 5%
CO2. The plates were incubated overnight with 100
μl of the solubilization solution in each well. For the
formation of purple formazan crystals, proportional to the number
of metabolically active viable cells, the absorbance was measured
using a microplate reader (DTX880; Beckman Coulter, Inc., Brea, CA,
USA) at a wavelength of 570 nm.
Assessment of cell viability by confocal
microscopy of fluorescein diacetate (FDA)/propidium iodide
(PI)-stained cells
The microspheres were washed twice with DMEM medium
without phenol red and then stained for 2 min with 5 mg/ml FDA and
10 mg/ml PI in Opti-MEM medium without phenol red (Gibco-Invitrogen
Life Technologies, Paisley, Scotland). The stained beads were
washed twice with Opti-MEM medium and examined using a Bio-Rad
Radiance 2100 confocal microscope (Bio-Rad Laboratories, Inc.,
Hertfordshire, UK). Images were captured and analyzed using
LaserSharp 2000 software (Bio-Rad Laboratories, Inc.).
Metabolic activity assay of CYP450
enzymes
Various groups of cell culture models were used in
our study that included: 2D, cells were cultured in a dish in a
routine two-dimensional manner; 3D, cells were encapsulated and
cultured under static conditions; and 3D-F, cells were encapsulated
and cultured in the fluidized bed bioreactor. CYP450 1A2, 3A4, 1A1
and 1B1 enzyme activity assays were carried out directly in 24-well
plates. The measurement of luciferase activity was performed with a
P450-Glo CYP1A2 assay (V8422), a CYP3A4 assay (V9002), a CYP1A1
assay (V8752) and a CYP1B1 assay (V8762) (all from Promega,
Madison, WI, USA). In brief, the cells were incubated at 37°C in
Krebs-Henseleit buffer (K3753; Sigma-Aldrich, St. Louis, MO, USA)
containing luciferin-1A2, fresh medium containing luciferin-IPA or
luciferin-CEE. After 1 or 4 h of incubation, 50 μl of buffer
or culture medium from each well were passed to a 96-well opaque
white plate by mixing with an equal volume of the luciferin
detection reagent to initiate a luminescent reaction. After 20 min
of shaking at room temperature, luminescence was measured using a
microplate reader (DTX880; Beckman Coulter, Inc.).
Ureogenesis and albumin synthesis
determination
After running the experiments, all media were
collected and stored at −80°C. The urea concentration was measured
using the urea assay kit (DIUR-500; Bioassay Systems LLC, Hayward,
CA, USA). The albumin concentration was assayed with the human
albumin ELISA quantitation set (E80-129; Bethyl Laboratories,
Montgomery, TX, USA). All results were analyzed with CurveExpert
1.3 software and fitted with logistic model with r2>0.99.
Immunofluorescence microscopy
At day 2 of culture, the cells plated in 24-well
plates were fixed with 90% ethanol and washed with saline. The
fixed cells were incubated with the indicated primary antibodies
followed by 2 U/ml of Alexa Fluor® 488 AffiniPure Rabbit
Anti-Goat IgG (H+L) (305-545-003; Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA). The nuclei were stained
with Hoechst 33342 (Hoechst 33342 Trihydrochloride, Trihydrate -
FluoroPure™ Grade, H21492; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). The slides were incubated at room temperature in the dark
for 20 min, rinsed with saline, and mounted in Vectashield (Vector
Laboratories, Peterborough, UK) prior to examination with a
confocal microscope.
Western blot analysis
After washing 3 times with phosphate buffered saline
(PBS), the cells were lysed in lysis buffer (no. 9803; Cell
Signaling Technology, Inc., Danvers, MA, USA) containing protease
inhibitors (Complete Protease Inhibitor Cocktail) and phosphatase
inhibitors (PhosSTOP) (both from Roche Diagnostics). Protein
concentrations were measured using the BCA assay kit (Thermo Fisher
Scientific, Inc.). Proteins in the lysates were separated using
SDS-PAGE and immunoblotted with respective antibodies.
Quantitative PCR (qPCR)
The microspheres were dissolved in 55 mM sodium
citrate. After washing with PBS twice, RNA was extracted using the
RNeasy Mini kit (15596026; Qiagen, Hilden, Germany) according to
the manufacturer's instructions. cDNA was synthesized using oligo
primers and a reverse transcription kit (037A; both from Takara
Bio, Inc., Shiga, Japan). The Bio-Rad Universal SYBR Supermix
(72-5121) was used to perform qPCR assays on a Bio-Rad Cycler
(C1000) (both from Bio-Rad Laboratories, Inc., Hercules, CA, USA)
with various sequences.
DNA microarray
Briefly, total RNA was extracted from the C3A cells
treated with 0.2 μM ITE using TRIzol reagent (Cat. no.
15596-108; Life Technologies, Carlsbad, CA, USA) following the
manufacturer's instructions, and the RIN was tested to inspect RNA
integrity using an Agilent Bioanalyzer 2100 (Agilent Technologies,
Inc., Santa Clara, CA, USA). Qualified total RNA was further
purified using the RNeasy micro kit (Cat. no. 74004) and the
RNase-Free DNase Set (Cat. no. 79254) (both from Qiagen). Total RNA
was amplified, labeled and purified using the GeneChip 3′ IVT PLUS
Reagent kit to obtain biotin-labeled cRNAs (Cat. no. 902416;
Affymetrix, Inc., Santa Clara, CA, USA) following the
manufacturer's instructions. Array hybridization and washes were
performed using the GeneChip® Hybridization, Wash and
Stain kit (Cat. no. 900720) in a hybridization oven 645 (Cat. no.
00-0331-220V) and Fluidics Station 450 (Cat. no. 00-0079) (all from
Affymetrix, Inc.) according to the manufacturer's instructions.
Gene expression profiling was conducted by Shanghai
Biotechnology Corporation using the Affymetrix PrimeView™ Human
Gene Expression Array (Affymetrix, Inc.). All data were analyzed
according to the manufacturer's instructions. Raw data originated
from Affymetrix CEL files were normalized by RMA background
correction and values were log2 transformed. Comparison of the data
sets by the t-test showed that 1,472 of the total of 49,293
probe-sets (2.99%) were differentially expressed (|fold changes|
≥2). In order to analyze the enrichment of P-values of each GO
term, we used Fisher's exact test to calculate P-values and R
package stats to calculate FDR (q-value) by the BH method
(www.r-project.org).
Statistical analysis
Statistical analysis was performed using the
Student's t-test and one-way analysis of variance (ANOVA) with SPSS
for Windows version 20.0 (SPSS, Inc., Chicago, IL, USA). Data from
representative experiments are presented as the means ± standard
deviation (SD). Differences were considered statistically
significant with a P-value <0.05. The experiments were repeated
at least 2 to 3 times in duplicate or triplicate for each
condition.
Results
Cytotoxicity of ITE on Huh7 and C3A
cells
To explore the potential toxicity of ITE on Huh7 and
C3A cells cultured in a monolayer, we examined cell viability and
morphology in the presence or absence of ITE (Fig. 1). The use of ITE at a
concentration of up to 1 μM and treatment for 48 h did not
affect the growth of the Huh7 cells. When the C3A cells were
treated for 48 h with ITE at a concentration of 0.5 μM and
above, ITE slightly yet significantly inhibited the growth of the
C3A cells (Fig. 1A). Phase
contrast microscopy revealed that treatment with 0.2 μM ITE
for 48 h did not have marked effects on the general morphology of
either the Huh7 or the C3A cells grown as monolayer cultures
(Fig. 1B). As ITE was dissolved
in DMSO, the control group (0 μM ITE) control media thus
also contained the same quantity of DMSO. Therefore, 0 μM
ITE as the control group was selected for the use in the
experiments described below.
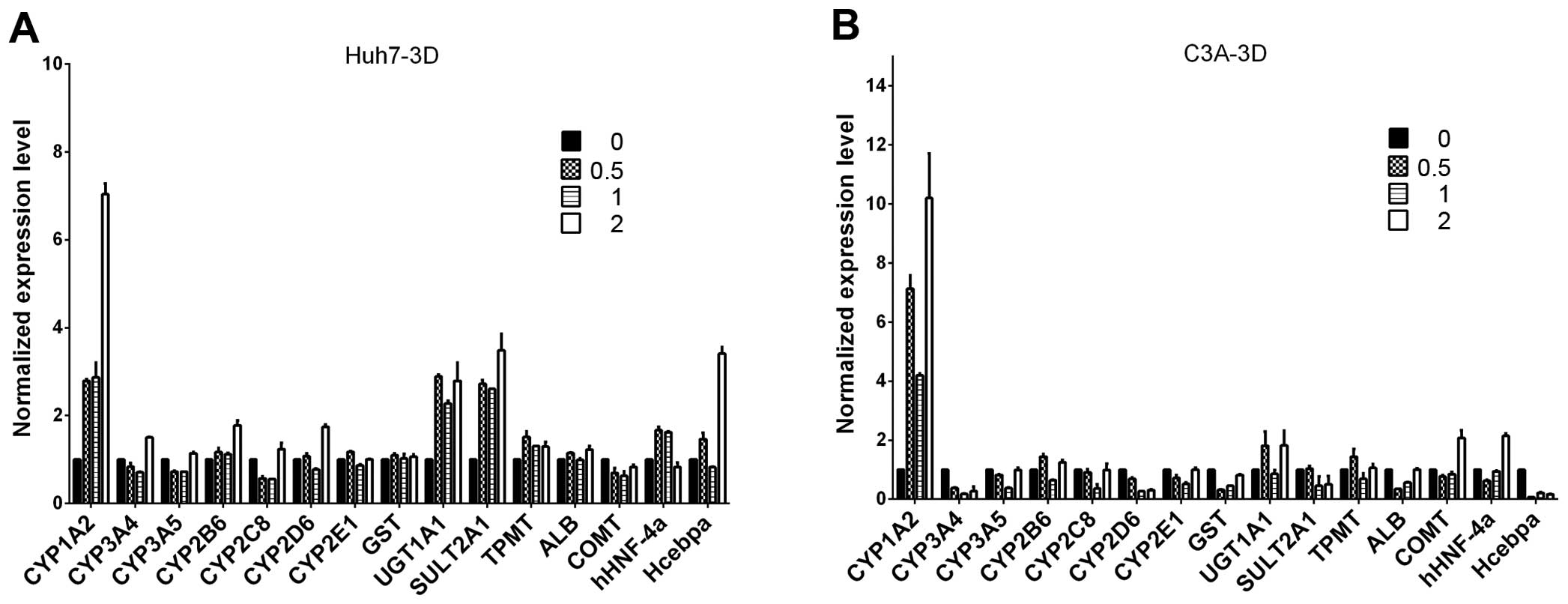
Liver-specific gene expression in Huh7
and C3A cells
Following a previous publication (22), we evaluated whether ITE in culture
enhances the transcription of several selected
metabolism-associated genes, including CYP450 phase I and II
enzymes, nuclear receptors and specific proteins. We focused on
comparing the expression profiles of Huh7 and C3A cells cultured
with ITE versus normal culture (Fig.
2). Our results indicated that the transcription levels of some
CYP450-related genes and nuclear receptors were elevated in the
cultures containing ITE. After being exposed to ITE, the liver
cells may modulate the CYP450-related genes of 1A2, 3A5, UGT1A1,
SULT2A1 and nuclear receptors of hHNF-4a in Huh7 cells, and 1A2,
3A4, 3A5, 2B6, 2C8, 2D6, 2E1 and nuclear receptors gene of hHNF-4a
in C3A cells.
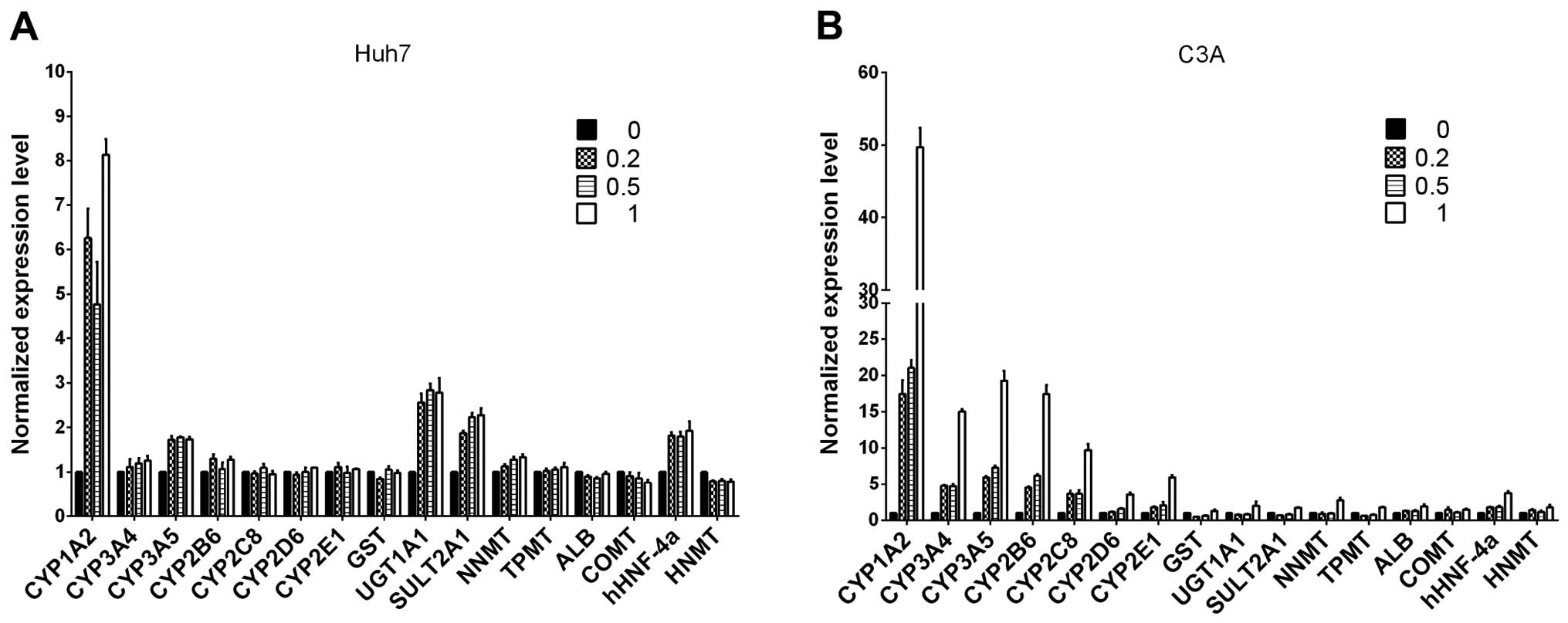 | Figure 2The mRNA levels of cytochrome P450
(CYP450s), phase II enzymes, nuclear receptors and specific
proteins were analyzed by RT-qPCR for (A) Huh7 and (B) C3A cells on
monolayer cultures treated with or without
2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester
(ITE) for 48 h. The values of 0, 0.2, 0.5, 1 indicate ITE molarity
0, 0.2, 0.5 and 1 μM, respectively. UGT, UDP
glycosyltransferase; SULT, sulfotransferase; COMT,
catechol-O-methyltransferase; HNMT, histamine
N-methyltransferase; NNMT, nicotinamide N-methyltransferase; CAR,
constitutive androstane receptor; ALB, albumin; GST, glutathione
S-transferase. |
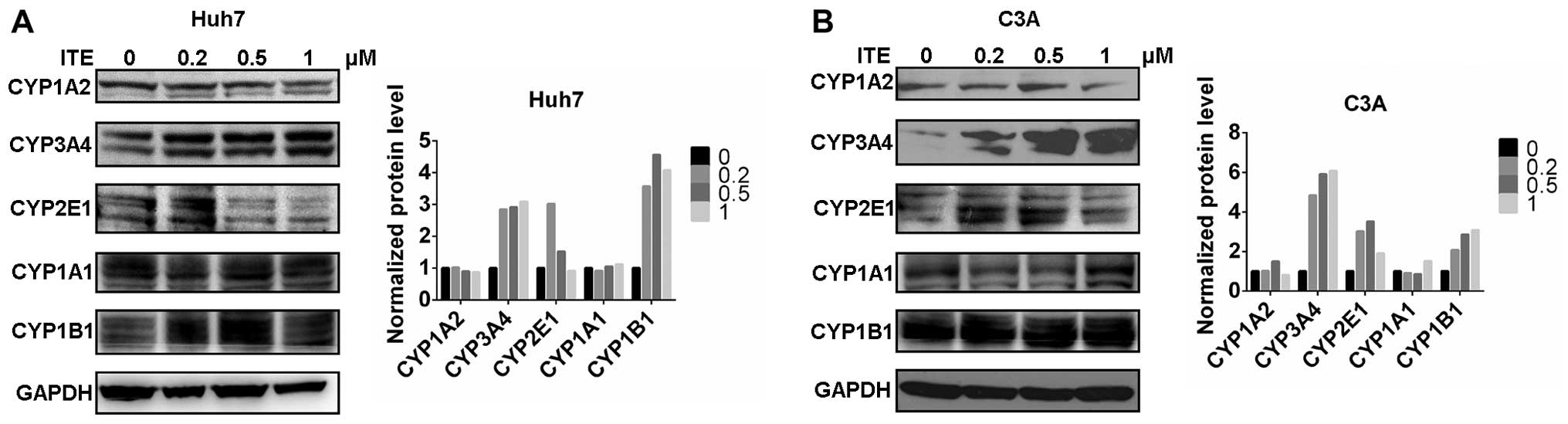
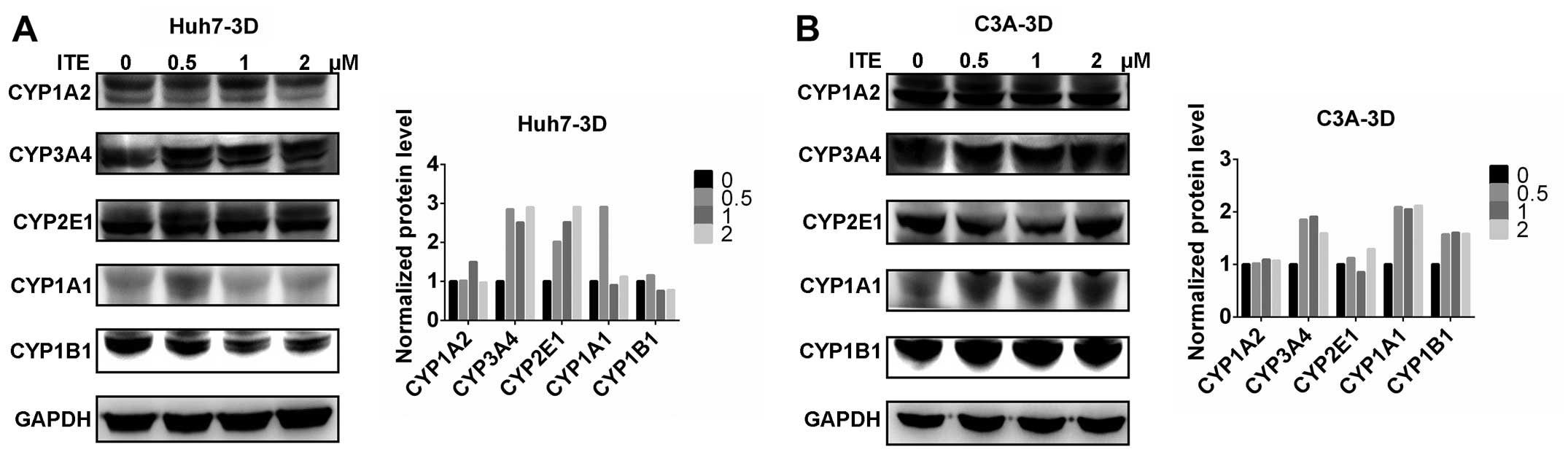
CYP450 protein expression in Huh7 and C3A
cells
We then measured the expression of proteins in the
CYP450 system in the Huh7 and C3A cells. The CYP1A2, CYP3A4,
CYP2E1, CYP1A1 and CYP1B1 protein levels were detected by western
blot analysis. The protein levels of CYP3A4, CYP2E1 and CYP1B1 were
increased by varying degrees in the presence of 0.2 μM ITE
in the culture medium compared to the cells cultured in normal
medium; however, CYP1A2 was not affected by ITE treatment (Fig. 3). Amongst the examined CYP450s,
the protein levels of CYP3A4 markedly increased in accordance with
the metabolic activities (shown below).
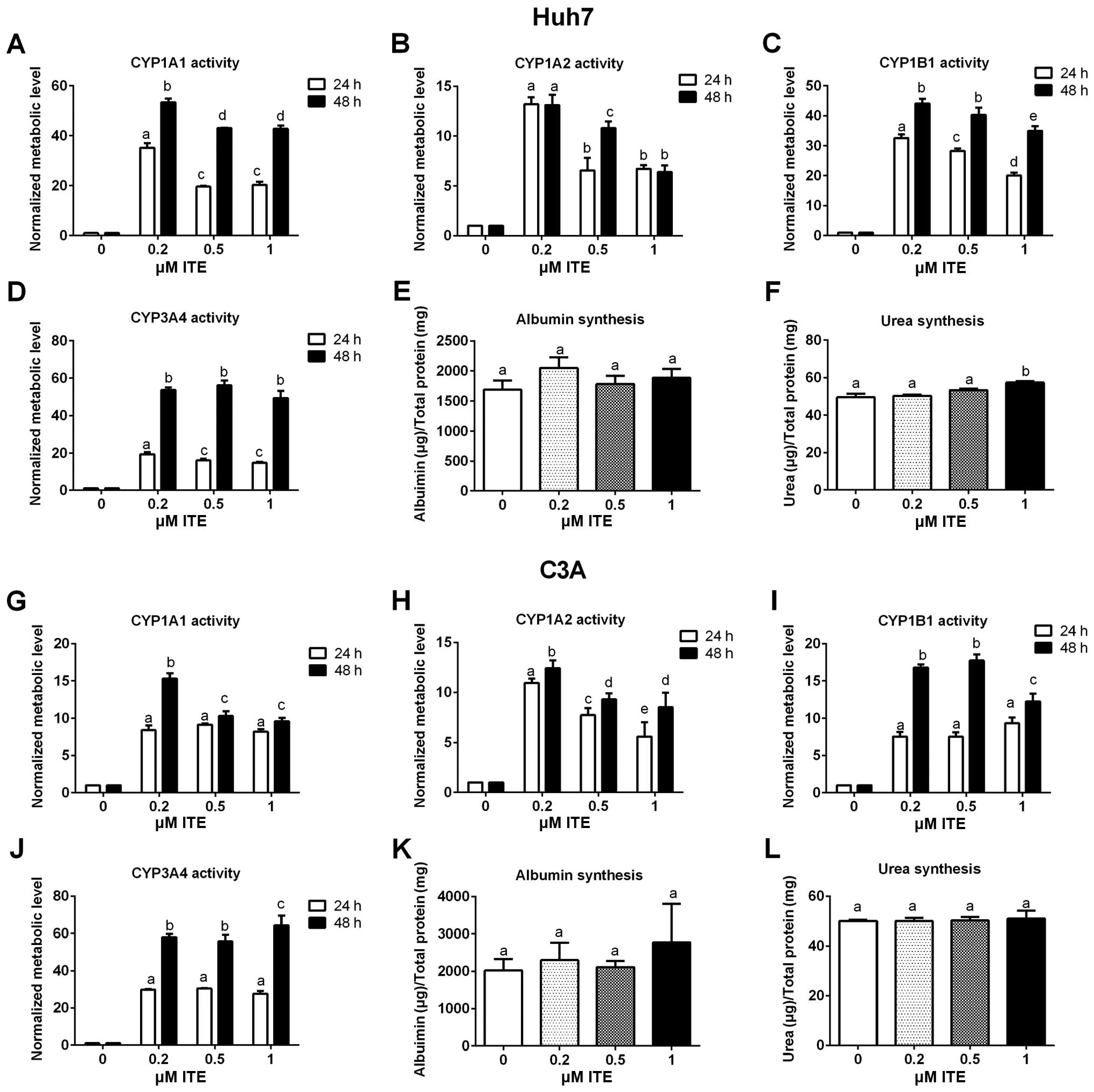
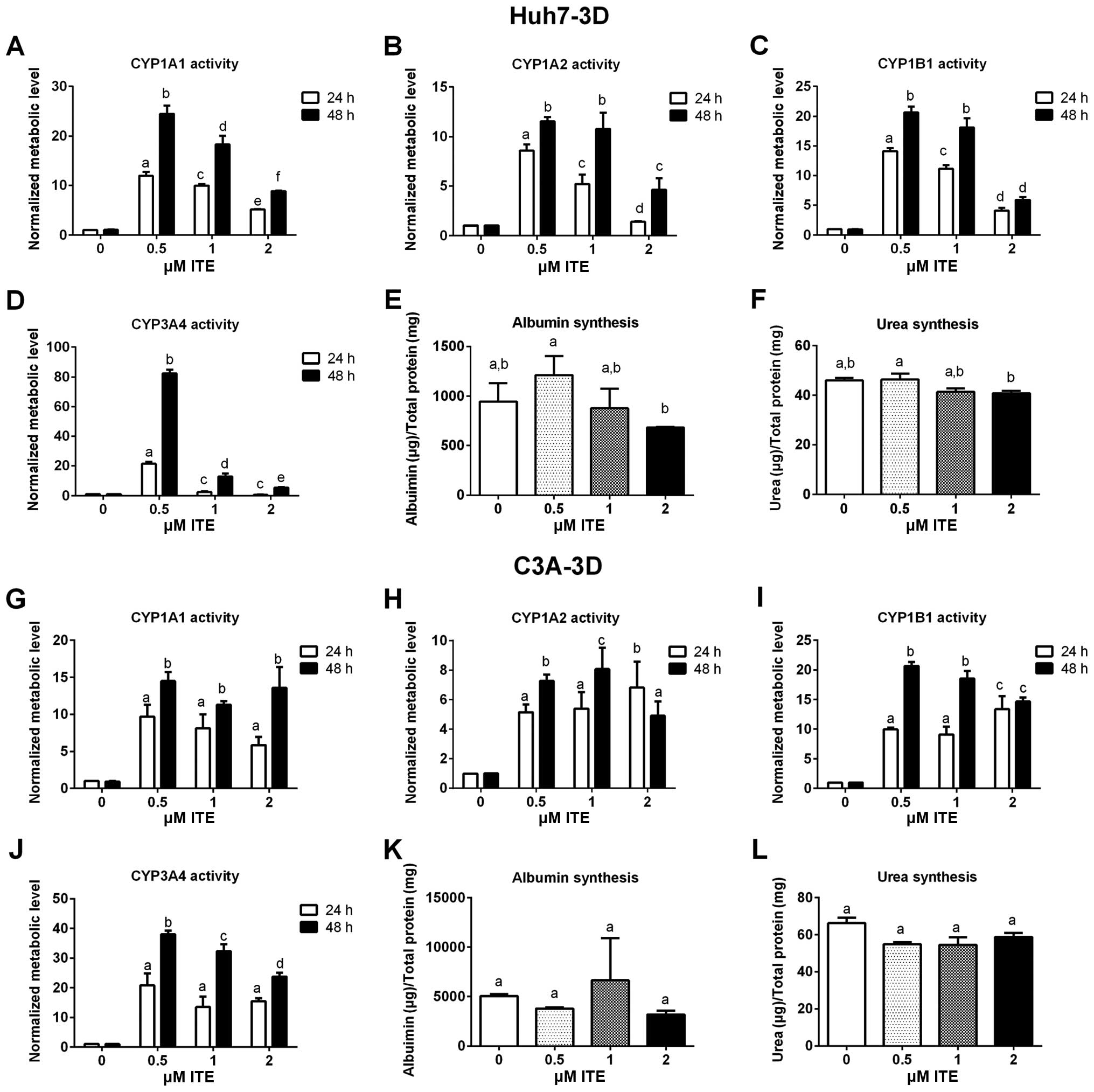
Metabolic functions of Huh7 and C3A
cells
Metabolic functions were examined to assess the
biotransformation and synthesis capacity of the Huh7 and C3A cells
in the presence of ITE in culture (Fig. 4). As expected, the CYP450 activity
levels (CYP1A1, CYP1A2, CYP1B1 and CYP3A4) were all significantly
increased in the Huh7 and C3A cells cultured with ITE. Of note, 0.2
μM ITE was the optimal concentration for most CYP450s, but
the activity of CYP3A4 in the C3A cells was higher at 1 μM
ITE (Fig. 4J). The changes in the
kinetics of CYP450 activity in response to ITE treatment were
different between Huh7 cells and C3A cells (Fig. 4A vs. C and G vs. I). In the Huh7
cells, the activities of both CYP1A1 and CYP1B1 reached the highest
levels at 0.2 μM ITE (Fig. 4A
and C). By contrast, in the C3A cells (Fig. 4G and I), the activity of CYP1A1
reached the highest level at 0.2 μM ITE and then decreased
at 0.5 and 1 μM ITE, while the activity of CYP1B1 kept
increasing until 0.5 μM ITE and then started to decrease at
1 μM ITE. Amongst the four CYP450s examined, CYP3A4
exhibited the highest increase in metabolic activity in both cell
lines (Fig. 4D and J). The
enhancement of CYP450 activity levels indicate a higher
detoxification capacity in liver cells maintained in our new
culture condition. By contrast, albumin secretion (Fig. 4E and K) and urea synthesis
(Fig. 4F and L), were only
slightly increased by ITE under these conditions and did not reach
statistical significance.
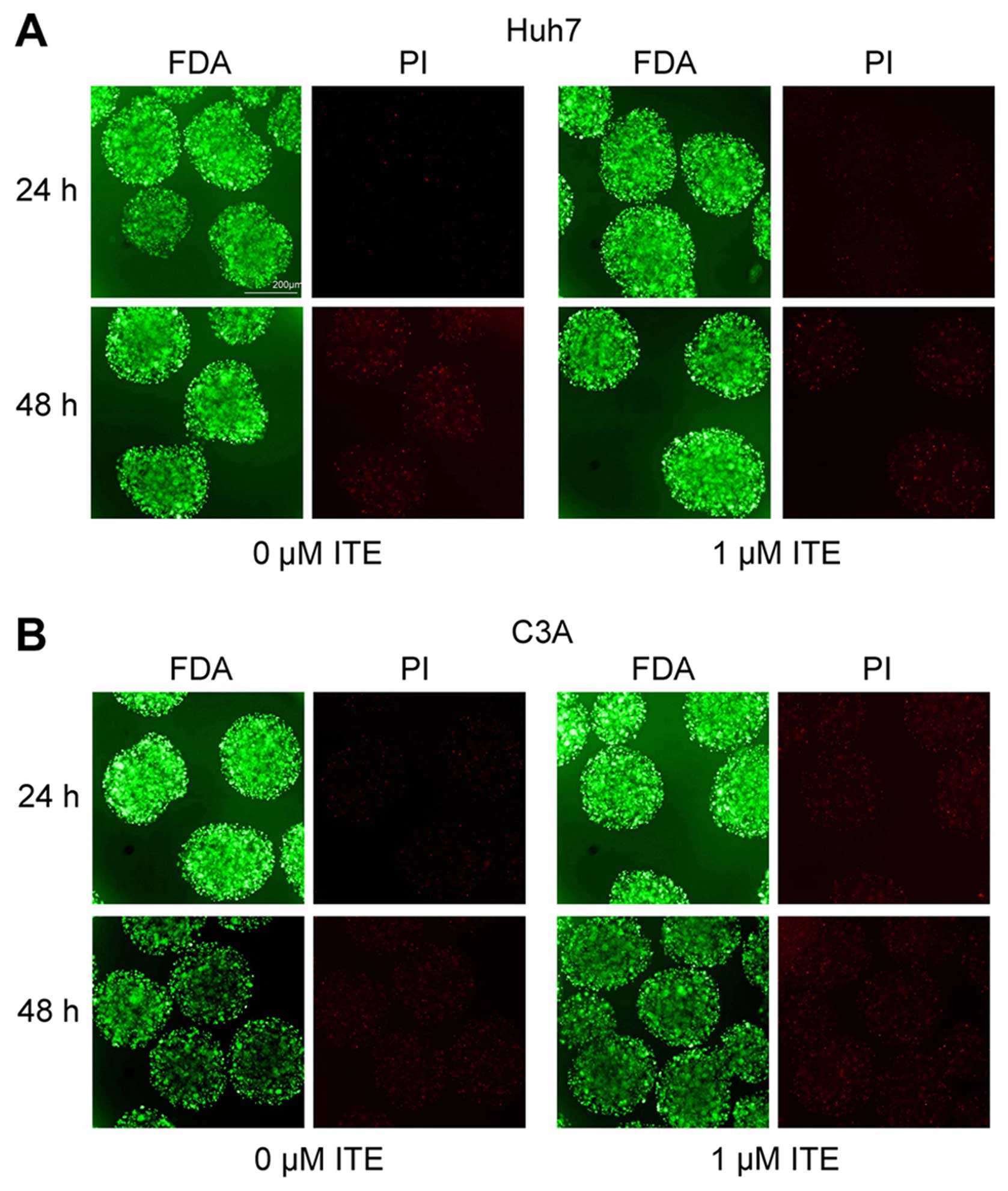
Toxic effect of ITE on Huh7 and C3A cells
in alginate microspheres
To explore the potential toxic effect of ITE on Huh7
and C3A cells cultured in alginate microspheres, we determined the
cellular morphology and viability in the presence or absence of ITE
in culture (Fig. 5). The
morphology of the Huh7 and C3A cells cultured in alginate
microspheres was not affected by ITE, and viability measured by
FDA/PI staining indicated that ITE (0 and 1 μM) was
minimally toxic to the growth of cells in alginate
microspheres.
Expression of liver-specific genes in
Huh7 and C3A cells in alginate microspheres
To explore whether ITE was able to further improve
the transcription of metabolism-associated genes in the Huh7 and
C3A cells cultured in microspheres, we examined the transcription
levels of CYP450-related genes. Our results revealed that ITE
increased the mRNA expression levels of CYP450-related genes, such
as 1A2, 2B6, 2D6, UGT1A1, SUL1A2 and nuclear receptor Hecbpa with a
>2-fold increase compared with the control group in the Huh7
microspheres, and 1A2, UGT1A1, COMT and hHNF-4a in the C3A
microspheres (Fig. 6).
CYP450 protein expression in Huh7 and C3A
cells in alginate microspheres
The CYP450 protein levels were then measured in the
Huh7 and C3A cells cultured in microspheres. The CYP1A2, CYP3A4,
CYP2E1, CYP1A1 and CYP1B1 proteins were detected by western blot
analysis, which confirmed that ITE increased the protein levels of
CYP450 enzymes in both the Huh7 and C3A cells (Fig. 7). The CYP3A4 and CYP2E1 protein
levels were increased by ITE treatment both in Huh7 microspheres
and C3A microspheres. The CYP1A1 protein level in C3A microspheres
was increased after ITE treatment. By contrast, in Huh7
microspheres, the CYP1A1 protein level was increased only with 0.5
μM ITE, but decreased with 1 and 2 μM ITE. However
CYP1A2 and CYP1B1 were not affected by ITE, which was inconsistent
with their metabolic activities (shown below). Again, amongst the
examined CYP450s, the greatest increase was observed in the protein
levels of CYP3A4.
Huh7 and C3A cell functions in alginate
microspheres
We evaluated the metabolic functions and synthesis
capacity of the Huh7 and C3A cells cultured in alginate
microspheres in the presence of ITE (Fig. 8). As already demonstrated in Huh7
and C3A cells on monolayer cultures, the metabolic activities of
the main CYP450s (CYP1A1, CYP1A2, CYP1B1 and CYP3A4) in the Huh7
and C3A cells cultured in alginate microspheres significantly
increased with ITE treatment (Fig.
8A–D and G–J). In the C3A microspheres, albumin secretion and
urea synthesis were not affected by ITE (Fig. 8K and L). However, albumin
secretion and urea synthesis in the Huh7 microspheres seemed to be
slightly affected. In contrast to 0.5 μM ITE treatment which
slightly increased the albumin secretion (Fig. 8E) and urea synthesis (Fig. 8F), higher concentrations (1 and 2
μM) of ITE decreased both albumin secretion (Fig. 8E) and urea synthesis (Fig. 8F). All these results indicated
that ITE treatment led to significantly higher detoxification
capacities of the liver cells, but had a lesser effect on their
biological synthesis capabilities.
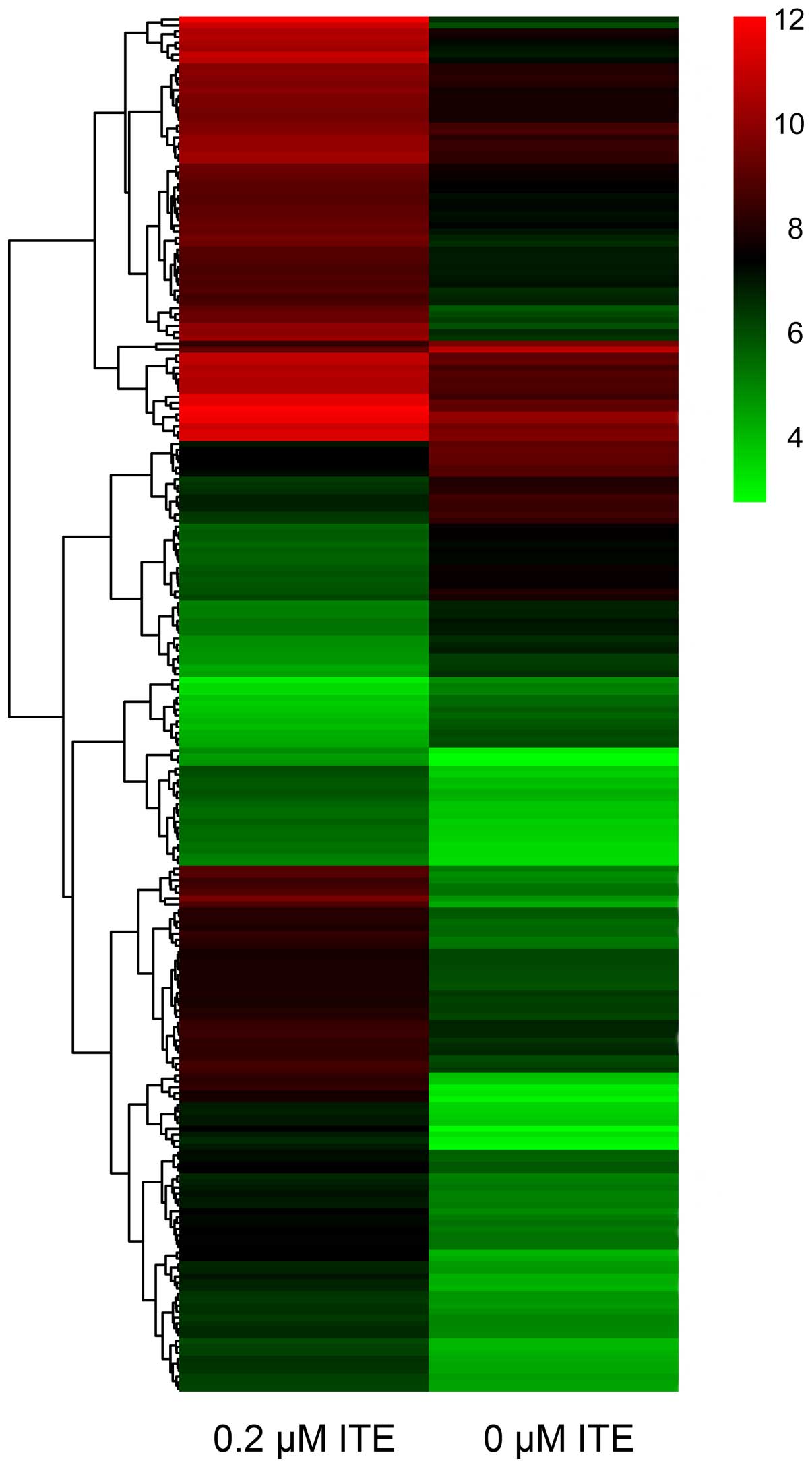
DNA microarray
In order to explore the transcriptional profile of
ITE-treated Huh7 and C3A cells on monolayer cultures, we performed
a DNA microarray. We found that the ITE-treated C3A cells elicited
quite different transcriptional profiles compared to the untreated
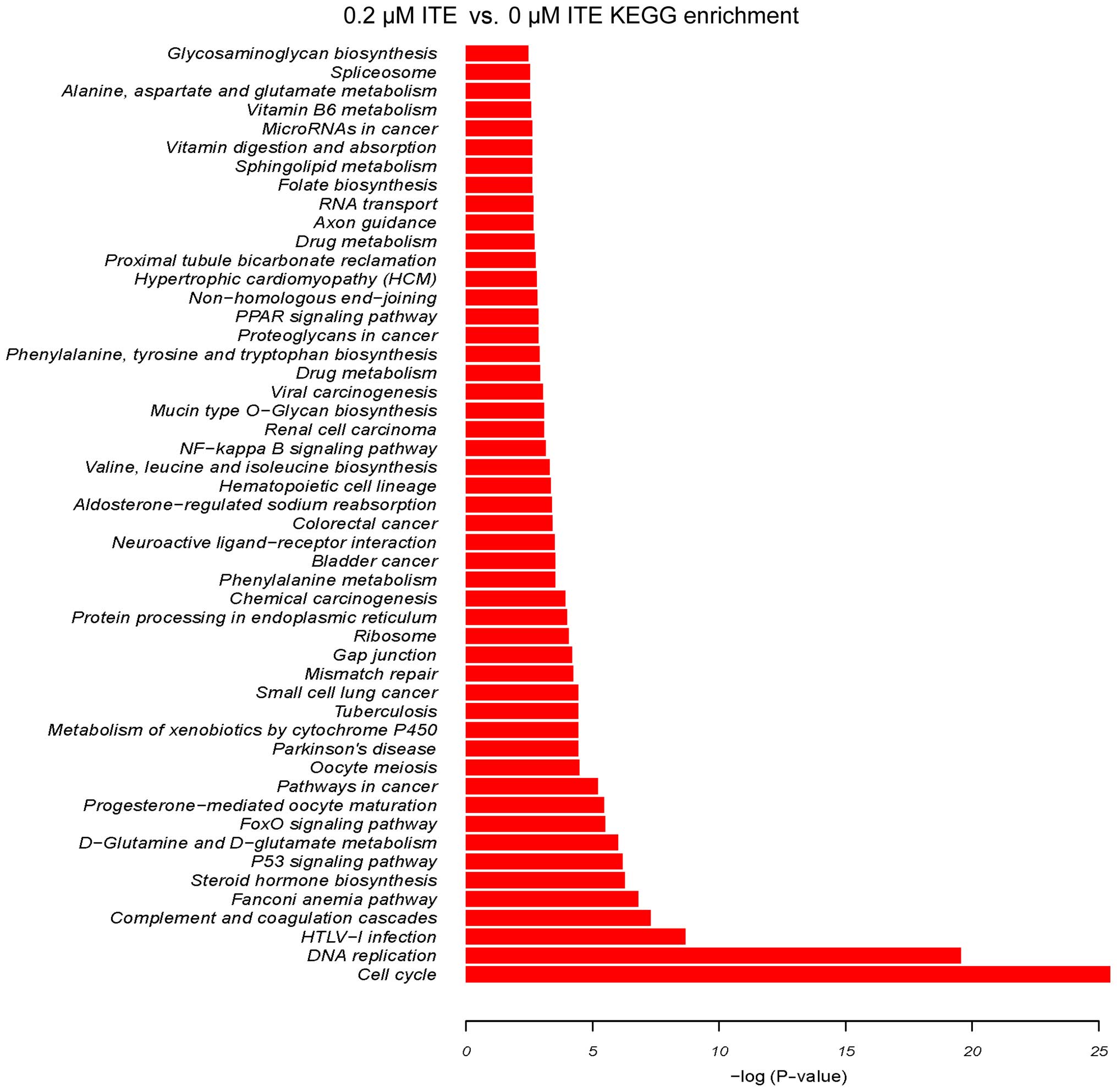
cells (Fig. 9). By contrast, 0.2
μM ITE markedly altered processes involved in cell cycle,
DNA replication and the metabolism of xenobiotics via the CYP450
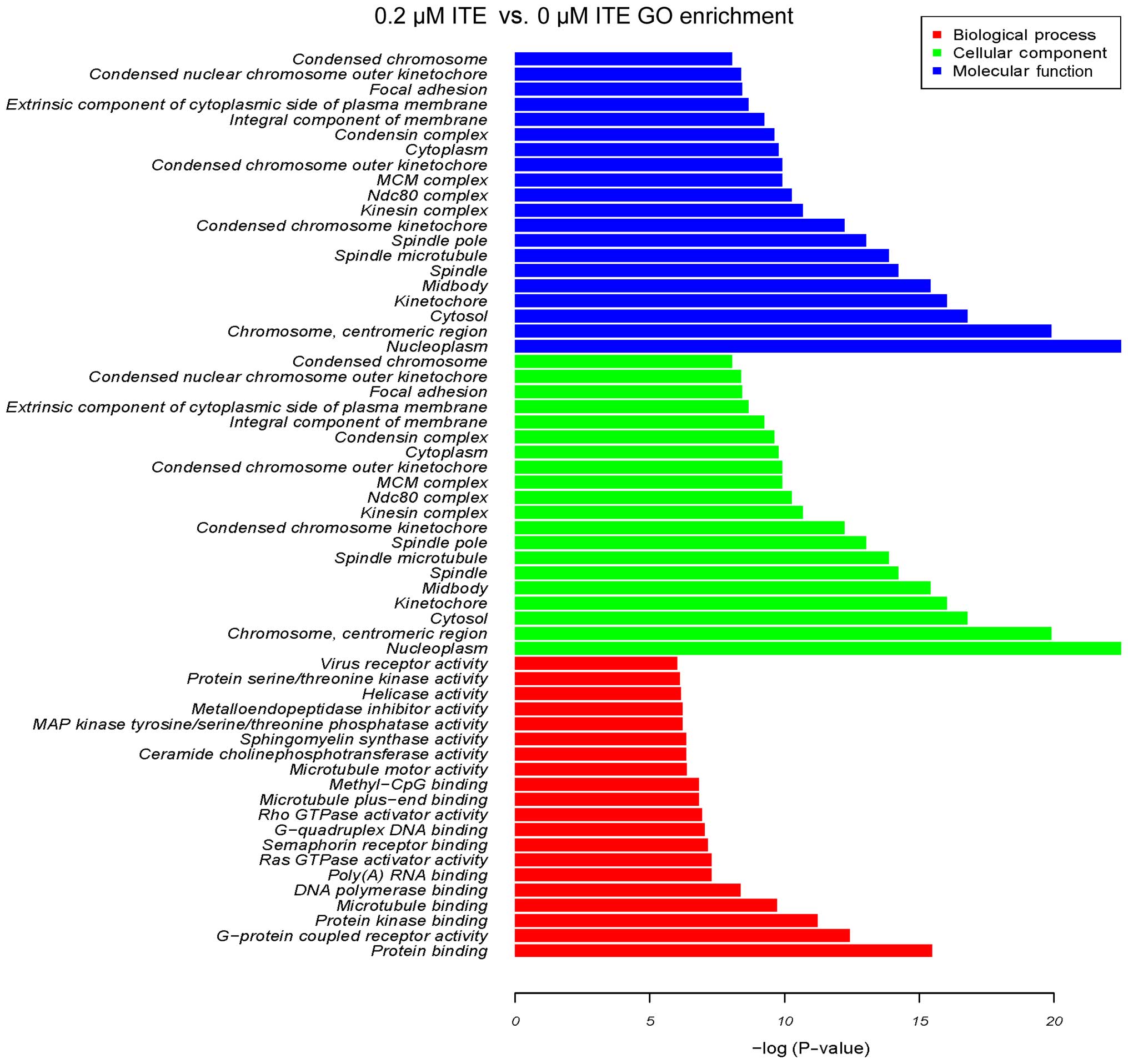
system (Fig. 10). Moreover GO
enrichment analysis further verified that processes, such as
mitotic cell cycle, mitotic nuclear division and DNA replication
were particularly altered by ITE treatment (Fig. 11), indicating a potential link
between P450-mediated metabolism activities and cell cycle
regulation.
Discussion
The liver plays many essential roles in maintaining
normal physiology. Cell availability, the maintenance of cell
viability and functionality are critical for the performance of
various purposes, such as bioartificial liver and fundamental cell
biology studies. It has been reported that the addition of low
concentrations of dexamethasone to hormone-defined medium is
beneficial for hepatocyte morphology, survival and liver-specific
functions (23–25). Nevertheless, these approaches are
not available in BAL design, as the continuous exposure of patients
to these specialized and non-physiological media components poses
major disadvantages (26).
However, low concentrations of ITE, a natural ligand isolated from
porcine lung tissue, has exhibited minimal toxicity (21). In this study, to the best of our
knowledge, we report for the first time, that liver cells cultured
in both monolayer or alginate microspheres in the presence of ITE
exhibited a significant enhancement in the maintenance of
hepatocyte metabolic functions without any marked toxicity. Hence,
our approach may be an optimal alternative for the enhancement of
the metabolic profile of C3A human-derived hepatocyte lines in
order to be used, not only in BALs, but also in clinical
trials.
The extensive use of human liver cell lines either
from tumoral origin or obtained by oncogenic immortalization is
hindered by the loss of various liver-specific functions,
particularly several CYP450-related enzyme activities (27). The CYP450 super-family is involved
in the metabolism of drugs, chemicals and endogenous substrates.
Among the CYP450 family, CYP1A2 and CYP3A4 are frequently
investigated in the human liver since they play essential roles in
drug clearance (7). Since
numerous toxic compounds accumulate in the circulation in patients
with ALF, a properly functioning detoxification system is a
prerequisite for a BAL (28). In
addition, as previously demonstrated, the treatment of experimental
ALF in dogs with bioreactors containing CYP3A4-overexpressing HepG2
cells improved survival compared to treatment with bioreactors
containing only HepG2 cells (29). During the process of hepatic
detoxification, which aims at the biotransformation of hydrophobic
toxins into water-soluble substances, intracellular modification is
specifically a hydroxylation reaction, conducted by the large
family of CYPs (28). Moreover,
among various factors involved in the regulation of hepatic
detoxification, the nuclear receptors are significantly important.
These include the AhR (dioxin), the hepatocyte nuclear factor 4α
(HNF4α), and the constitutive androstane receptor (CAR) (30–32). Thus, this study focused on the
activity of the P450s in order to optimize hepatocyte
functions.
AhR regulates hepatic detoxification at every level;
however, the binding to an activating ligand is needed in order to
exert its effects (33). Previous
studies have demonstrated that AhR regulates all CYP450 enzymes
that are also induced by aromatic hydrocarbons, such as TCDD
(34–36) and 3-methylcholanthrene (3-MC)
(37,38). In agreement with previous
observations in other cell types, our results indicated that ITE
may efficiently enhance the activity of CYP450 enzymes without any
observable toxic effect on liver cells. AhR is a cytosolic
transcription factor that is normally inactive, bound to several
co-chaperones. Upon binding to its ligands, including ITE, the
chaperones dissociate, resulting in the translocation of AhR to the
nucleus. This leads to the formation of a heterodimer with the
closely-related ARNT nuclear protein (19,39). Accordingly, the AhR/ARNT complex
can alter the transcription of the CYP1 enzymes and thereby
increase CYP450 activity (40).
Noticeably, ITE preferentially enhances the protein levels and
metabolic activity of CYP3A4 in both monolayer culture and in
alginate microspheres compared to other CYP450 family members.
Paradoxically, the transcription levels of CYP3A4 were not markedly
increased by ITE treatment in those settings. Thus, the molecular
mechanisms underlying the effects of ITE on CYP3A4 warrant further
investigation.
A previous study demonstrated that the expression
levels of CYP450 may negatively correlate with the hepatocyte
proliferation rate (4).
Specifically, cells trapped in microspheres are not able to
proliferate, possibly due to the structure of the gel itself and
the minimal interactions with the matrix (41). These data are in agreement with
other publications using alginate sponges as an approach to
facilitate hepatocyte aggregation and the re-expression of
differentiated functions prior to implantation (42). Therefore, we believe that one
potential mechanism through which ITE regulates CYP450 activity is
to reduce the interactions between the hepatocytes and the matrix
via the microspheres. Under such conditions, the proliferation rate
of hepatocytes is reduced compared to the activity of the CYP450
isoenzymes. Moreover, it has been demonstrated that AhR activation
can lead to G0–G1 cell cycle arrest, a decreased capacity for DNA
replication and to the inhibition of cell proliferation (43). On the other hand, the
transcriptionally active AhR/ARNT heterodimer initiates gene
transcription for many phase I drug metabolizing enzymes and
several phase II conjugating enzymes, thus controlling the
xenobiotic detoxification response (19,44). Furthermore, it has been confirmed
that augmenter of liver regeneration (ALR), a hepatotrophic factor,
downregulates CYP450 in human liver, thereby linking growth signals
with the regulation of hepatic metabolism (45). In addition, it has been
demonstrated that polychlorinated biphenyls (PCBs) used as AhR
agonists lead to cell proliferation associated with the mRNA
expression of CYP450 1A1 (46).
This suggests that there is an association between cell cycle
regulation and CYP450 induction by AhR ligands, such as ITE. Our
assumption was supported by the results of DNA microarray (Figs. 9Figure 10–11), which indicated that the presence
of ITE in the culture medium may have inhibited the cell cycle
progression of hepatocytes, while promoting their CYP450
activities.
In contrast to conventional monolayer culture,
artificial constructs of 3D multi-cellular spherical aggregates
manifest a high degree of cell-cell contact, which is of the utmost
importance for the communication signals necessary for coordinating
and integrating gene expression patterns (47). Indeed, 3D aggregates closely
resemble the in vivo situation regarding cell shape and
cellular environment (48), which
can in turn regulate gene expression and enhance the biological
behavior of cells (49).
Moreover, spherical microcapsules offer optimal surface-to-volume
ratio for protein and nutrient diffusion as well as cell viability.
3D aggregates allow cell survival along with protein secretion
activity upon appropriate host stimuli, without the deleterious
effects of immunosuppressant drugs (31). This study confirmd that the
treatment of 3D cultures of C3A cells/Huh7 cells with ITE improved
the CYP activity of these organotypic liver tissue structures. This
is of utmost importance for drug development and chemical testing.
In fact, these 3D C3A cells/Huh7 cells spheroids with enhanced CYP
activity may be of great value for the assessment of drug excretion
to the bile, efflux transporter-mediated drug-drug interactions and
toxicity of chemical compounds. Furthermore, among a multitude of
techniques conceived and developed to generate 3D aggregates in
vitro, the encapsulation of cells within the confines of
semi-permeable membranes is likely to become a promising method for
cell transplantation therapies and BALs (50). Thus, it is necessary to further
investigate the effects of ITE administration in microsphere
cultures of hepatocytes in a bioreactor. In the present study, we
found that the presence of ITE in culture increased CYP450
activities and functional gene expression, indicating that ITE is a
novel promoting factor for future clinical application of BALs to
improve metabolic functions of hepatocytes in microspheres.
In addition, it is acknowledged that the microsphere
diameter may affect the properties and/or biological
characteristics of cells (51).
In our previous studies (4,52),
we evaluated the permeability and viability of hepatocytes in
microspheres of different diameters (300 and 800 μm).
However, no significant differences between these two diameters
were found. Moreover, it has been shown that large microsphere
diameters can offer favorable permeability and maintain cell
viability by improving the exchange of nutrients and growth factors
(4). Furthermore, Gautier et
al (53) also demonstrated
that microspheres with a large diameter of 1,000 μm provided
a reliable entrapment process for hepatocytes to be used as a
bioartificial liver. Accordingly microspheres with a diameter of
800 μm were employed in our study.
Moreover, it is important to emphasize that the
CYP450 activities were higher in monolayer culture than in
microspheres in the presence of ITE. These results appear to
contradict those of previous studies showing that microspheres
providing 3D conditions exhibit enhanced functionality (47,54,55). Such a paradox may be attributed to
the hydrophobic properties of ITE. Furthermore, in the microsphere
setting, substances first need to cross the alginate porous
structure, where they have a lower diffusivity than that observed
in saline solution (41). The
lower diffusivity may result in the restricted and delayed
transport of ITE from the supernatant to the inner bead, thus
conferring a worse outcome for microspheres than for monolayer
cultures. Of note, we found that albumin synthesis and urea
synthesis in the C3A microspheres were higher than those observed
in monolayer culture, which is in agreement with the findings of
previous studies. Our explanation was that the lower diffusivity
caused by microspheres cultured under static conditions may affect
ITE efficiency; however, the synthesis functions which were a
result of accumulation were free from the lower diffusivity.
Therefore, further studies are required in order to investigate the
effects of ITE administration in microsphere cultures of
hepatocytes in a bioreactor to exclude the low diffusivity
distraction.
In conclusion, in this study, we examined the
effects of various concentrations of ITE on Huh7 and C3A cells
cultured in a monolayer and on microspheres. Our data indicated
that the addition of ITE to Huh7 and C3A cells in either a
monolayer or on microspheres significantly enhanced the protein
levels and metabolic activities of the major CYP450 enzymes and the
metabolic functions of Huh7 and C3A cells. Moreover, we verified
that the improved metabolic functions of the liver cells are
associated with the metabolism of xenobiotics via AhR-dependent
signaling pathways. Taken together, the improvement of liver cell
functions by ITE may be a promising approach for the treatment of
liver diseases.
Acknowledgments
This study was financially supported by the National
High Technology Research and Development Program of China (863
Program, grant nos. 2012AA020204 and 2013AA020102), the National
Natural Science Foundation of China (grant no. 31271465), and the
Zhejiang Provincial Natural Science Foundation of China for
Distinguished Young Scholars (grant no. R2100226). We would like to
thank Dr Jiasheng Song (AhR Pharmaceuticals, Inc.) for kindly
providing the ITE, and Jianzhou Li and Yini Wang for general
technical assistance and advice.
References
|
1
|
Bernal W and Wendon J: Acute liver
failure. N Engl J Med. 369:2525–2534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tostões RM, Leite SB, Miranda JP, Sousa M,
Wang DI, Carrondo MJ and Alves PM: Perfusion of 3D encapsulated
hepatocytes - a synergistic effect enhancing long-term
functionality in bioreactors. Biotechnol Bioeng. 108:41–49. 2011.
View Article : Google Scholar
|
|
3
|
Carpentier B, Gautier A and Legallais C:
Artificial and bioartificial liver devices: present and future.
Gut. 58:1690–1702. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang Y, Li J, Pan X, Zhou P, Yu X, Cao H,
Wang Y and Li L: Co-culture with mesenchymal stem cells enhances
metabolic functions of liver cells in bioartificial liver system.
Biotechnol Bioeng. 110:958–968. 2013. View Article : Google Scholar
|
|
5
|
Chamuleau RA, Deurholt T and Hoekstra R:
Which are the right cells to be used in a bioartificial liver?
Metab Brain Dis. 20:327–335. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakamura T, Tomita Y, Hirai R, Yamaoka K,
Kaji K and Ichihara A: Inhibitory effect of transforming growth
factor-beta on DNA synthesis of adult rat hepatocytes in primary
culture. Biochem Biophys Res Commun. 133:1042–1050. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zanger UM, Turpeinen M, Klein K and Schwab
M: Functional pharmacogenetics/genomics of human cytochromes P450
involved in drug biotransformation. Anal Bioanal Chem.
392:1093–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gebhardt R, Hengstler JG, Müller D,
Glöckner R, Buenning P, Laube B, Schmelzer E, Ullrich M, Utesch D,
Hewitt N, et al: New hepatocyte in vitro systems for drug
metabolism: metabolic capacity and recommendations for application
in basic research and drug development, standard operation
procedures. Drug Metab Rev. 35:145–213. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gómez-Lechón MJ, Donato MT, Castell JV and
Jover R: Human hepatocytes as a tool for studying toxicity and drug
metabolism. Curr Drug Metab. 4:292–312. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vermeir M, Annaert P, Mamidi RN, Roymans
D, Meuldermans W and Mannens G: Cell-based models to study hepatic
drug metabolism and enzyme induction in humans. Expert Opin Drug
Metab Toxicol. 1:75–90. 2005. View Article : Google Scholar
|
|
11
|
Filippi C, Keatch SA, Rangar D, Nelson LJ,
Hayes PC and Plevris JN: Improvement of C3A cell metabolism for
usage in bioartificial liver support systems. J Hepatol.
41:599–605. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sivertsson L, Ek M, Darnell M, Edebert I,
Ingelman-Sundberg M and Neve EP: CYP3A4 catalytic activity is
induced in confluent Huh7 hepatoma cells. Drug Metab Dispos.
38:995–1002. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sussman NL, Gislason GT, Conlin CA and
Kelly JH: The Hepatix extracorporeal liver assist device: initial
clinical experience. Artif Organs. 18:390–396. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ellis AJ, Hughes RD, Wendon JA, Dunne J,
Langley PG, Kelly JH, Gislason GT, Sussman NL and Williams R:
Pilot-controlled trial of the extracorporeal liver assist device in
acute liver failure. Hepatology. 24:1446–1451. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hughes RD, Nicolaou N, Langley PG, Ellis
AJ, Wendon JA and Williams R: Plasma cytokine levels and
coagulation and complement activation during use of the
extracorporeal liver assist device in acute liver failure. Artif
Organs. 22:854–858. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsay JJ, Tchou-Wong KM, Greenberg AK, Pass
H and Rom WN: Aryl hydrocarbon receptor and lung cancer. Anticancer
Res. 33:1247–1256. 2013.PubMed/NCBI
|
|
17
|
Cheng J, Li W, Kang B, Zhou Y, Song J, Dan
S, Yang Y, Zhang X, Li J, Yin S, et al: Tryptophan derivatives
regulate the transcription of Oct4 in stem-like cancer cells. Nat
Commun. 6:72092015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Minsavage GD, Park SK and Gasiewicz TA:
The aryl hydrocarbon receptor (AhR) tyrosine 9, a residue that is
essential for AhR DNA binding activity, is not a phosphoresidue but
augments AhR phosphorylation. J Biol Chem. 279:20582–20593. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hankinson O: The aryl hydrocarbon receptor
complex. Annu Rev Pharmacol Toxicol. 35:307–340. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Whitlock JP Jr: Induction of cytochrome
P4501A1. Annu Rev Pharmacol Toxicol. 39:103–125. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song J, Clagett-Dame M, Peterson RE, Hahn
ME, Westler WM, Sicinski RR and DeLuca HF: A ligand for the aryl
hydrocarbon receptor isolated from lung. Proc Natl Acad Sci USA.
99:14694–14699. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khetani SR and Bhatia SN: Microscale
culture of human liver cells for drug development. Nat Biotechnol.
26:120–126. 2008. View Article : Google Scholar
|
|
23
|
Isom HC, Secott T, Georgoff I, Woodworth C
and Mummaw J: Maintenance of differentiated rat hepatocytes in
primary culture. Proc Natl Acad Sci USA. 82:3252–3256. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dich J, Vind C and Grunnet N: Long-term
culture of hepatocytes: effect of hormones on enzyme activities and
metabolic capacity. Hepatology. 8:39–45. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kubota H and Reid LM: Clonogenic
hepatoblasts, common precursors for hepatocytic and biliary
lineages, are lacking classical major histocompatibility complex
class I antigen. Proc Natl Acad Sci USA. 97:12132–12137. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Allen JW, Hassanein T and Bhatia SN:
Advances in bioartificial liver devices. Hepatology. 34:447–455.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guillouzo A, Corlu A, Aninat C, Glaise D,
Morel F and Guguen-Guillouzo C: The human hepatoma HepaRG cells: a
highly differentiated model for studies of liver metabolism and
toxicity of xenobiotics. Chem Biol Interact. 168:66–73. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nibourg GA, Huisman MT, van der Hoeven TV,
van Gulik TM, Chamuleau RA and Hoekstra R: Stable overexpression of
pregnane X receptor in HepG2 cells increases its potential for
bioartificial liver application. Liver Transpl. 16:1075–1085. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang N, Tsuruoka S, Yamamoto H, Enosawa S,
Omasa T, Sata N, Matsumura T, Nagai H and Fujimura A: The
bioreactor with CYP3A4- and glutamine synthetase-introduced HepG2
cells: treatment of hepatic failure dog with diazepam overdosage.
Artif Organs. 29:681–684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fujii-Kuriyama Y and Mimura J: Molecular
mechanisms of AhR functions in the regulation of cytochrome P450
genes. Biochem Biophys Res Commun. 338:311–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rabanel JM, Banquy X, Zouaoui H, Mokhtar M
and Hildgen P: Progress technology in microencapsulation methods
for cell therapy. Biotechnol Prog. 25:946–963. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jover R, Moya M and Gómez-Lechón MJ:
Transcriptional regulation of cytochrome p450 genes by the nuclear
receptor hepatocyte nuclear factor 4-alpha. Curr Drug Metab.
10:508–519. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Puga A, Ma C and Marlowe JL: The aryl
hydrocarbon receptor cross-talks with multiple signal transduction
pathways. Biochem Pharmacol. 77:713–722. 2009. View Article : Google Scholar :
|
|
34
|
Marie S, Anderson A and Cresteil T:
Transplacental induction of cytochromes P-450IA1 and P-450IA2 by
polycyclic aromatic carcinogens: TCDD-binding protein level as the
rate-limiting step. Carcinogenesis. 9:2059–2063. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tritscher AM, Goldstein JA, Portier CJ,
McCoy Z, Clark GC and Lucier GW: Dose-response relationships for
chronic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin in a rat
tumor promotion model: quantification and immunolocalization of
CYP1A1 and CYP1A2 in the liver. Cancer Res. 52:3436–3442.
1992.PubMed/NCBI
|
|
36
|
Döhr O, Vogel C and Abel J: Different
response of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-sensitive
genes in human breast cancer MCF-7 and MDA-MB 231 cells. Arch
Biochem Biophys. 321:405–412. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gonzalez FJ, Tukey RH and Nebert DW:
Structural gene products of the Ah locus. Transcriptional
regulation of cytochrome P1-450 and P3-450 mRNA levels by
3-methylcholanthrene. Mol Pharmacol. 26:117–121. 1984.PubMed/NCBI
|
|
38
|
Goasduff T, Menez JF, Dreano Y and Berthou
F: CYP1A2 and 2E1 expression in rat liver treated with combined
inducers (3-methylcholanthrene and ethanol). Biochem Biophys Res
Commun. 211:497–503. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mimura J and Fujii-Kuriyama Y: Functional
role of AhR in the expression of toxic effects by TCDD. Biochim
Biophys Acta. 1619:263–268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kinasiewicz A, Gautier A, Lewinska D,
Bukowski J, Legallais C and Weryński A: Culture of C3A cells in
alginate beads for fluidized bed bioartificial liver. Transplant
Proc. 39:2911–2913. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
David B, Dufresne M, Nagel MD and
Legallais C: In vitro assessment of encapsulated C3A hepatocytes
functions in a fluidized bed bioreactor. Biotechnol Prog.
20:1204–1212. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Glicklis R, Shapiro L, Agbaria R, Merchuk
JC and Cohen S: Hepatocyte behavior within three-dimensional porous
alginate scaffolds. Biotechnol Bioeng. 67:344–353. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fan Y, Boivin GP, Knudsen ES, Nebert DW,
Xia Y and Puga A: The aryl hydrocarbon receptor functions as a
tumor suppressor of liver carcinogenesis. Cancer Res. 70:212–220.
2010. View Article : Google Scholar
|
|
44
|
Schnekenburger M, Peng L and Puga A: HDAC1
bound to the Cyp1a1 promoter blocks histone acetylation associated
with Ah receptor-mediated trans-activation. Biochim Biophys Acta.
1769:569–578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Thasler WE, Dayoub R, Mühlbauer M,
Hellerbrand C, Singer T, Gräbe A, Jauch KW, Schlitt HJ and Weiss
TS: Repression of cytochrome P450 activity in human hepatocytes in
vitro by a novel hepatotrophic factor, augmenter of liver
regeneration. J Pharmacol Exp Ther. 316:822–829. 2006. View Article : Google Scholar
|
|
46
|
Vondrácek J, Machala M, Bryja V,
Chramostová K, Krcmár P, Dietrich C, Hampl A and Kozubík A: Aryl
hydrocarbon receptor-activating polychlorinated biphenyls and their
hydroxylated metabolites induce cell proliferation in
contact-inhibited rat liver epithelial cells. Toxicol Sci.
83:53–63. 2005. View Article : Google Scholar
|
|
47
|
Curcio E, Salerno S, Barbieri G, De
Bartolo L, Drioli E and Bader A: Mass transfer and metabolic
reactions in hepatocyte spheroids cultured in rotating wall
gas-permeable membrane system. Biomaterials. 28:5487–5497. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Beningo KA, Dembo M and Wang YL: Responses
of fibroblasts to anchorage of dorsal extracellular matrix
receptors. Proc Natl Acad Sci USA. 101:18024–18029. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang X, Wang W, Yu W, Xie Y, Zhang X,
Zhang Y and Ma X: Development of an in vitro multicellular tumor
spheroid model using microencapsulation and its application in
anticancer drug screening and testing. Biotechnol Prog.
21:1289–1296. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Orive G, Tam SK, Pedraz JL and Hallé JP:
Biocompatibility of alginate-poly-L-lysine microcapsules for cell
therapy. Biomaterials. 27:3691–3700. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Neubauer MP, Poehlmann M and Fery A:
Microcapsule mechanics: from stability to function. Adv Colloid
Interface Sci. 207:65–80. 2014. View Article : Google Scholar
|
|
52
|
Lv G, Zhao L, Zhang A, Du W, Chen Y, Yu C,
Pan X, Zhang Y, Song T, Xu J, et al: Bioartificial liver system
based on choanoid fluidized bed bioreactor improve the survival
time of fulminant hepatic failure pigs. Biotechnol Bioeng.
108:2229–2236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gautier A, Carpentier B, Dufresne M, Vu
Dinh Q, Paullier P and Legallais C: Impact of alginate type and
bead diameter on mass transfers and the metabolic activities of
encapsulated C3A cells in bioartificial liver applications. Eur
Cell Mater. 21:94–106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hoshikawa A, Nakayama Y, Matsuda T, Oda H,
Nakamura K and Mabuchi K: Encapsulation of chondrocytes in
photopolymerizable styrenated gelatin for cartilage tissue
engineering. Tissue Eng. 12:2333–2341. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bazou D, Coakley WT, Hayes AJ and Jackson
SK: Long-term viability and proliferation of alginate-encapsulated
3-D HepG2 aggregates formed in an ultrasound trap. Toxicol In
Vitro. 22:1321–1331. 2008. View Article : Google Scholar : PubMed/NCBI
|