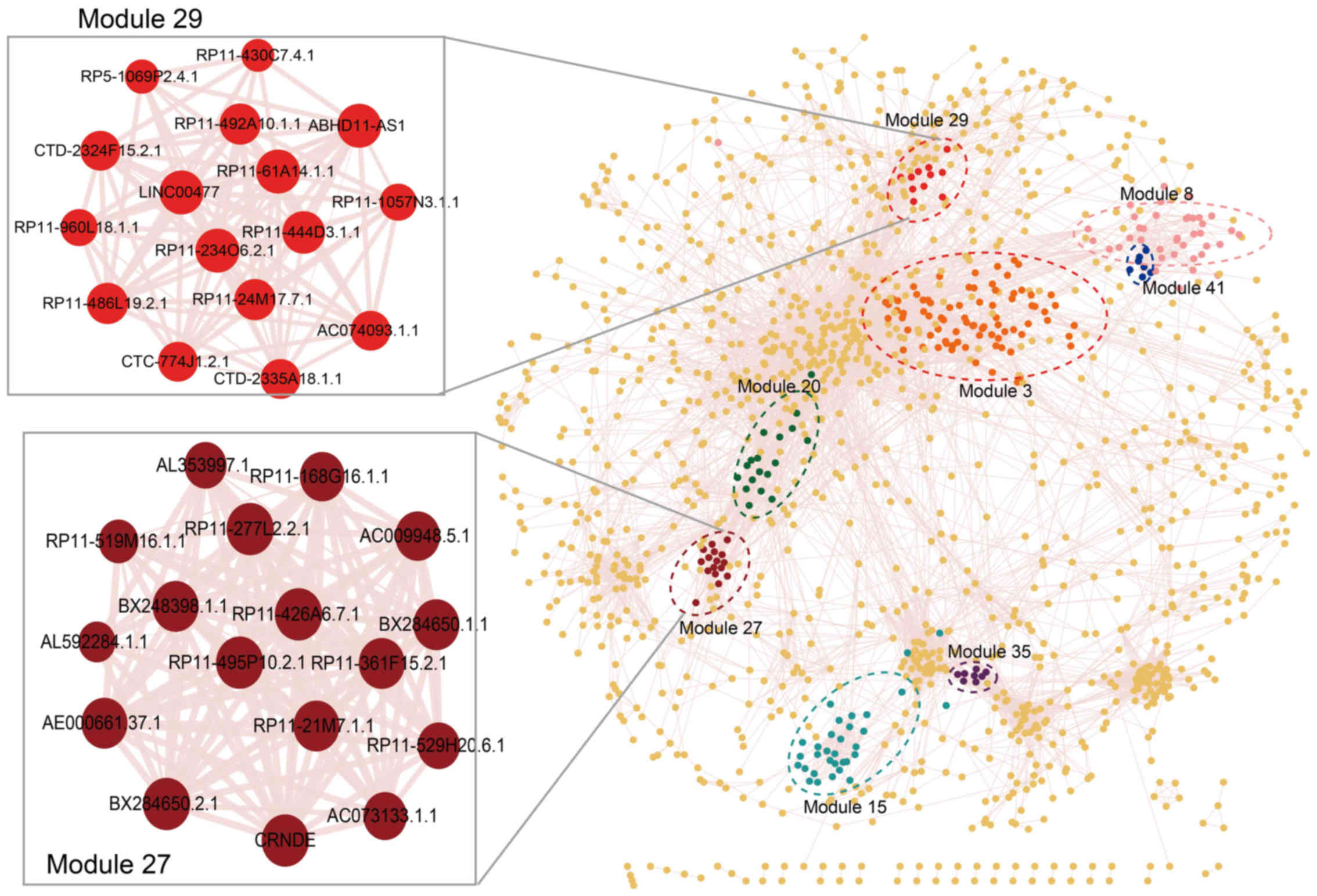
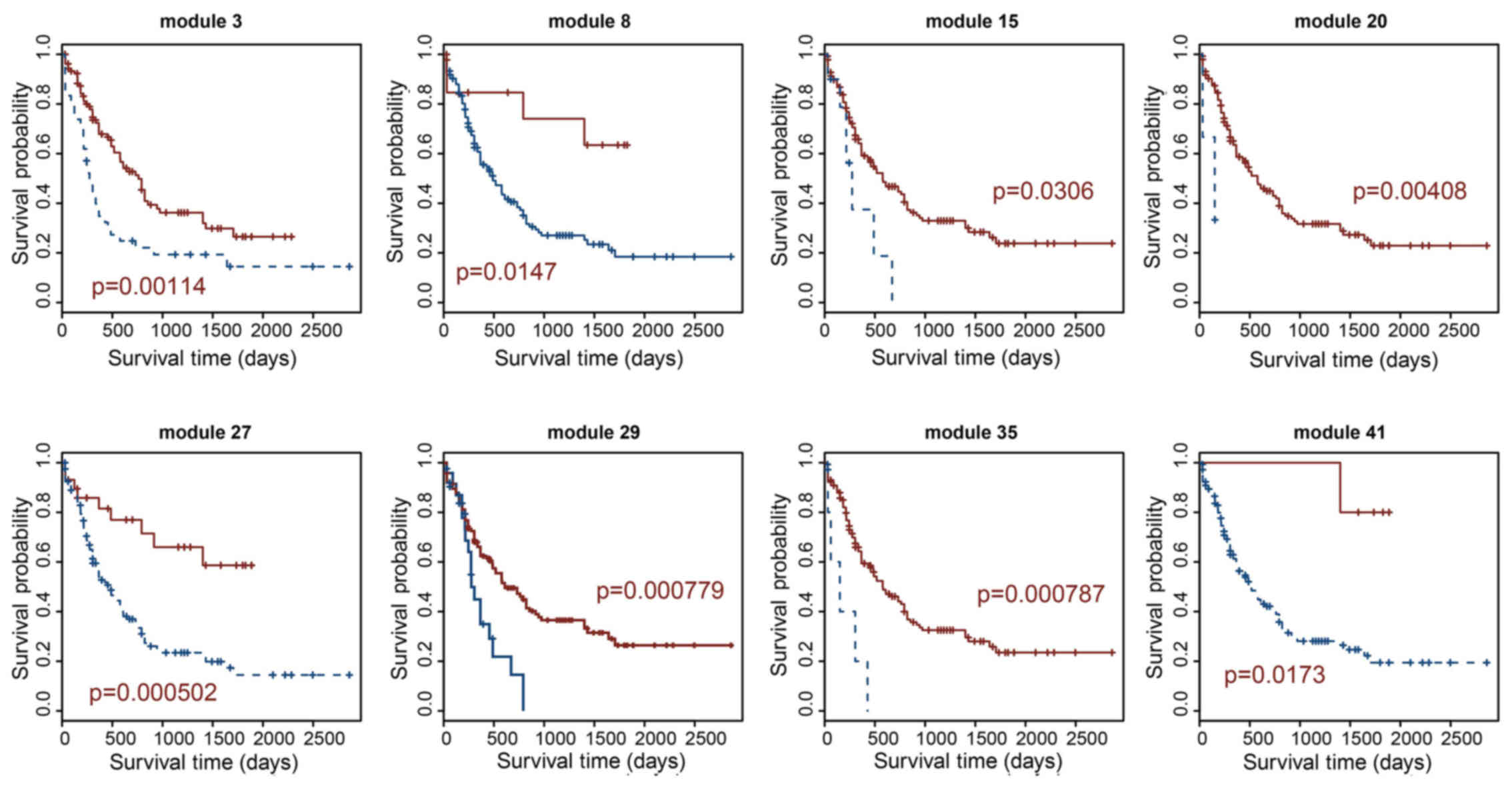
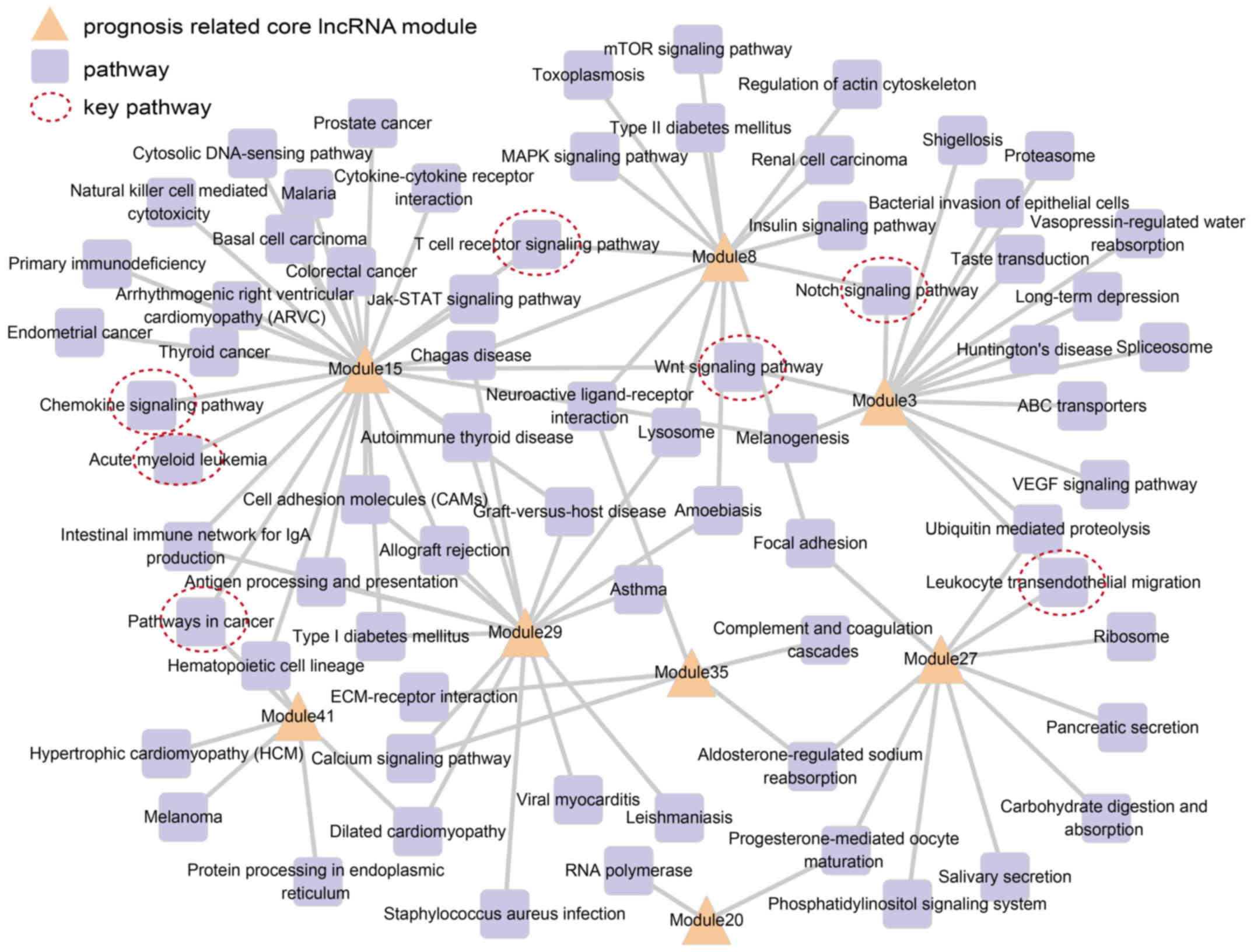
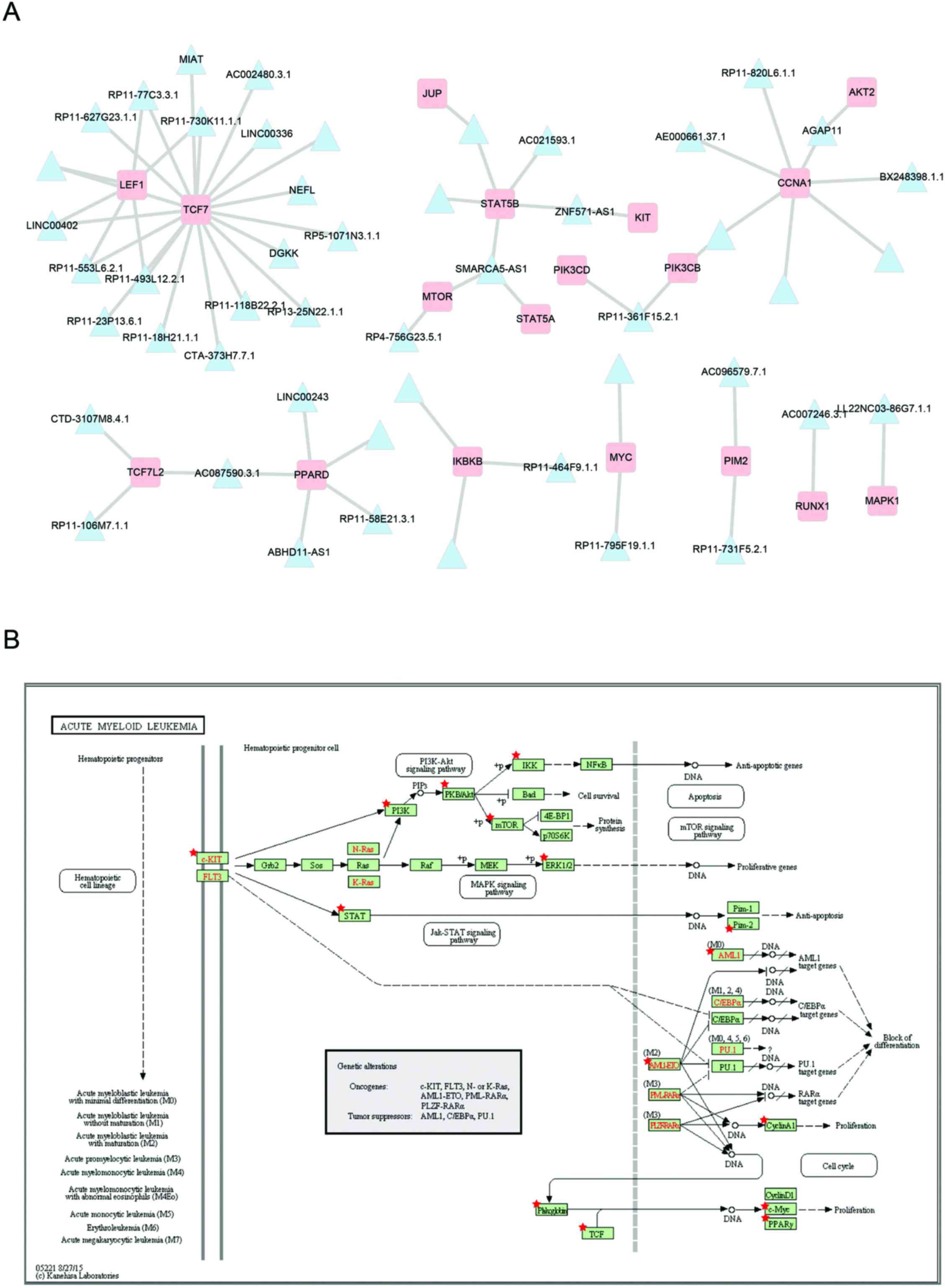
|
1
|
Döhner H, Estey EH, Amadori S, Appelbaum
FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson
RA, et al: European LeukemiaNet: Diagnosis and management of acute
myeloid leukemia in adults: Recommendations from an international
expert panel, on behalf of the European LeukemiaNet. Blood.
115:453–474. 2010. View Article : Google Scholar
|
|
2
|
Schlenk RF, Döhner K, Krauter J, Fröhling
S, Corbacioglu A, Bullinger L, Habdank M, Späth D, Morgan M, Benner
A, et al: German-Austrian Acute Myeloid Leukemia Study Group:
Mutations and treatment outcome in cytogenetically normal acute
myeloid leukemia. N Engl J Med. 358:1909–1918. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marcucci G, Maharry KS, Metzeler KH,
Volinia S, Wu YZ, Mrózek K, Nicolet D, Kohlschmidt J, Whitman SP,
Mendler JH, et al: Clinical role of microRNAs in cytogenetically
normal acute myeloid leukemia: miR-155 upregulation independently
identifies high-risk patients. J Clin Oncol. 31:2086–2093. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Díaz-Beyá M, Navarro A, Ferrer G, Díaz T,
Gel B, Camós M, Pratcorona M, Torrebadell M, Rozman M, Colomer D,
et al: Acute myeloid leukemia with translocation (8;16)(p11;p13)
and MYST3-CREBBP rearrangement harbors a distinctive microRNA
signature targeting RET proto-oncogene. Leukemia. 27:595–603. 2013.
View Article : Google Scholar
|
|
5
|
Díaz-Beyá M, Brunet S, Nomdedéu J, Tejero
R, Díaz T, Pratcorona M, Tormo M, Ribera JM, Escoda L, Duarte R, et
al: Cooperative AML group CETLAM (Grupo Cooperativo Para el Estudio
y Tratamiento de las Leucemias Agudas y Mielodisplasias): MicroRNA
expression at diagnosis adds relevant prognostic information to
molecular categorization in patients with intermediate-risk
cytogenetic acute myeloid leukemia. Leukemia. 28:804–812. 2014.
View Article : Google Scholar
|
|
6
|
Schwind S, Maharry K, Radmacher MD, Mrózek
K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler
KH, et al: Prognostic significance of expression of a single
microRNA, miR-181a, in cytogenetically normal acute myeloid
leukemia: A Cancer and Leukemia Group B study. J Clin Oncol.
28:5257–5264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hirano T, Yoshikawa R, Harada H, Harada Y,
Ishida A and Yamazaki T: Long noncoding RNA, CCDC26, controls
myeloid leukemia cell growth through regulation of KIT expression.
Mol Cancer. 14:902015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT,
Xu LD, Wang YY, Du L, Zhang YP, et al: MiRNA-miRNA synergistic
network: Construction via co-regulating functional modules and
disease miRNA topological features. Nucleic Acids Res. 39:825–836.
2011. View Article : Google Scholar
|
|
11
|
Yoshimura K, Okanoue T, Ebise H, Iwasaki
T, Mizuno M, Shima T, Ichihara J and Yamazaki K: Identification of
novel noninvasive markers for diagnosing nonalcoholic
steatohepatitis and related fibrosis by data mining. Hepatology.
63:462–473. 2016. View Article : Google Scholar
|
|
12
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ivliev AE, 't Hoen PA and Sergeeva MG:
Coexpression network analysis identifies transcriptional modules
related to proastrocytic differentiation and sprouty signaling in
glioma. Cancer Res. 70:10060–10070. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clarke C, Madden SF, Doolan P, Aherne ST,
Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown
J, et al: Correlating transcriptional networks to breast cancer
survival: A large-scale coexpression analysis. Carcinogenesis.
34:2300–2308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Tang H, Sun Z, Bungum AO, Edell ES,
Lingle WL, Stoddard SM, Zhang M, Jen J, Yang P and Wang L:
Network-based approach identified cell cycle genes as predictor of
overall survival in lung adenocarcinoma patients. Lung Cancer.
80:91–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Udyavar AR, Hoeksema MD, Clark JE, Zou Y,
Tang Z, Li Z, Li M, Chen H, Statnikov A, Shyr Y, et al:
Co-expression network analysis identifies Spleen Tyrosine Kinase
(SYK) as a candidate oncogenic driver in a subset of small-cell
lung cancer. BMC Syst Biol. 7(Suppl 5): S12013. View Article : Google Scholar
|
|
17
|
Chou WC, Cheng AL, Brotto M and Chuang CY:
Visual gene-network analysis reveals the cancer gene co-expression
in human endometrial cancer. BMC Genomics. 15:3002014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu R, Cheng Y, Yu J, Lv QL and Zhou HH:
Identification and validation of gene module associated with lung
cancer through coexpression network analysis. Gene. 563:56–62.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu J, Yu J, Cordero KE, Johnson MD, Ghosh
D, Rae JM, Chinnaiyan AM and Lippman ME: A transcriptional
fingerprint of estrogen in human breast cancer predicts patient
survival. Neoplasia. 10:79–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
22
|
Kremer KN, Peterson KL, Schneider PA, Meng
XW, Dai H, Hess AD, Smith BD, Rodriguez-Ramirez C, Karp JE,
Kaufmann SH and Hedin KE: CXCR4 chemokine receptor signaling
induces apoptosis in acute myeloid leukemia cells via regulation of
the Bcl-2 family members Bcl-XL, Noxa, and Bak. J Biol Chem.
288:22899–22914. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gallay N, Anani L, Lopez A, Colombat P,
Binet C, Domenech J, Weksler BB, Malavasi F and Herault O: The role
of platelet/endothelial cell adhesion molecule 1 (CD31) and CD38
antigens in marrow microenvironmental retention of acute
myelogenous leukemia cells. Cancer Res. 67:8624–8632. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Staal FJT, Famili F, Garcia Perez L and
Pike-Overzet K: Aberrant Wnt signaling in leukemia. Cancers
(Basel). 8:782016. View Article : Google Scholar
|
|
25
|
Heidel FH, Arreba-Tutusaus P, Armstrong SA
and Fischer T: Evolutionarily conserved signaling pathways: acting
in the shadows of acute myelogenous leukemia's genetic diversity.
Clin Cancer Res. 21:240–248. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y: Alterations in the expression
pattern of TCR zeta chain in T cells from patients with
hematological diseases. Hematology. 13:267–275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Le Dieu R, Taussig DC, Ramsay AG, Mitter
R, Miraki-Moud F, Fatah R, Lee AM, Lister TA and Gribben JG:
Peripheral blood T cells in acute myeloid leukemia (AML) patients
at diagnosis have abnormal phenotype and genotype and form
defective immune synapses with AML blasts. Blood. 114:3909–3916.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi L, Chen S, Lu Y, Wang X, Xu L, Zhang
F, Yang L, Wu X, Li B and Li Y: Changes in the MALT1-A20-NF-κB
expression pattern may be related to T cell dysfunction in AML.
Cancer Cell Int. 13:372013. View Article : Google Scholar
|
|
29
|
Fu Y, Zhu H, Wu W, Xu J, Chen T, Xu B,
Qian S, Li J and Liu P: Clinical significance of lymphoid
enhancer-binding factor 1 expression in acute myeloid leukemia.
Leuk Lymphoma. 55:371–377. 2014. View Article : Google Scholar
|
|
30
|
Metzeler KH, Heilmeier B, Edmaier KE,
Rawat VP, Dufour A, Döhner K, Feuring-Buske M, Braess J,
Spiekermann K, Büchner T, et al: High expression of lymphoid
enhancer-binding factor-1 (LEF1) is a novel favorable prognostic
factor in cytogenetically normal acute myeloid leukemia. Blood.
120:2118–2126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yohe S: Molecular genetic markers in acute
myeloid leukemia. J Clin Med. 4:460–478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garg S, Shanmukhaiah C, Marathe S, Mishra
P, Babu Rao V, Ghosh K and Madkaikar M: Differential antigen
expression and aberrant signaling via I3/AKT, MAP/ERK, JAK/STAT,
and Wnt/beta catenin pathways in Lin-/CD38-/CD34 cells in acute
myeloid leukemia. Eur J Haematol. 96:309–317. 2016. View Article : Google Scholar
|