Introduction
Acinetobacter baumannii (A. baumanii) is a
notable pathogen that causes hospital-acquired infections among
immune-compromised patients, accounting for 5% of Gram-negative
infections (1). Due to a strong
resistance to desiccation and multiple antibacterial agents, the
widespread dissemination of multidrug-resistant (MDR) A.
baumanii strains has been a threat to hospitalized patients in
recent years (2).
Antibiotic resistance determinants play pivotal
roles in whether or not A. baumannii will flourish in the
host (3). For instance, the
expression of β-lactamase genes [e.g., oxacillinase (OXA)-235 gene,
blaOXA-51 and blaTEM-1] has
been shown to be involved in antibiotic resistance (4–6).
An 86-kb region AbaR resistant to heavy metal and antibiotics has
been found in a MDR isolate AYE (7), indicating the important role of AbaR
in the spread of A. baumannii in hospitals (8,9).
Furthermore, other resistance determinants, such as macrolide
(msrA/msrB), aminoglycoside (e.g., aacC1,
armA, and aphA1) and tetracycline [e.g., tet
(39), tet (A), and tet (B)] have
been identified in various A. baumannii isolates (10).
Bacterial whole-genome sequencing (WGS) has enhanced
our ability to evaluate antibiotic resistance determinants. For
example, a AbaR-type genomic resistance island, AbaR22, has been
identified in the MDR A. baumannii strain, MDR-ZJ06, via WGS
(11). A whole-genome comparison
detected 18 putative single nucleotide polymorphisms (SNPs) between
2 pre- and post-therapy A. baumannii isolates (12). Furthermore, 10 types of AbaR
resistance islands were identified in 2 A. baumannii
isolates using WGS (13). Despite
increased research on antibiotic resistance determinants in A.
baumannii, the molecular mechanisms of antibiotic resistance in
MDR A. baumannii are not yet fully understood, and various
antibiotic resistance genes have not been detected.
In the present study, we applied WGS to obtain the
whole genomic sequence of the MDR A. baumannii strain,
MDR-SHH02, isolated from a patient with breast cancer. Furthermore,
the antibiotic resistance of MDR-SHH02 to multiple antibiotics was
determined, and potential antibiotic resistance genes in MDR-SHH02
were predicted. The results of our study may enhance our
understanding of the molecular mechanisms of antibiotic resistance
in MDR A. baumannii, and provide a clinical guidance for the
therapy of A. baumannii-infected patients.
Materials and methods
Isolation and identification of A.
baumanii strain
The clinical MDR A. baumanii strain, named
MDR-SHH02, was isolated from the blood obtained from a 65-year-old
woman with terminal-stage breast cancer at Shanghai Sixth People's
Hospital Affiliated to Shanghai Jiaotong University, Shanghai,
China. This patient had received a double mastectomy and nearby
lymph node excision. After being discharged from the hospital, this
patient was hospitalized again due to symptoms of fever, cough
(lasting for days) and shortness of breath. During her second
hospital administration, she received several antimicrobial
therapies, including maxipime, impenem, methylprednisolone,
levofloxacin and cefoperazone-sulbactam sodium. A. baumanii
was positive in the blood culture and sputum culture. The results
of the antimicrobial susceptibility test revealed that the A.
baumanii strain was resistant to multiple commonly used
antibiotics. This study was approved by the Shanghai Health and
Family Planning Commission Foundation (Shanghai, China), and
informed consent was obtained from the patient.
The isolated strain was inoculated onto blood agar
plates and then incubated in an atmosphere of 5% CO2 at
35°C for 48 h. Afterwards, this strain was identified using
morphological and biochemical tests according to standard methods
(14). Colonies with typical
morphological and biochemical characteristics of
Acinetobacter were cultivated on 5% sheep blood agar and
identified using an automated Microscan® system (Dade
Behring, Inc., West Sacramento, CA, USA). The A. baumanii
strain was stored at −70°C in skim milk for further analyses.
Antibiotic resistance test for A.
baumanii MDR-SHH02
According to the Clinical and Laboratory Standards
Institute (CLSI) guidelines (15), disc diffusion assay (DDA) with dry
wafers saturated by 19 types of antibiotics, including gentamicin
(10 µg/wafer), tobramycin (30 µg/wafer), amikacin (30
µg/wafer), ampicillin-sulbactam (10/10 µg/wafer),
ceftazidime (30 µg/wafer), ciprofloxacin (5
µg/wafer), levofloxacin (5 µg/wafer), imipenem (10
µg/wafer), meropenem (10 µg/wafer),
piperacillin/tazobactam (100/10 µg/wafer),
ticarcillin/clavulanic acid (75/10 µg/wafer), cefepime (30
µg/wafer), cefotaxime (30 µg/wafer), ceftriaxone (30
µg/wafer), doxycycline (30 µg/wafer), minocyline (30
µg/wafer), tetracycline (30 µg/wafer), piperacillin
(100 µg/wafer) and trimethoprim-sulfamethoxazole (1.25/23.75
µg/wafer) (all from Oxoid, Ltd., Basingstoke, UK), were
carried out using the Kirby-Bauer (KB) method, as previously
described (16). Briefly,
Mueller-Hinton agar (Oxoid, Ltd.) plates were overlaid with the
inocula of the clinical A. baumanii strain, and the
turbidity of the inocula was equivalent to the 0.5 McFarland
standard. Subsequently, dry wafers saturated by antibiotics were
placed on the surface of the agar, and plates were placed in an
atmosphere of 5% CO2 at 35°C. Following 24 h of culture,
the diameter of the inhibition zone around each wafer was measured
according to the CLSI criteria (15). In this test, Escherichia
coli ATCC 25922, ATCC 35218 and Pseudomonas aeruginosa
ATCC 27853 obtained from the Clinical Laboratory Center of the
Ministry of Health were used as reference strains.
DNA preparation, library construction and
sequencing
The genomic DNA of A. baumanii MDR-SHH02 was
extracted using a bacterial genomic DNA purification kit (Tiangen
Biotech Co., Ltd., Beijing, China) according to the manufacturer's
instructions. The Illumina sequencing library was then prepared
using the Nextera™ DNA Sample Preparation kit
(Illumina®-Compatible). Paired-end dual index 2×90 bp
sequencing was fulfilled following the Illumina HiSeq 2000.
Sequencing was performed by Beijing Genomics Institute (BGI;
Shenzhen, China). The sequencing data were uploaded to the public
database the National Center for Biotechnology Information (NCBI;
http://www.ncbi.nlm.nih.gov/) under the
BioProject PRJNA256112 with BioSample accession no.
SAMN02991371.
Quality control
For the raw sequencing data, the reads were cleaned
by removing the empty reads, adapter sequences and reads with n≥10%
using the SeqPrep program (https://github.com/jstjohn/SeqPrep) and Sickle
(https://github.com/najoshi/sickle).
In addition, the reads were trimmed by discarding the reads
containing >30% bases with a Q-value ≤20 in the 3′ terminal, as
well as reads with adaptor sequences (the length of overlapping
sequences between adaptor and read was at least >15 bp, and the
number of mismatch bases was <3 bp).
Genome assembly
The clean reads were assembled using the short
oligonucleotide analysis package SOAPdenovo (version 2.04;
http://soap.genomics.org.cn/). To
determine whether the GC content has a significant effect on
sequencing randomness or not, the GC content and average depth of
the genomic sequence were calculated without repetition as a unit
of 500 bp.
Genome annotation
Genes in the assembled genomic sequence were
predicted using Glimmer 3.0 (http://www.cbcb.umd.edu/software/glimmer/) (17), which is a system for identifying
genes in DNA sequences of microorganism, particularly bacteria,
archaea and viruses.
Furthermore, tRNA and rRNA (5S, 16S, and 23S rRNA)
in the genomic sequence were searched using tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/)
(18) and RNAmmer 1.2 (http://www.cbs.dtu.dk/services/RNAmmer/)
(19), respectively.
Additionally, tandem repeat sequences and clustered
regularly interspaced short palindromic repeats (CRISPR) in the
genomic sequence were predicted using the Tandem Repeat Finder
(http://tandem.bu.edu/trf/trf.html)
and CRISPR Finder (http://crispr.u-psud.fr/Server/) software,
respectively. Insertion sequences (ISs) were characterized using
the IS Finder database (https://www-is.biotoul.fr//), and the parameter -e was
set as 1e-5, identity set as 35%. Besides, protein domains
associated with the genomic sequence were predicted using the
InterPro database (https://www.ebi.ac.uk/interpro/), and the parameter
was set as -appl PfamA.
Functional annotation of genes
Sequence alignment of the amino acid sequences of
genes to the Cluster of Orthologous Groups (COGs) of proteins
database (http://www.ncbi.nlm.nih.gov/COG/) (20) was performed using the Basic Local
Alignment Search Tool (BLASTP; version 2.0) program from NCBI
(E-value ≤10–4) (21). We also
performed sequence alignment of the amino acid sequences of genes
to the NCBI non-redundant (NR) database (E-value ≤10–10, identity
score ≥35%, and coverage length ≥80%). If the amino acid sequence
of a gene was aligned to multiple sequences in the databases, the
optimal result was retained.
Construction of phylogenetic tree
Based on the NCBI 16S rRNA gene database, 16S rRNA
gene sequences of 5 A. baumanii strains, including ATCC
17978, ATCC 19606, CIP 70.34, DSM 30007 and A. baumanii JCM
68415, as well as 2 species belonging to Acinetobacter
(A. haemolyticus ATCC 17906 and A. bereziniae ATCC
17924), were used to construct the phylogenetic tree, along with
MDR-SHH02. Briefly, multiple sequence alignment was performed using
ClustalW-2.1 (22). Subsequently,
the software package PHYLIP 3.695 (http://evolution.genetics.washington.edu/phylip.html),
along with the bootstrap algorithm, was used to construct the
maximum likelihood phylogenetic tree, and the phylogenetic tree was
visualized by FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Prediction of pathogenicity islands
(PAIs)
The Pathogenicity Island database (PAIDB; http://www.paidb.re.kr/about_paidb.php),
which is a web-based user-friendly resource and widely used for
detecting PAIs in newly sequenced genomes (23), was utilized to predict PAIs in the
genomic sequence of MDR-SHH02.
Identification of antibiotic resistance
genes
To identify potential antibiotic resistance genes in
the genomic sequence of MDR-SHH02, sequence alignment of the
protein sequences of antibiotic resistance genes in the Antibiotic
Resistance Genes database (ARDB; http://ardb.cbcb.umd.edu/) (24) and MDR-SHH02 genomic sequence was
conducted using BLASTP (E-value ≤10–10, identity score ≥90%, and
coverage length ≥80%).
Results
Antibiotic-resistance of A. baumanii
MDR-SHH02
The antibiotic-resistance assay revealed that the
diameter of the inhibition zone for 17 types of antibiotics on
MDR-SHH02 plates was 6 mm, apart from levofloxacin and minocyline
(diameter, 10 mm) (Table I).
According to the CLSI criteria, MDR-SHH02 was resistant to all of
the tested antibiotics.
 | Table IThe results of antibiotic-resistance
assay for A. baumannii MDR-SHH02. |
Table I
The results of antibiotic-resistance
assay for A. baumannii MDR-SHH02.
| Antibiotic
name | Diameter of
inhibition zone on MDR-SHH02 plate (mm) |
Antibiotic-resistance of MDR-SHH02 |
|---|
|
Ampicillin-sulbactam | 6 | R |
| Ceftazidime | 6 | R |
| Ciprofloxacin | 6 | R |
| Levofloxacin | 10 | R |
| Imipenem | 6 | R |
| Meropenem | 6 | R |
| Gentamicin | 6 | R |
| Tobramycin | 6 | R |
| Amikacin | 6 | R |
|
Piperacillin/tazobactam | 6 | R |
|
Ticarcillin/clavulanic acid | 6 | R |
| Cefepime | 6 | R |
| Cefotaxime | 6 | R |
| Ceftriaxone | 6 | R |
| Doxycycline | 6 | R |
| Minocyline | 10 | R |
| Tetracycline | 6 | R |
| Piperacillin | 6 | R |
|
Trimethoprim-sulfamethoxazole | 6 | R |
Assembly and annotation of the genomic
sequence of A. baumanii MDR-SHH02
During the genome assembly, a total of 85 scaffolds
were generated, and the scaffold N50 length was 131,822 bp. The
total draft genome length of MDR-SHH02 was 4,003,808 bp, with
38.99% of GC content. There were 3,787 coding sequences, 62 tRNA
sequences and 3 rRNA sequences in the genomic sequence. Moreover, 2
CRISPR and 36 tandem repeat sequences, as well as 29 ISs were
predicted in the genomic sequence (Table II).
 | Table IIThe results of genome assembly and
annotation of A. baumannii MDR-SHH02. |
Table II
The results of genome assembly and
annotation of A. baumannii MDR-SHH02.
| Feature | Statistics |
|---|
| Length of total
draft genome length (bp) | 4,003,808 |
| No. of
scaffolds | 85 |
| Length of scaffold
N50 (bp) | 131,822 |
| GC content (%) | 38.99 |
| No. of coding
sequences | 3,787 |
| No. of tRNAs | 62 |
| No. of rRNAs | 3 |
| No. of CRISPR | 2 |
| No. of tandem
repeat sequences | 36 |
| No. of insertion
sequences | 29 |
Furthermore, numerous protein domains were predicted
in the genomic sequence of MDR-SHH02, such as the AcrB/AcrD/AcrF
family (Table III).
 | Table IIIThe top 10 predicted protein domains
in the genomic sequence of A. baumannii MDR-SHH02. |
Table III
The top 10 predicted protein domains
in the genomic sequence of A. baumannii MDR-SHH02.
| Gene ID | Protein domain | P-value |
|---|
| MDRSHH02002231 | AcrB/AcrD/AcrF
family | <1.0E-300 |
| MDRSHH02000985 | Monomeric
isocitrate dehydrogenase | <1.0E-300 |
| MDRSHH02003056 | AcrB/AcrD/AcrF
family | <1.0E-300 |
| MDRSHH02000840 | AcrB/AcrD/AcrF
family | <1.0E-300 |
| MDRSHH02003594 | AcrB/AcrD/AcrF
family | <1.0E-300 |
| MDRSHH02000667 | AcrB/AcrD/AcrF
family | <1.0E-300 |
| MDRSHH02000153 | Phosphoenolpyruvate
carboxylase | 2.70E-292 |
| MDRSHH02000199 | Urocanase | 5.20E-287 |
| MDRSHH02000831 | Phosphoenolpyruvate
carboxykinase | 2.70E-279 |
| MDRSHH02003134 | AcrB/AcrD/AcrF
family | 5.50E-266 |
Functional annotation of the genomic
sequence of A. baumanii MDR-SHH02
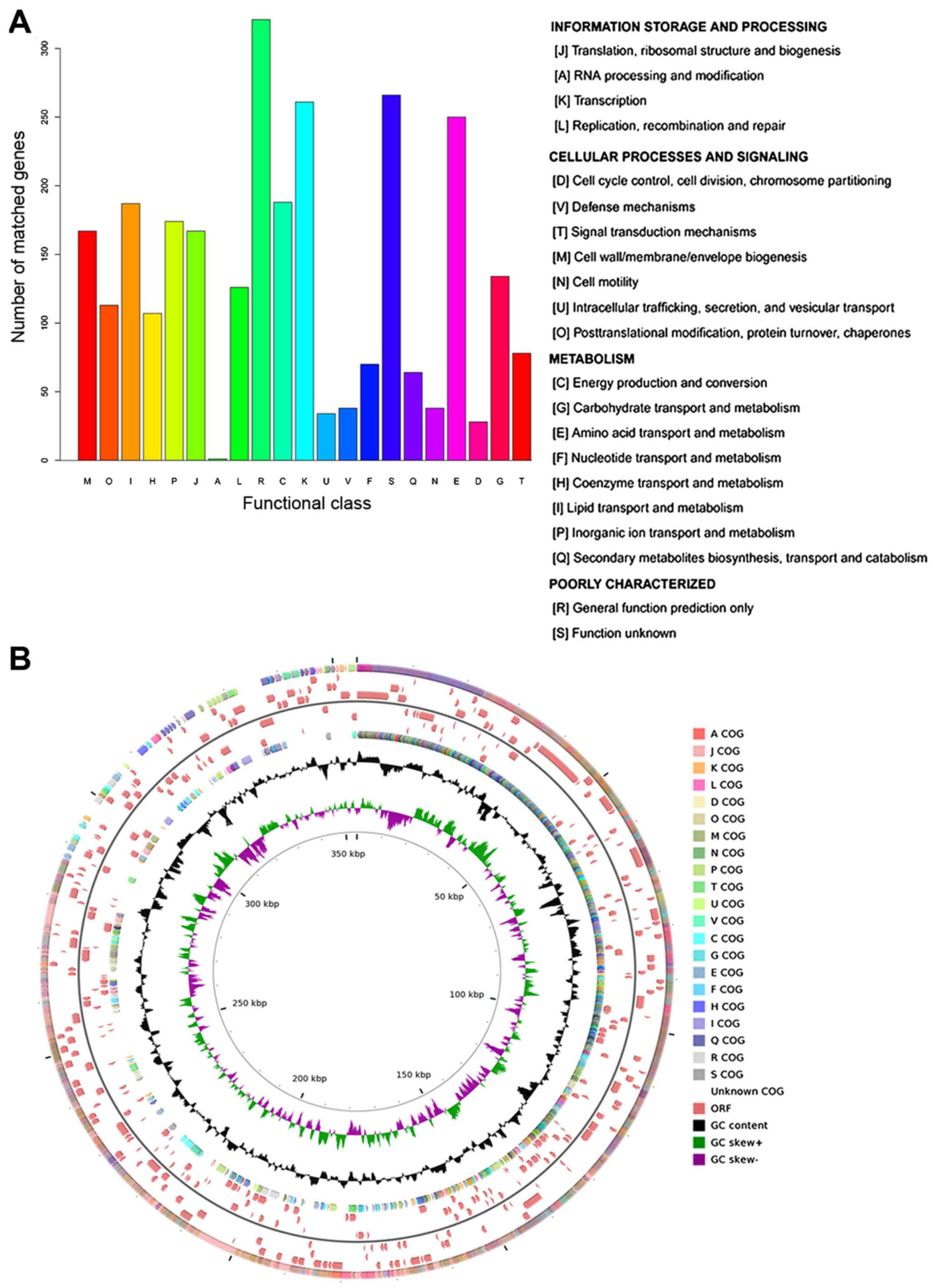
According to the COG annotation, 74.25%
(2,812/3,787) of coding sequences were annotated into 21 COG terms,
which were divided into 3 categories: information storage of
processing, cellular processes and signaling, and metabolism. Apart
from the category of poorly characterized, most o the coding
sequences were annotated into 'transcription' (number of coding
sequences, 261) and 'amino acid transport and metabolism' (number
of coding sequences, 250) (Fig.
1A). The distribution of COG categories in the genomic sequence
of A. baumannii MDR-SHH02 is shown in Fig. 1B.
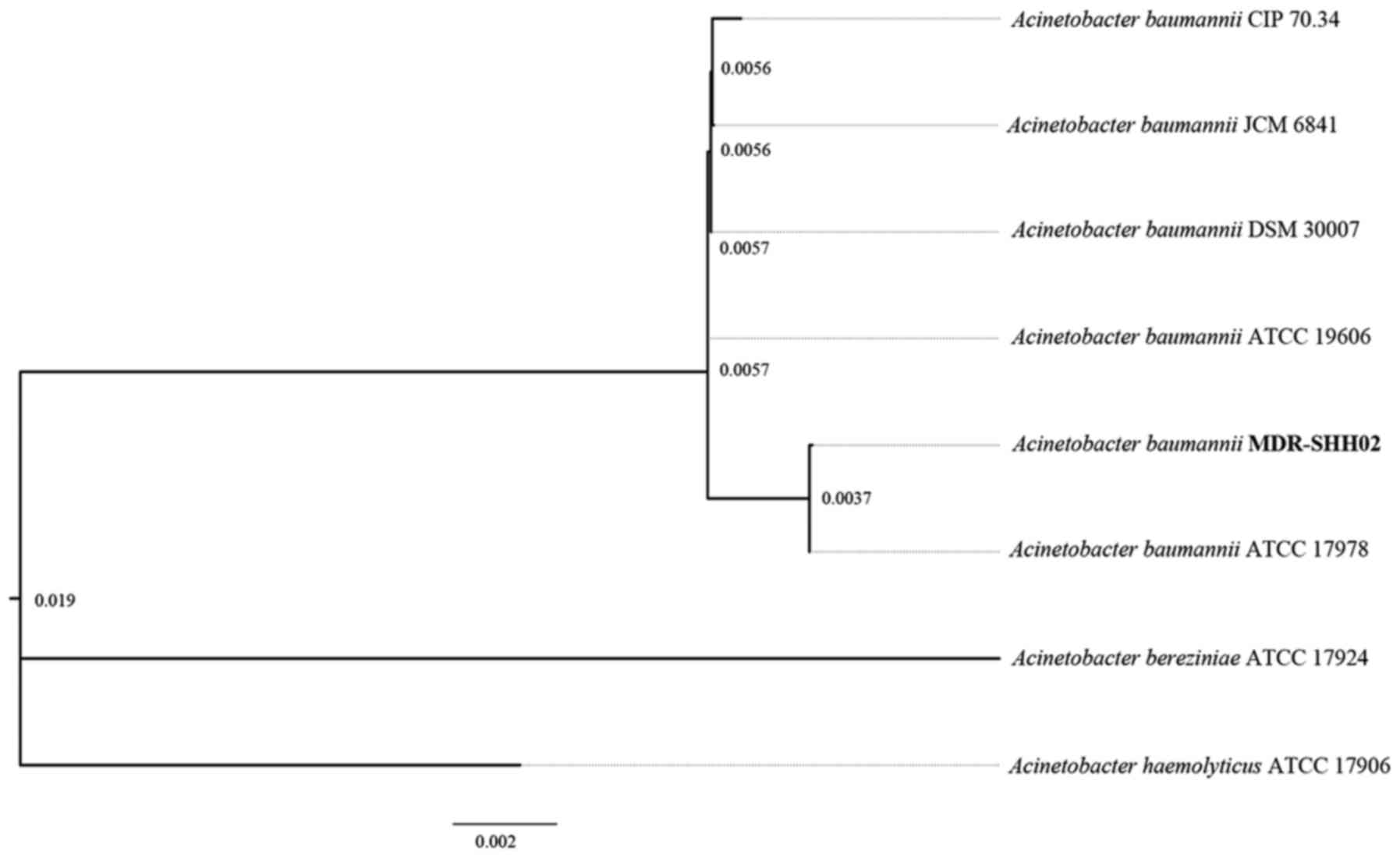
Analysis of the phylogenetic tree
Based on the 16S rRNA gene sequences of A.
baumanii in NCBI, the phylogenetic tree revealed that the
genomic sequence of MDR-SHH02 was most similar to the 16S rRNA gene
sequence of A. baumanii ATCC 17978 (Fig. 2).
Analysis of PAIs
During the process of bacterial infection, PAIs play
pivotal roles in the evolution of pathogens and the development of
diseases. In the genomic sequence of MDR-SHH02, a total of 45 PAIs
homologous to the sequence MDRSHH02000806 were detected (Table IV). Most of the PAIs were
previously identified from Escherichia coli [e.g., locus of
enterocyte effacement (LEE)] and Pseudomonas (e.g., PAPI-1
and T-PAI).
 | Table IVPathogenicity islands homologous to a
region in the genomic sequence of A. baumannii
MDR-SHH02. |
Table IV
Pathogenicity islands homologous to a
region in the genomic sequence of A. baumannii
MDR-SHH02.
| Gene ID | Start | End | Size (bp) | no. of ORFs | PAIs homologous to
this region |
|---|
| MDRSHH02000806 | 332 | 21071 | 20740 | 18 | PAPI-1
(Pseudomonas aeruginosa PA14) |
| | | | | PAGI-2(C)
(Pseudomonas aeruginosa C) |
| | | | | PAGI-3(SG)
(Pseudomonas aeruginosa SG17M) |
| | | | | PAGI-5
(Pseudomonas aeruginosa PSE9) |
| | | | | PPHGI-1
(Pseudomonas syringae pv. phaseolicola 1302A) |
| | | | | SPI-7
(Salmonella enterica subsp. enterica serovar Typhi str.
CT18) |
| | | | | SPI-7
(Salmonella enterica subsp. enterica serovar Typhi Ty2) |
| | | | | AbaR25
(Acinetobacter baumannii BJAB07104) |
| | | | | AbaR26
(Acinetobacter baumannii BJAB0868) |
| | | | | Hrp PAI (Erwinia
amylovora 321) |
| | | | | S-PAI
(Pseudomonas cichorii 83-1) |
| | | | | Hrp PAI
(Pseudomonas syringae pv. tomato DC3000) |
| | | | | Hrp PAI
(Pseudomonas syringae pv. tomato str. DC3000) |
| | | | | T-PAI
(Pseudomonas viridiflava LP23.1a) |
| | | | | T-PAI
(Pseudomonas viridiflava PNA3.3a) |
| | | | | S-PAI
(Pseudomonas viridiflava RMX23.1a) |
| | | | | S-PAI
(Pseudomonas viridiflava ME3.1b) |
| | | | | S-PAI
(Pseudomonas viridiflava RMX3.1b) |
| | | | | Not named
(Corynebacterium urealyticum DSM 7109) |
| | | | | LEE (Citrobacter
rodentium DBS100) |
| | | | | PAI I 536
(Escherichia coli 536) |
| | | | | LEE (Escherichia
coli E2348/69) |
| | | | | LEE (Escherichia
coli O157:H7 str. EDL933 ATCC43895) |
| | | | | LEE (Escherichia
coli 71074) |
| | | | | LEE (Escherichia
coli 83/39) |
| | | | | LEE (Escherichia
coli REPEC 84/110-1) |
| | | | | LEE (Escherichia
coli RW1374) |
| | | | | LEE (Escherichia
coli RDEC-1) |
| | | | | LEE (Escherichia
coli O157:H7 EDL933) |
| | | | | LEE (Escherichia
coli O157:H7 str. Sakai) |
| | | | | LEE (Escherichia
coli O103:H2 str. 12009) |
| | | | | LEE (Escherichia
coli O26:H11 str. 11368) |
| | | | | LEE (Escherichia
coli O111:H-str. 11128) |
| | | | | LEE II
(Escherichia coli 413/89-1) |
| | | | | AGI-3
(Escherichia coli BEN2908) |
| | | | | TAI (Escherichia
coli O157:H7 EDL933) |
| | | | | OI-122
(Escherichia coli O157:H7 EDL933) |
| | | | | PAI I CFT073
(Escherichia coli CFT073) |
| | | | | Not named
(Escherichia coli UMN026) |
| | | | | SESS LEE
(Salmonella enterica subsp. salamae serovar Sofia
S1296) |
| | | | | SESS LEE
(Salmonella enterica subsp. salamae serovar Sofia
S1635) |
| | | | | SHI-1 (Shigella
flexneri 2a str. 301) |
| | | | | SHI-1 (Shigella
flexneri 2a str. 2457T) |
| | | | | SRL (Shigella
flexneri 2a YSH6000) |
| | | | | Not named
(Yersinia pestis CO92) |
Screening of antibiotic resistance
genes
To reveal the genes relevant to the antibiotic
resistance of MDR-SHH02, sequence alignment of the protein
sequences of antibiotic resistance genes in ARDB and MDR-SHH02
genomic sequence was performed. Based on the selection criteria, a
total of 12 gene sequences (e.g., MDRSHH02002408, MDRSHH02000600
and MDRSHH02000597) in MDR-SHH02 were highly similar to the
sequences of antibiotic resistance genes in ARDB, such as
aac(3)-Ia,
aac(6′)-Ib, ant(2″)-Ia and
aph(3′)-Ia. According to the antibiotics that were
resistant by the 12 gene sequences, MDR-SHH02 was resistant to
multiple antibiotics, such as gentamicin, amikacin, tobramycin,
spectinomycin, streptomycin and neomycin (Table V), which was partly consistent
with the aforementioned results of antibiotic-resistance assay.
 | Table VThe potential gene sequences in A.
baumannii MDR-SHH02 relevant to antibiotic resistance. |
Table V
The potential gene sequences in A.
baumannii MDR-SHH02 relevant to antibiotic resistance.
| Gene ID | Resistance gene
type from ARDB | Resistance gene
class | Antibiotics |
|---|
| MDRSHH02002408 |
aac(3)-Ia |
Aminoglycoside
N-acetyltransferase, which modifies aminoglycosides by
acetylation | Astromicin,
gentamicin, sisomicin |
| MDRSHH02000600 |
aac(6′)-Ib |
Aminoglycoside
N-acetyltransferase, which modifies aminoglycosides by
acetylation | Amikacin,
dibekacin, isepamicin, netilmicin, sisomicin, tobramycin |
| MDRSHH02000597 |
ant(2″)-Ia |
Aminoglycoside
O-nucleotidylyltransferase, which modifies aminoglycosides by
adenylylation | Dibekacin,
gentamicin, kanamycin, sisomicin, tobramycin |
| MDRSHH02000611 |
ant(3″)-Ia |
Aminoglycoside
O-nucleotidylyltransferase, which modifies aminoglycosides by
adenylylation | Spectinomycin,
streptomycin |
| MDRSHH02001946 | aph33ib |
Aminoglycoside
O-phosphotransferase, which modifies aminoglycosides by
phosphorylation | Streptomycin |
| MDRSHH02002406 |
aph(3′)-Ia |
Aminoglycoside
O-phosphotransferase, which modifies aminoglycosides by
phosphorylation | Gentamincin
b, kanamycin, neomycin, paromomycin, lividomycin, ribostamycin |
| MDRSHH02001945 |
aph(6)-Id |
Aminoglycoside
O-phosphotransferase, which modifies aminoglycosides by
phosphorylation | streptomycin |
| MDRSHH02000608 | bl2b_tem,
bl2b_tem1 | Class A
β-lactamase. This enzyme breaks the β-lactam antibiotic ring open
and deactivates the molecule's antibacterial properties | Cephalosporin,
penicillin, cephalosporin i, cephalosporin ii |
| MDRSHH02000599 | catb3 | Group B
chloramphenicol, acetyltransferase which can inactivate
chloramphenicol. Also referred to as xenobiotic
acetyltransferase |
Chloramphenicol |
| MDRSHH02000610 | sul1 |
Sulfonamide-resistant dihydropteroate
synthase, which can not be inhibited by sulfonamide | Sulfonamide |
| MDRSHH02001738 | sul2 |
Sulfonamide-resistant dihydropteroate
synthase, which can not be inhibited by sulfonamide | Sulfonamide |
| MDRSHH02001941 | tetb | Major facilitator
superfamily transporter, tetracycline efflux pump | Tetracycline |
Discussion
In the present study, we reported the draft genomic
sequence of the clinical MDR A. baumannii strain, MDR-SHH02,
and predicted one gene seuqence homologous to 45 PAIs and 12
potential gene sequences relevant to antibiotic resistance. The
antibiotic-resistance assay and the high similarity between the 12
gene sequences in MDR-SHH02 and the sequences of antibiotic
resistance genes in ARDB, revealed that MDR-SHH02 was resistant to
multiple antibiotics.
According to the prediction of PAIs, the gene
sequence MDRSHH02000806 was homologous to 45 PAIs, such as LEE and
PAPI-1. LEE PAIs were previously identified from multiple
enteropathogenic Escherichia coli strains, and they are
highly conserved in gene order and nucleotide sequence (25,26). PAPI-1 was previously identified
from the P. aeruginosa strain, PA14, and it contributes
directly and synergistically along with PAPI-2 to the virulence of
PA14 (27). Therefore, the
virulence of MDR-SHH02 may be due to the presence of MDRSHH02000806
homologous to these PAIs.
In this study, we discovered a set of gene sequences
that were highly similar to the sequences of multiple genes
encoding aminoglycoside-modifying enzymes (AMEs), including 2
aminoglycoside N-acetyltransferase genes [aac(3)-Ia and aac(6′)-Ib],
2 aminoglycoside O-nucleotidylyltransferase genes
[ant(2″)-Ia and ant(3″)-Ia], and 3 aminoglycoside
O-phosphotransferase genes [aph33ib,
aph(3′)-Ia and aph(6)-Id]. The expression of AMEs
enables bacteria to catalyze the modification of amino and hydroxyl
groups on sugar moieties, such as aminoglycosides (28), which is a major cause of
aminoglycoside resistance in many bacteria (29). The majority of
aminoglycoside-resistant Acinetobacter isolates have the
ability of enzymatic modification of aminoglycosides by
acetyltransferases, nucleotidyltransferases and/or
phosphotransferases (30).
Previous studies have reported the prevalence of multiple AME genes
[e.g., aac(3)-Ia, and
aac(6′)-Ib] in a set of A. baumannii isolates
that are resistant to various aminogl ycosides (e.g., amikacin,
gentamicin and tobramycin) (31,32). Besides, Aph(6)-Id and ant(3″)-Ia have been
detected in the A. baumannii strain, MRSN 12227, which is
resistant to various antibiotics, such as amikacin, tobramycin and
cefotaxime (33). Another study
reported that ant(2″)-Ia present in a group of A.
baumannii isolates (62.6%) was associated with resistance to
the tested aminoglycosides (amikacin, tobramycin and gentamicin)
(34). Furthermore, in this
study, we found that MDR-SHH02 was resistant to 19 antibiotics,
such as several types of aminoglycosides (amikacin, gentamicin and
tobramycin), indicating that the resistance of MDR-SHH02 to
aminoglycosides likely resulted from the coding sequences highly
similar to AME genes. However, aph33ib has not been
previously detected in A. baumannii isolates, and it is
worthy of further study. For example, following knockout and
complementation of the gene sequence that is highly similar to
aph33ib, the resistance of MDR-SHH02 to aminoglycosides is
determined.
In this study, several gene sequences in MDR-SHH02
had a high similarity to class A β-lactamase genes (bl2b_tem
and bl2b_tem1), group B chloramphenicol acetyltransferase
gene (catb3), sulfonamide-resistant dihydropteroate synthase
genes (sul1 and sul2) and tetracycline efflux pump
gene (tetb). The gene bl2b_tem has been detected in
Staphylococcus aureus (35), and bl2b_tem1 was detected
in a series of marine bacteria, such as Pelagibacter,
Polaribacter and Roseobacter (36) . However, there is no evidence to
support that bl2b_tem and bl2b_tem1 are carried by
A. baumannii isolates. All other genes (catb3,
sul1, sul2 and tetb) have been found in A.
baumannii isolates (37–40).
Despite the aforementioned results, there were still
several limitations to this study. The association of
MDRSHH02000806 with the virulence of MDR-SHH02 needs to be
validated in further studies. Besides, the associations between the
12 gene sequences similar to AME genes and the antibiotic
resistance of MDR-SHH02 are also needs to be confirmed in further
studies. We aim to conduct such experiments in our future
studies.
In conclusion, this study fulfilled the draft
genomic sequence of the clinical MDR A. baumannii strain,
MDR-SHH02, and 12 gene sequences in MDR-SHH02 had a highly
similarity to the sequences of genes encoding AMEs [e.g.,
aac(3)-Ia,
ant(2″)-Ia, aph33ib and aph(3′)-Ia],
β-lactamase genes (bl2b_ tem and bl2b_tem1),
sulfonamide-resistant dihydropteroate synthase genes (sul1
and sul2), catb3 and tetb. Of these genes,
aph33ib, bl2b_tem and bl2b_tem1 were potential
new antibiotic resistance genes. Furthermore, the
antibiotic-resistance assay revealed that MDR-SHH02 was resistant
to multiple antibiotics, such as amikacin, gentamicin and
tobramycin. These findings were expected to enrich the data of
antibiotic resistance genes in MDR A. baumannii, and provide
a clinical guidance for the personalized therapy of A.
baumannii-infected patients.
Acknowledgments
This study was supported by a project supported by
Shanghai Health and Family Planning Commission Foundation, China
(grant no. 20134010) and a project supported by the Natural Science
Foundation of Shanghai, China (grant no. 15ZR1436100).
References
|
1
|
Lockhart SR, Abramson MA, Beekmann SE,
Gallagher G, Riedel S, Diekema DJ, Quinn JP and Doern GV:
Antimicrobial resistance among Gram-negative bacilli causing
infections in intensive care unit patients in the United States
between 1993 and 2004. J Clin Microbiol. 45:3352–3359. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Visca P, Seifert H and Towner KJ:
Acinetobacter infection - an emerging threat to human health. IUBMB
Life. 63:1048–1054. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mortensen BL and Skaar EP: Host-microbe
interactions that shape the pathogenesis of Acinetobacter baumannii
infection. Cell Microbiol. 14:1336–1344. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Higgins PG, Pérez-Llarena FJ, Zander E,
Fernández A, Bou G and Seifert H: OXA-235, a novel class D
β-lactamase involved in resistance to carbapenems in Acinetobacter
baumannii. Antimicrob Agents Chemother. 57:2121–2126. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zander E, Nemec A, Seifert H and Higgins
PG: Association between β-lactamase-encoding bla DiversiLab
rep-PCR-based typing of Acinetobacter (OXA-51) variants and
baumannii isolates. J Clin Microbiol. 50:1900–1904. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krizova L, Poirel L, Nordmann P and Nemec
A: TEM-1 β-lactamase as a source of resistance to sulbactam in
clinical strains of Acinetobacter baumannii. J Antimicrob
Chemother. 68:2786–2791. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fournier PE, Vallenet D, Barbe V, Audic S,
Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, et
al: Comparative genomics of multidrug resistance in Acinetobacter
baumannii. PLoS Genet. 2:e72006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ramírez MS, Vilacoba E, Stietz MS, Merkier
AK, Jeric P, Limansky AS, Márquez C, Bello H, Catalano M and
Centrón D: Spreading of AbaR-type genomic islands in multidrug
resistance Acinetobacter baumannii strains belonging to different
clonal complexes. Curr Microbiol. 67:9–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Šeputienė V, Povilonis J and Sužiedėlienė
E: Novel variants of AbaR resistance islands with a common backbone
in Acinetobacter baumannii isolates of European clone II.
Antimicrob Agents Chemother. 56:1969–1973. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taitt CR, Leski T, Stockelman MG, Craft
DW, Zurawski DV, Kirkup BC and Vora GJ: Antimicrobial resistance
determinants in Acinetobacter baumannii isolates taken from
military treatment facilities. Antimicrob Agents Chemother.
58:767–781. 2014. View Article : Google Scholar :
|
|
11
|
Zhou H, Zhang T, Yu D, Pi B, Yang Q, Zhou
J, Hu S and Yu Y: Genomic analysis of the multidrug-resistant
Acinetobacter baumannii strain MDR-ZJ06 widely spread in China.
Antimicrob Agents Chemother. 55:4506–4512. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hornsey M, Loman N, Wareham DW, Ellington
MJ, Pallen MJ, Turton JF, Underwood A, Gaulton T, Thomas CP,
Doumith M, et al: Whole-genome comparison of two Acinetobacter
baumannii isolates from a single patient, where resistance
developed during tigecycline therapy. J Antimicrob Chemother.
66:1499–1503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Liu F, Zhang Y, Wang X, Zhao C, Chen
H, Zhang F, Zhu B, Hu Y and Wang H: Evolution of
carbapenem-resistant Acinetobacter baumannii revealed through
whole-genome sequencing and comparative genomic analysis.
Antimicrob Agents Chemother. 59:1168–1176. 2015. View Article : Google Scholar :
|
|
14
|
Atlas RM, Brown AE and Parks LC:
Laboratory Manual of Experimental Microbiology. Mosby; St. Louis,
MO: pp. 119–127. 1995
|
|
15
|
Watts JL: Performance Standards For
Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria
Isolated From Animals: Approved Standard. Clinical and Laboratory
Standards Institute; 2008
|
|
16
|
Bauer AW, Kirby WM, Sherris JC and Turck
M: Antibiotic susceptibility testing by a standardized single disk
method. Am J Clin Pathol. 45:493–496. 1966.PubMed/NCBI
|
|
17
|
Delcher AL, Bratke KA, Powers EC and
Salzberg SL: Identifying bacterial genes and endosymbiont DNA with
Glimmer. Bioinformatics. 23:673–679. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schattner P, Brooks AN and Lowe TM: The
tRNAscan-SE, snoscan and snoGPS web servers for the detection of
tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server): W686–W689.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lagesen K, Hallin P, Rødland EA,
Staerfeldt H-H, Rognes T and Ussery DW: RNAmmer: consistent and
rapid annotation of ribosomal RNA genes. Nucleic Acids Res.
35:3100–3108. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tatusov RL, Fedorova ND, Jackson JD,
Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov
SL, Nikolskaya AN, et al: The COG database: an updated version
includes eukaryotes. BMC Bioinformatics. 4:412003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mount DW: Using the basic local alignment
search tool (BLAST). CSH Protoc. 2007:pdb. top172007.PubMed/NCBI
|
|
22
|
Chenna R, Sugawara H, Koike T, Lopez R,
Gibson TJ, Higgins DG and Thompson JD: Multiple sequence alignment
with the Clustal series of programs. Nucleic Acids Res.
31:3497–3500. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoon SH, Park Y-K, Lee S, Choi D, Oh TK,
Hur C-G and Kim JF: Towards pathogenomics: a web-based resource for
pathogenicity islands. Nucleic Acids Res. 35(Database): D395–D400.
2007. View Article : Google Scholar
|
|
24
|
Liu B and Pop M: ARDB - Antibiotic
Resistance Genes Database. Nucleic Acids Res. 37(Database):
D443–D447. 2009. View Article : Google Scholar
|
|
25
|
Pósfai G, Koob MD, Kirkpatrick HA and
Blattner FR: Versatile insertion plasmids for targeted genome
manipulations in bacteria: isolation, deletion, and rescue of the
pathogenicity island LEE of the Escherichia coli O157:H7 genome. J
Bacteriol. 179:4426–4428. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tauschek M, Strugnell RA and Robins-Browne
RM: Characterization and evidence of mobilization of the LEE
pathogenicity island of rabbit-specific strains of enteropathogenic
Escherichia coli. Mol Microbiol. 44:1533–1550. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harrison EM, Carter ME, Luck S, Ou H-Y, He
X, Deng Z, O'Callaghan C, Kadioglu A and Rajakumar K: Pathogenicity
islands PAPI-1 and PAPI-2 contribute individually and
synergistically to the virulence of Pseudomonas aeruginosa strain
A14. Infect Immun. 78:1437–1446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Towner KJ: Acinetobacter: an old friend,
but a new enemy. J Hosp Infect. 73:355–363. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ramirez MS and Tolmasky ME: Aminoglycoside
modifying enzymes. Drug Resist Updat. 13:151–171. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nemec A, Dolzani L, Brisse S, van den
Broek P and Dijkshoorn L: Diversity of aminoglycoside-resistance
genes and their association with class 1 integrons among strains of
pan-European Acinetobacter baumannii clones. J Med Microbiol.
53:1233–1240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC
and Lee JC: Genetic basis of resistance to aminoglycosides in
Acinetobacter spp. and spread of armA in Acinetobacter baumannii
sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis.
64:185–190. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ploy MC, Denis F, Courvalin P and Lambert
T: Molecular characterization of integrons in Acinetobacter
baumannii: description of a hybrid class 2 integron. Antimicrob
Agents Chemother. 44:2684–2688. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Waterman PE, McGann P, Snesrud E, Clifford
RJ, Kwak YI, Munoz-Urbizo IP, Tabora-Castellanos J, Milillo M,
Preston L, Aviles R, et al: Bacterial peritonitis due to
Acinetobacter baumannii sequence type 25 with plasmid-borne new
delhi metallo-β-lactamase in Honduras. Antimicrob Agents Chemother.
57:4584–4586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akers KS, Chaney C, Barsoumian A, Beckius
M, Zera W, Yu X, Guymon C, Keen EF III, Robinson BJ, Mende K, et
al: Aminoglycoside resistance and susceptibility testing errors in
Acinetobacter baumannii-calcoaceticus complex. J Clin Microbiol.
48:1132–1138. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kostić T, Ellis M, Williams MR, Stedtfeld
TM, Kaneene JB, Stedtfeld RD and Hashsham SA: Thirty-minute
screening of antibiotic resistance genes in bacterial isolates with
minimal sample preparation in static self-dispensing 64 and 384
assay cards. Appl Microbiol Biotechnol. 99:7711–7722. 2015.
View Article : Google Scholar
|
|
36
|
Hatosy SM and Martiny AC: The ocean as a
global reservoir of antibiotic resistance genes. Appl Environ
Microbiol. 81:7593–7599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chang-Tai Z, Yang L, Zhong-Yi H,
Chang-Song Z, Yin-Ze K, Yong-Ping L and Chun-Lei D: High frequency
of integrons related to drug-resistance in clinical isolates of
Acinetobacter baumannii. Indian J Med Microbiol. 29:118–123. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Martí S, Fernández-Cuenca F, Pascual A,
Ribera A, Rodríguez-Baño J, Bou G, Miguel Cisneros J, Pachón J,
Martínez-Martínez L and Vila J; Grupo de Estudio de Infección
Hospitalaria (GEIH): Prevalence of the tetA and tetB genes as
mechanisms of resistance to tetracycline and minocycline in
Acinetobacter baumannii clinical isolates. Enferm Infecc Microbiol
Clin. 24:77–80. 2006.In Spanish. View Article : Google Scholar
|
|
39
|
Post V and Hall RM: AbaR5, a large
multiple-antibiotic resistance region found in Acinetobacter
baumannii. Antimicrob Agents Chemother. 53:2667–2671. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agersø Y and Petersen A: The tetracycline
resistance determinant Tet 39 and the sulphonamide resistance gene
sulII are common among resistant Acinetobacter spp. isolated from
integrated fish farms in Thailand. J Antimicrob Chemother.
59:23–27. 2007. View Article : Google Scholar
|