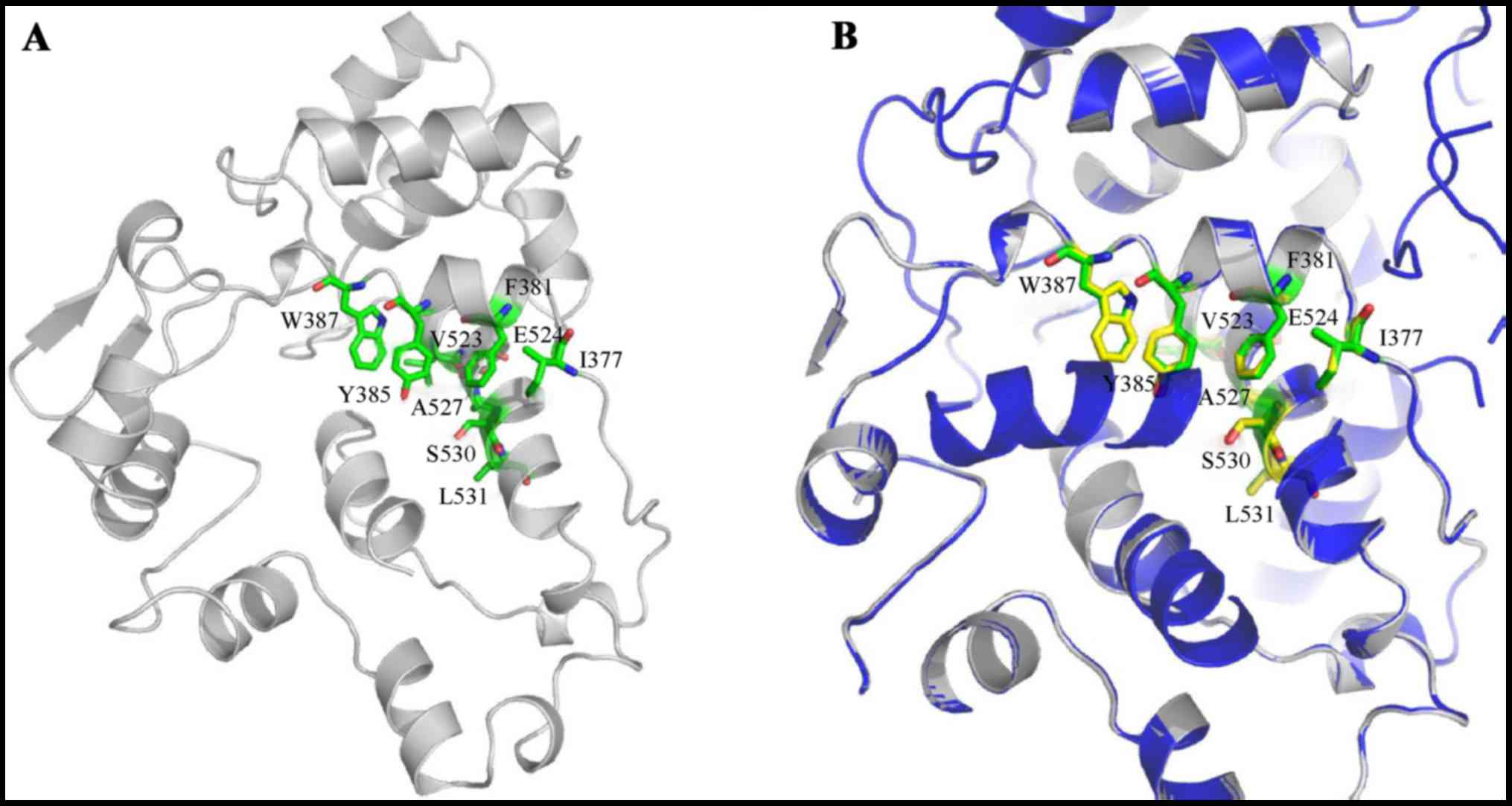
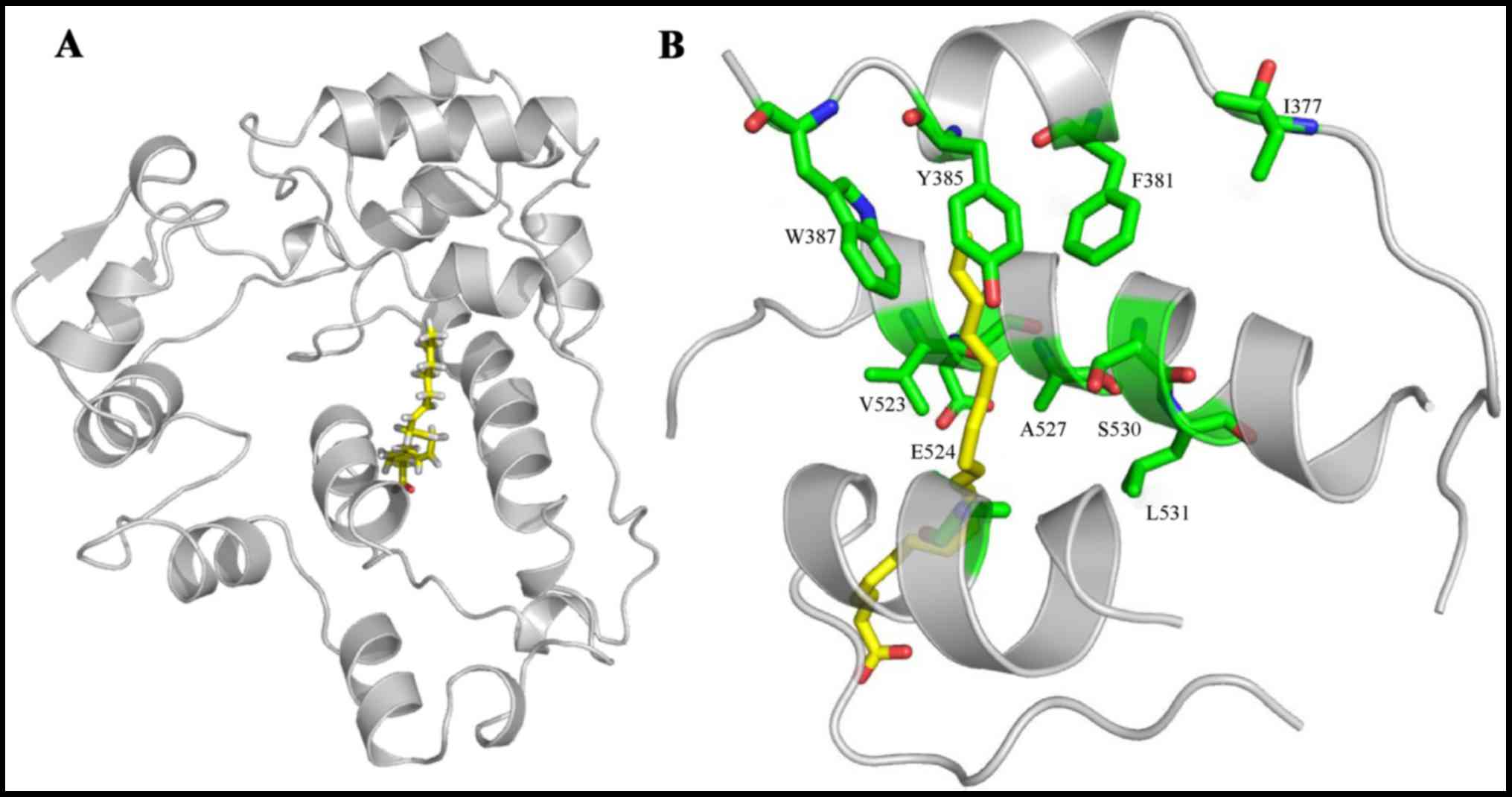
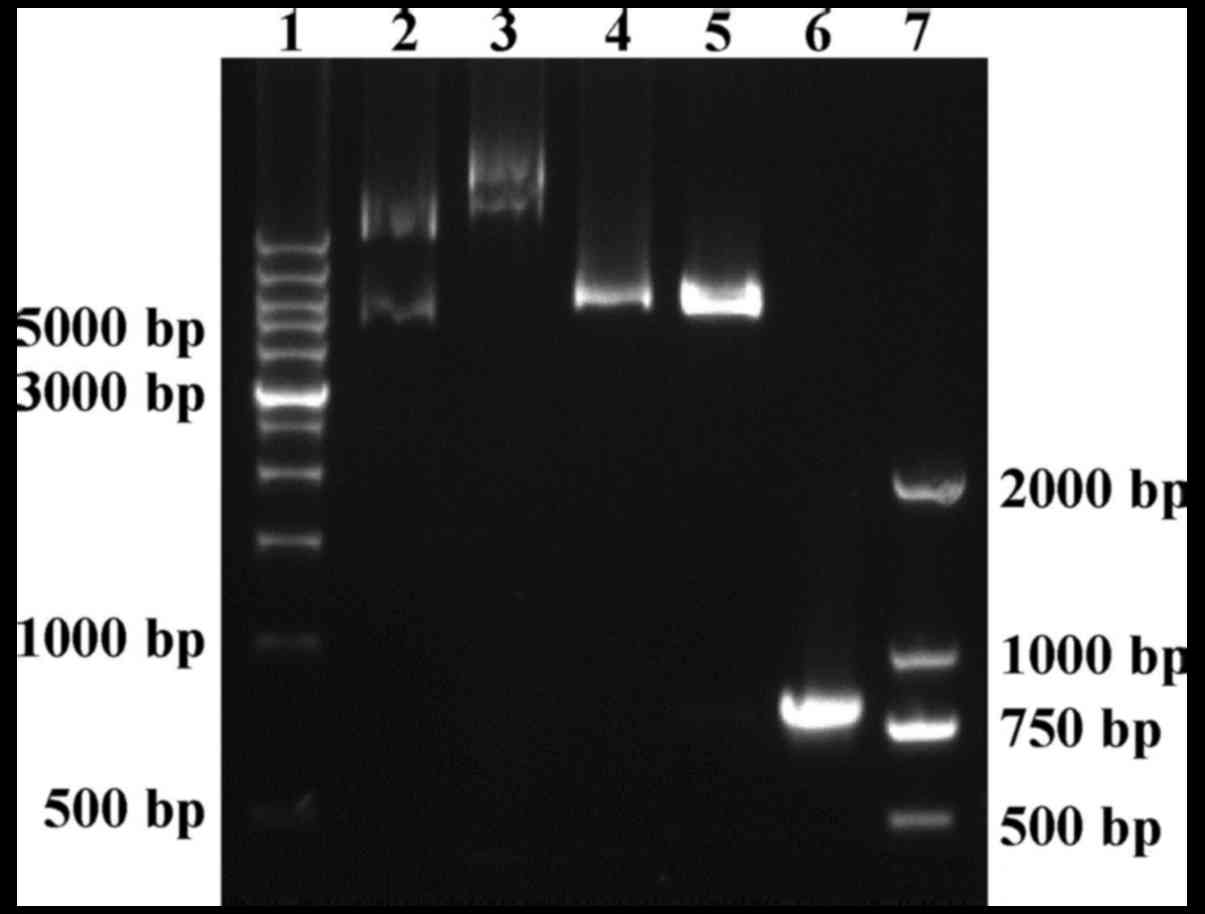
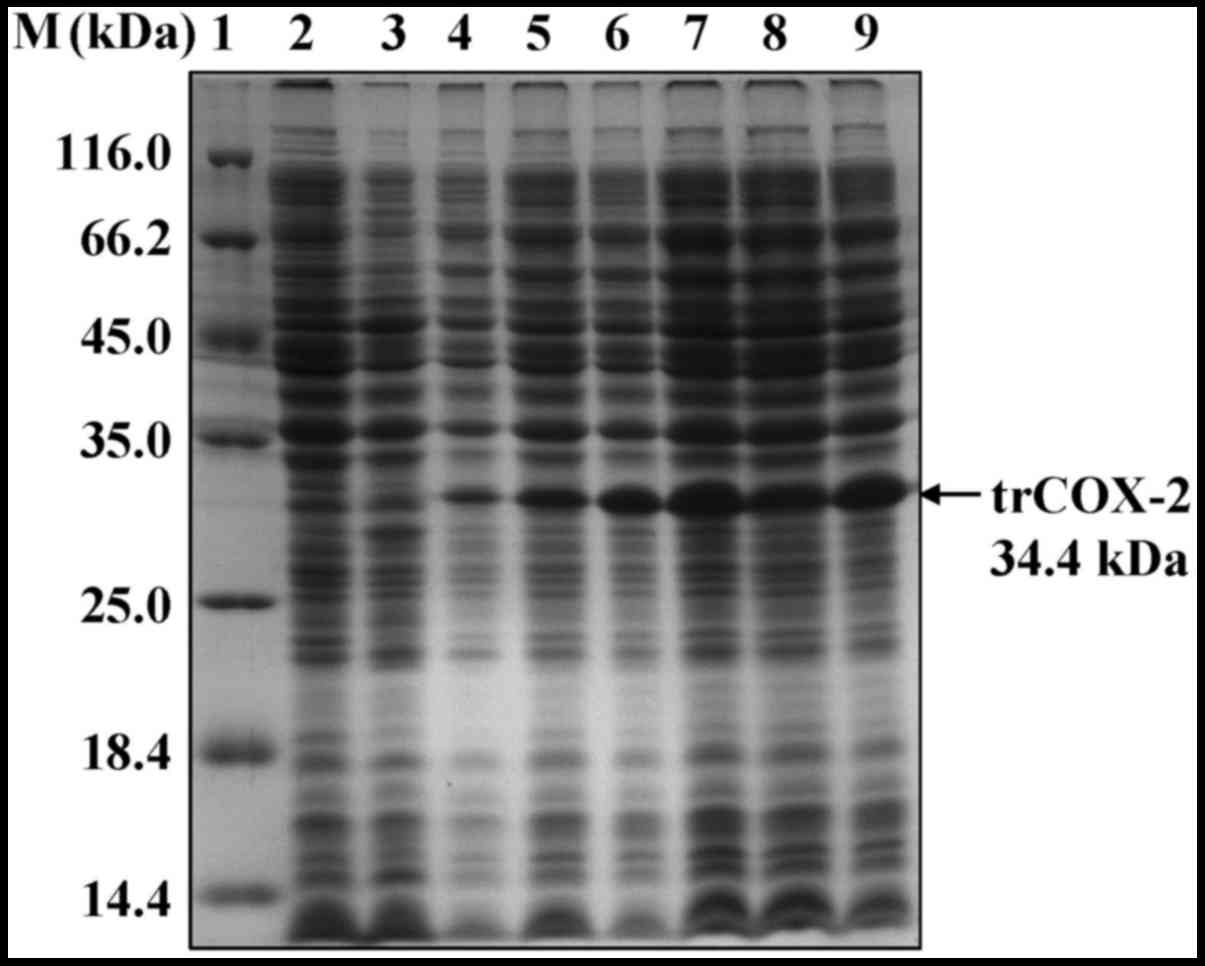
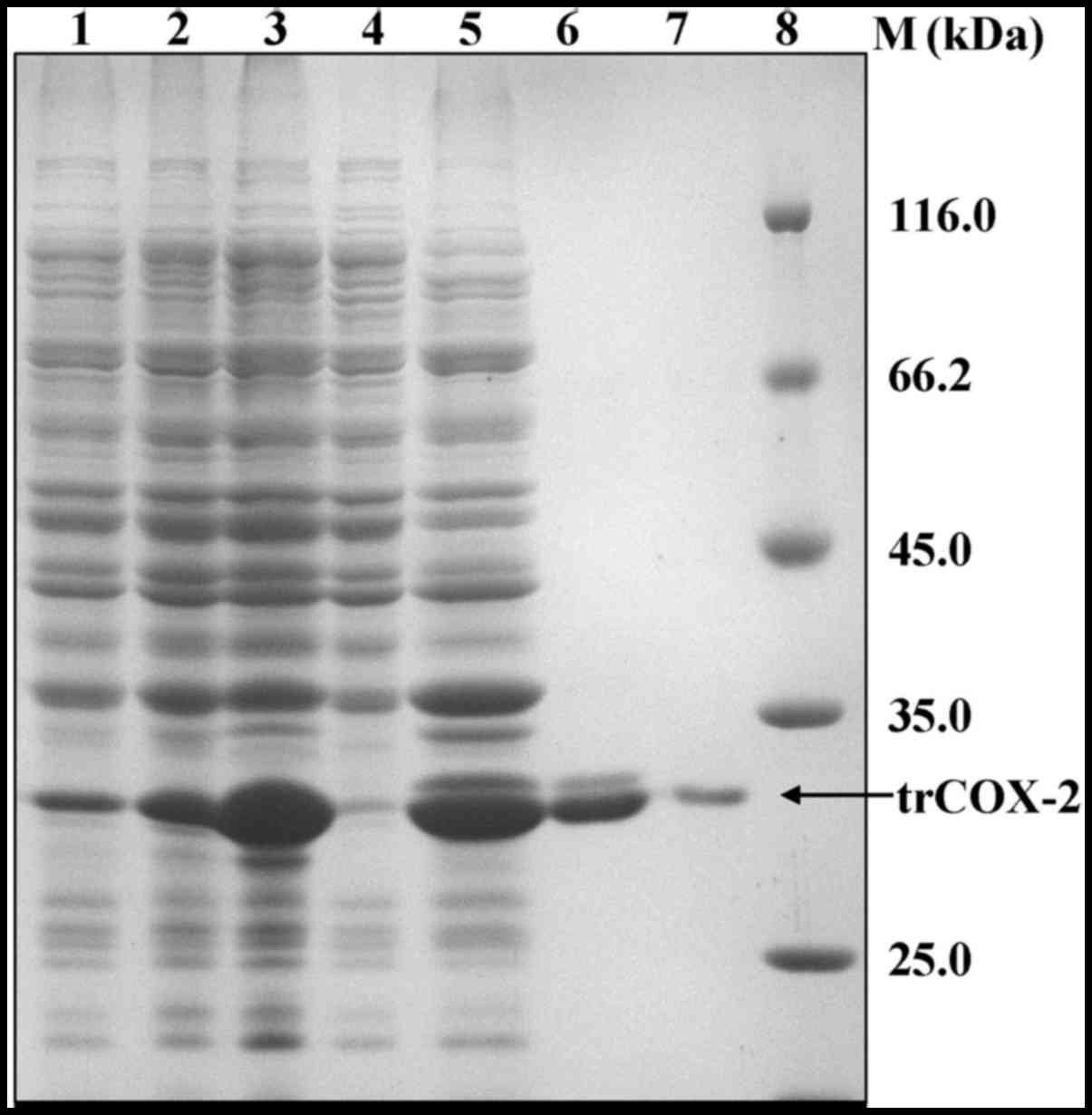
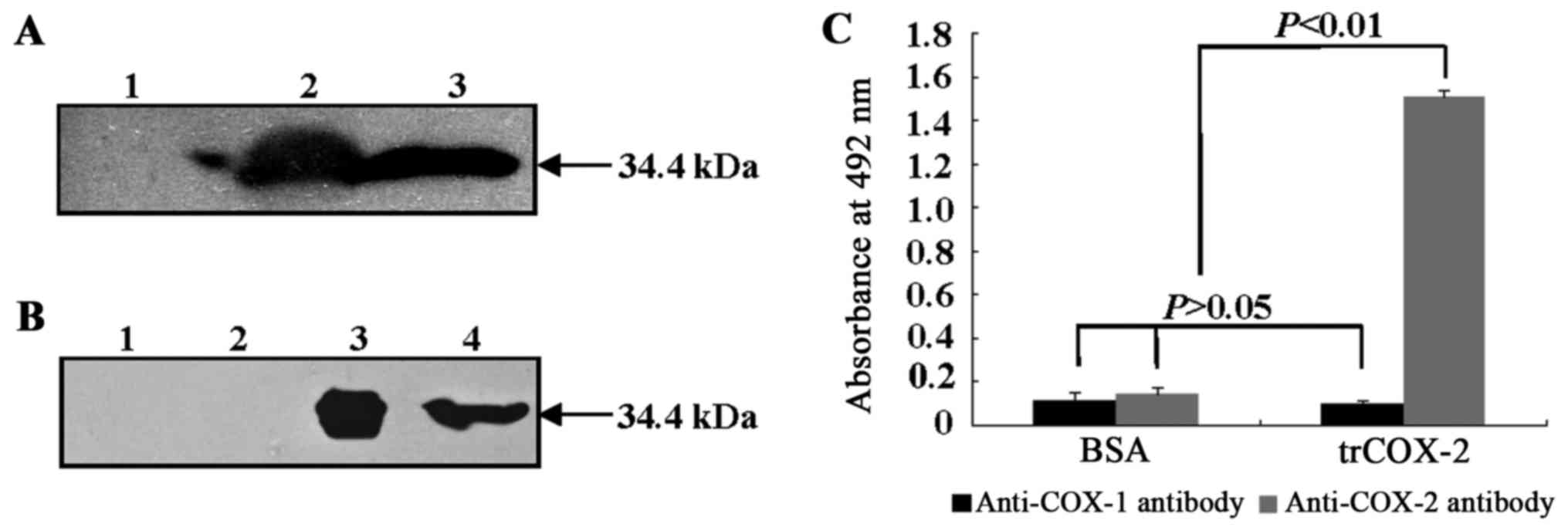
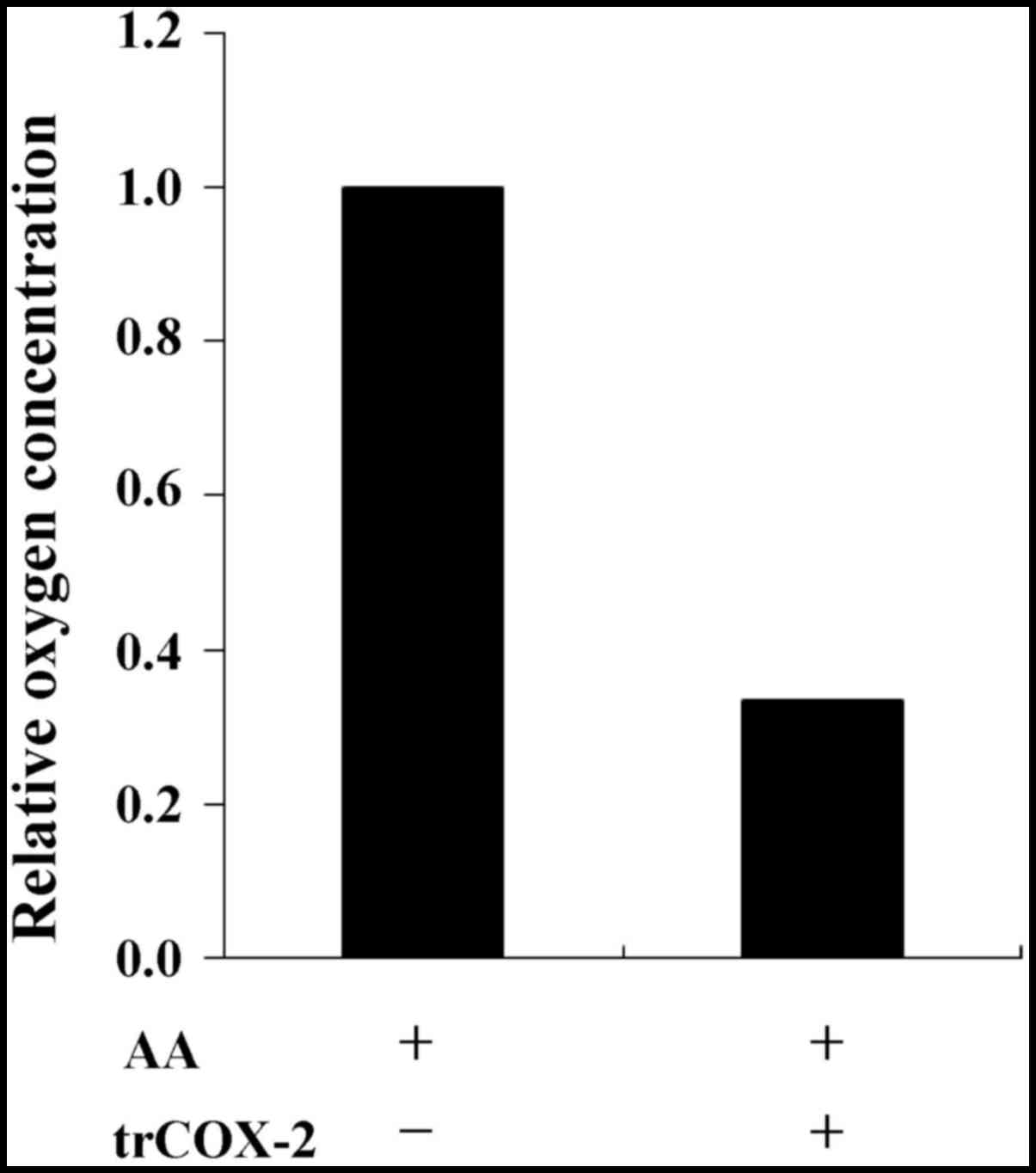
|
1
|
Chandrasekharan V and Simmons DL: The
cyclooxygenases. Genome Biol. 5:2412004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rowlinson SW, Crews BC, Lanzo CA and
Marnett LJ: The binding of arachidonic acid in the cyclooxygenase
active site of mouse prostaglandin endoperoxide synthase-2 (COX-2).
A putative L-shaped binding conformation utilizing the top channel
region. J Biol Chem. 274:23305–23310. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vecchio AJ and Malkowski MG: The
structural basis of endocannabinoid oxygenation by
cyclooxygenase-2. J Biol Chem. 286:20736–20745. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vecchio AJ, Orlando BJ, Nandagiri R and
Malkowski MG: Investigating substrate promiscuity in
cyclooxygenase-2: the role of Arg-120 and residues lining the
hydrophobic groove. J Biol Chem. 287:24619–24630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dong L, Vecchio AJ, Sharma NP, Jurban BJ,
Malkowski MG and Smith WL: Human cyclooxygenase-2 is a sequence
homodimer that functions as a conformational heterodimer. J Biol
Chem. 286:19035–19046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vecchio AJ, Simmons DM and Malkowski MG:
Structural basis of fatty acid substrate binding to
cyclooxygenase-2. J Biol Chem. 285:22152–22163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kudalkar SN, Nikas SP, Kingsley PJ, Xu S,
Galligan JJ, Rouzer CA, Banerjee S, Ji L, Eno MR, Makriyannis A, et
al: 13-Methylarachidonic acid is a positive allosteric modulator of
endocannabinoid oxygenation by cyclooxygenase. J Biol Chem.
290:7897–7909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tanabe T and Tohnai N: Cyclooxygenase
isozymes and their gene structures and expression. Prostaglandins
Other Lipid Mediat. 68–69:95–114. 2002. View Article : Google Scholar
|
|
9
|
Li G, Han C, Xu L, Lim K, Isse K and Wu T:
Cyclooxygenase-2 prevents Fas-induced liver injury through
up-regulation of epidermal growth factor receptor. Hepatology.
50:834–843. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghosh N, Chaki R, Mandal V and Mandal SC:
COX-2 as a target for cancer chemotherapy. Pharmacol Rep.
62:233–244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khan Z, Khan N, Tiwari RP, Sah NK, Prasad
GB and Bisen PS: Biology of Cox-2: an application in cancer
therapeutics. Curr Drug Targets. 12:1082–1093. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Misra S and Sharma K: COX-2 signaling and
cancer: new players in old arena. Curr Drug Targets. 15:347–359.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kurtova AV, Xiao J, Mo Q, Pazhanisamy S,
Krasnow R, Lerner SP, Chen F, Roh TT, Lay E, Ho PL, et al: Blocking
PGE2-induced tumour repopulation abrogates bladder
cancer chemoresistance. Nature. 517:209–213. 2015. View Article : Google Scholar
|
|
14
|
Yin J, Li G, Ren X and Herrler G: Select
what you need: a comparative evaluation of the advantages and
limitations of frequently used expression systems for foreign
genes. J Biotechnol. 127:335–347. 2007. View Article : Google Scholar
|
|
15
|
Wu Y, Zou D, Cao Y, Yao N, Wang J, Wang W,
Jiang H and Li G: Expression and purification of a human
anti-cyclin D1 single-chain variable fragment antibody AD5 and its
characterization. Int J Mol Med. 32:1451–1457. 2013.PubMed/NCBI
|
|
16
|
Percival MD, Bastien L, Griffin PR,
Kargman S, Ouellet M and O'Neill GP: Investigation of human
cyclooxygenase-2 glycosylation heterogeneity and protein expression
in insect and mammalian cell expression systems. Protein Expr
Purif. 9:388–398. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gierse JK: Purification of recombinant
human COX-1 and COX-2. Methods Mol Biol. 644:21–29. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gierse JK, McDonald JJ, Hauser SD,
Rangwala SH, Koboldt CM and Seibert K: A single amino acid
difference between cyclooxygenase-1 (COX-1) and -2 (COX-2) reverses
the selectivity of COX-2 specific inhibitors. J Biol Chem.
271:15810–15814. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cromlish WA, Payette P, Culp SA, Ouellet
M, Percival MD and Kennedy BP: High-level expression of active
human cyclooxygenase-2 in insect cells. Arch Biochem Biophys.
314:193–199. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gul N, Linares DM, Ho FY and Poolman B:
Evolved Escherichia coli strains for amplified, functional
expression of membrane proteins. J Mol Biol. 426:136–149. 2014.
View Article : Google Scholar
|
|
21
|
Liu B, Li G, Sui X, Yin J, Wang H and Ren
X: Expression and functional analysis of porcine aminopeptidase N
produced in prokaryotic expression system. J Biotechnol. 141:91–96.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arbabi-Ghahroudi M, Tanha J and MacKenzie
R: Prokaryotic expression of antibodies. Cancer Metastasis Rev.
24:501–519. 2005. View Article : Google Scholar
|
|
23
|
Sheibani N: Prokaryotic gene fusion
expression systems and their use in structural and functional
studies of proteins. Prep Biochem Biotechnol. 29:77–90. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim CS and Lee EK: Effects of operating
parameters in in vitro renaturation of a fusion protein of human
growth hormone and glutathione S transferase from inclusion body.
Process Biochem. 36:111–117. 2000. View Article : Google Scholar
|
|
25
|
Li GY, Zou DS, Zhou LH and Cao YH:
Expression and purification of recombinant human cyclin D1 in E.
coli BL21. J Jilin Univ. 44:839–843. 2006.
|
|
26
|
Cao YH, Xu JJ, Chen Y, Wang Q, Feng J, Hao
DY and Li GY: Prokaryotic expression, purification and renaturation
of recombinant human CDK4. J Jilin Univ. 46:992–996. 2008.
|
|
27
|
Xiang Y, Wang HY, Lei W and Sun M: The
expression and purification of COX-2 in Escherichia coli. J
Southwest Univ. 12:121–125. 2008. View Article : Google Scholar
|
|
28
|
Blobaum AL, Xu S, Rowlinson SW, Duggan KC,
Banerjee S, Kudalkar SN, Birmingham WR, Ghebreselasie K and Marnett
LJ: Action at a distance: mutations of peripheral residues
transform rapid reversible inhibitors to slow, tight binders of
cyclooxygenase-2. J Biol Chem. 290:12793–12803. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arnold K, Bordoli L, Kopp J and Schwede T:
The SWISS-MODEL workspace: a web-based environment for protein
structure homology modelling. Bioinformatics. 22:195–201. 2006.
View Article : Google Scholar
|
|
30
|
Guex N, Peitsch MC and Schwede T:
Automated comparative protein structure modeling with SWISS-MODEL
and Swiss-PdbViewer: a historical perspective. Electrophoresis.
30(Suppl 1): S162–S173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kiefer F, Arnold K, Künzli M, Bordoli L
and Schwede T: The SWISS-MODEL repository and associated resources.
Nucleic Acids Res. 37(Database): D387–D392. 2009. View Article : Google Scholar :
|
|
32
|
Biasini M, Bienert S, Waterhouse A, Arnold
K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M,
Bordoli L, et al: SWISS-MODEL: modelling protein tertiary and
quaternary structure using evolutionary information. Nucleic Acids
Res. 42:W252–W258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Grosdidier A, Zoete V and Michielin O:
Fast docking using the CHARMM force field with EADock DSS. J Comput
Chem. 32:2149–2159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grosdidier A, Zoete V and Michielin O:
SwissDock, a protein-small molecule docking web service based on
EADock DSS. Nucleic Acids Res. 39:W270–W277. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nandana V, Singh S, Singh AN and Dubey VK:
Procerain B, a cysteine protease from Calotropis procera, requires
N-terminus pro-region for activity: cDNA cloning and expression
with pro-sequence. Protein Expr Purif. 103:16–22. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Doray B, Chen CD and Kemper B: N-terminal
deletions and His-tag fusions dramatically affect expression of
cytochrome 450 2C2 in bacteria. Arch Biochem Biophys. 393:143–153.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Housaindokhta MR, Bozorgmehr MR, Hosseini
HE, Jalal R, Asoodeh A, Saberi M, Haratipour Z and Monhemi H:
Structural properties of the truncated and wild types of
Taka-amylase: a molecular dynamics simulation and docking study. J
Mol Catal B Enzym. 95:36–40. 2013. View Article : Google Scholar
|
|
38
|
Wilkins MR, Gasteiger E, Bairoch A,
Sanchez JC, Williams KL, Appel RD and Hochstrasser DF: Protein
identification and analysis tools in the ExPASy server. Methods Mol
Biol. 112:531–552. 1999.PubMed/NCBI
|
|
39
|
Kim SF, Huri DA and Snyder SH: Inducible
nitric oxide synthase binds, S-nitrosylates, and activates
cyclooxygenase-2. Science. 310:1966–1970. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gierse JK, Hauser SD, Creely DP, Koboldt
C, Rangwala SH, Isakson PC and Seibert K: Expression and selective
inhibition of the constitutive and inducible forms of human
cyclooxygenase. Biochem J. 305:479–484. 1995. View Article : Google Scholar
|