Introduction
Breast cancer is the most frequently diagnosed
cancer in both more and less economically developed countries and
remains the leading cause of cancer-related death in women
worldwide (1). Although the
breast cancer incidence and death rate have been greatly reduced in
developed countries such as the US and the UK due to early
detection and improved treatment, its incidence and mortality rate
in South America, Africa and Asia have been rising possibly due to
the reproductive pattern change, increased obesity or lack of
physical activity and delayed application of early screening
(1). According to the latest data
released by the National Cancer Center of China, the crude
mortality rate of female breast cancer in registration areas was
10.24/100,000 in 2009, accounting for 54% of all female cancer
mortality (2). To provide early
detection and more effective therapy for breast cancer,
comprehensive understanding of the molecular mechanisms mediating
breast cell transformation is crucial.
It has been shown that X-linked genes are involved
in the development of breast cancer (3). Among these, the X-linked ribosomal
S6 kinase RPS6KA6 (RSK4) was first identified as a commonly deleted
gene in complex X-linked mental retardation patients (4) and its ubiquitous expression in a
number of cell types suggested its involvement in multiple
biological and pathological processes such as murine embryogenesis,
fibroblast growth and diabetes development (5–8).
RSK4 expression was found to be downregulated in several types of
tumors such as colon carcinomas, renal cell carcinomas, colorectal
adenomas, endometrial cancer and ovarian cancer (9–11).
Furthermore, overexpression of RSK4 was found to induce cell cycle
arrest and senescence features while RSK4 inhibition resulted in
bypass of stress or oncogene-induced senescence, suggesting its
function as a potential tumor-suppressor gene (9,12).
More importantly, the expression of RSK4 was also found to be
aberrantly regulated in breast cancer cells (3). In a recent study of breast cancer
patient specimens, RSK4 expression was found to be significantly
decreased in breast cancer tissues in comparison with non-cancerous
tissues (13). Further
investigation also showed that the downregulation of RSK4
expression in breast cancer cells is mediated by gene expression
inhibition through promoter hypermethylation, which frequently
occurs in breast cancer cells (13). Consistently, RSK4 knockdown in
human breast adenocarcinoma cells significantly promoted
proliferation and migration, which further confirms the role of
RSK4 in the development and progression of breast cancer (14).
In human breast adenocarcinoma cells with silenced
RSK4 gene expression, the mRNA expression levels of RSK4 and
E-cadherin were significantly downregulated while the expression of
Ki-67, cyclin D1 and CXCR4 was markedly increased. These results
suggest that these genes are in the downstream signaling during
RSK4-regulated cell fate decision (14). The 90-kDa ribosomal S6 kinases
(RSKs) belong to serine kinases that are activated by growth
factors through the Ras-dependent mitogen-activated protein (MAP)
kinase cascade including MEK and extracellular signal-regulated
kinase (ERK) (7). Previous
studies have also shown that RSK4 is one of the ERK substrates and
functions as a target of the ERK signaling pathway (7,15).
Moreover, it has also been demonstrated that RSK4 inhibits the
transcriptional activation of specific targets of RTK signaling, as
well as the activation of ERK in mice, and RSK4 expression
inversely correlates with the presence of activated ERK 1/2
(6). Although the expression and
function of RSK4 in tumorigenesis have been confirmed by several
studies, its expression during breast cancer development and
potential application as a breast cancer biomarker need further
validation.
In this study, we showed that RSK4 expression was
significantly suppressed in cell lines (MCF-7, T47D, ZR-75-1,
MDA-MB-436, SK-BR-3 and MDA-MB-231) and breast cancer tissues. In
addition, RSK4 overexpression in MDA-MB-231 cells effectively
inhibited proliferation, migration and invasion. Moreover,
overexpression of RSK4 in a breast cancer mouse model significantly
suppressed breast cancer progression. Furthermore, we also
demonstrated that the AKT and ERK signaling pathways and epithelial
mesenchymal transition (EMT) were involved in the RSK4-regulated
breast cancer development, and the reduced expression of RSK4 in
MDA-MB-231 cells may be attributed to DNA methylation in its
promoter region. These results indicate that RSK4 may function as a
valuable biomarker for the study of breast cancer progression and
may have translational application for the early detection and
novel therapy of breast cancer.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of Guangxi Medical University, and we received informed
consent forms from all patients.
Cell lines and tissues
MCF-10A (non-malignant) and MCF-7, T47D, ZR-75-1,
MDA-MB-436, SK-BR-3 and MDA-MB-231 human breast cancer cell lines
were kindly provided by the Affiliated Tumor Hospital of Guangxi
Medical University. The non-cancerous matched tissue and cancerous
tissue samples were obtained from 8 patients with breast cancer
after breast-reductive surgery in the Affiliated Tumor Hospital of
Guangxi Medical University, and detailed information (including
age, diagnosis, TNM, stage and histological grade) on the 8
patients is shown in Table I.
 | Table IDetailed information of the eight
breast cancer patients. |
Table I
Detailed information of the eight
breast cancer patients.
| No. | Age (years) | Diagnosis | TNM | Stage | Histological
grade |
|---|
| 1 | 52 | Invasive ductal
carcinoma | T1N1M0 | I | I |
| 2 | 54 | Invasive ductal
carcinoma | T1N3M0 | I | I |
| 3 | 72 | Invasive ductal
carcinoma | T2N1M1 | II | II |
| 4 | 63 | Carcinosarcoma | T2N2M1 | IV | III |
| 5 | 73 | Invasive ductal
carcinoma | T1N0M0 | I | I |
| 6 | 31 | Invasive ductal
carcinoma | T2N1M0 | II | I |
| 7 | 58 | Invasive ductal
carcinoma | T2N1M1 | III | II |
| 8 | 60 | Invasive ductal
carcinoma | T1N1M0 | I | I |
Gel-based RT-PCR and real-time
RT-PCR
Firstly, we detected the mRNA expression level in
cell lines and four tissue samples (normal and cancerous) from 8
patients with breast cancer. Total RNA of the cells and tissue
samples was extracted using TRIzol reagent according to the
manufacturer's instructions (Thermo Fisher Scientific, Carlsbad,
CA, USA). Firstly, total RNA clean up was performed with the RNeasy
Mini kit (Qiagen, Inc., Valencia, CA, USA), then 210 µl of
100% EtOH and 50 µl of RNase/DNase-free water were added to
40 µl of the RNA sample followed by the addition of buffer
RLT containing 1% β-mercaptoethanol. Prior to RT-PCR, per sample, 5
µl of the RNA sample prepared above was diluted 100-fold
using RNase-free TE buffer (5 µl of RNA + 95 µl of
RNase-free TE buffer; Takara Bio, Inc., Otsu, Japan) in a 96-well
plate. The reaction system of RT-PCR was conducted in a final
volume of 20 µl containing 8 ng of RNA (2 ng/µl) and
12 µl of 1× concentration of Bio-Rad iScript Onestep RT-PCR
mix with SYBR-Green (Bio-Rad, Berkeley, CA, USA) and 4 µl of
primer mix. The RT-PCR reaction condition was 94°C for 2 min,
followed by 30 cycles of 94°C for 30 sec, 55°C for 1 min and 68°C
for 2 min and a final extension cycle was carried out at 68°C for 5
min. The GAPDH gene was used as an internal control. The RT-PCR
product was separated on 2% agarose gels and the gel image was
analyzed and quantified using Image-Quant software version 5.2
(Molecular Dynamics, Sunnyvale, CA, USA). Secondly, after
measurement of the RNA concentration and reverse transcription,
mRNA expression levels were determined by real-time RT-PCR using an
ABI PRISM 7900HT sequence detection system (Applied Biosystems Life
Technologies, Foster City, CA, USA). Three independent biological
repeats were performed and the GAPDH gene was used as an internal
control. The primers used for RSK4 gene and GAPDH were RSK4 F,
5′-TGAGTGGTGGAAACTGGGACA ATA-3′ and R,
5′-TGGCATGGACTGTGGTCATGAGTC-3′; GAPDH F, 5′-GCACCGTCAAGGCTGAGAAC-3′
and R, 5′-TGGTGAAGACGCCAGTGGA-3′. First-strand cDNA was reversely
transcribed using Primerscript II 1st Strand cDNA Synthesis kit
(no. 6210A; Takara Bio, Inc.). The Takara kit (no. RR390A; Takara
Bio, Inc.) was used for real-time RT PCR, which was conducted in a
20 µl reaction system with 10 µl 2× mix buffer,
forward and reverse primers (0.4 µl each), 1 µl cDNA,
and 15.4 µl double distilled water. The real-time RT-PCR was
performed using the following conditions: 94°C for 5 min, 30 cycles
of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and
extension at 72°C for 40 sec. The 2−ΔΔCq method was
applied for quantification of the relative expression of target
genes.
Western blot analysis
For protein extraction, cultured breast cancer cell
lines were first lysed in cell lysis buffer (140 mM NaCl, 10 mM
Tris-HCl, 1% Triton X-100, 1 mM EDTA, 1× protease inhibitor
cocktail). For protein extraction in breast tissues, collected
breast tissues were first homogenized in liquid nitrogen, and then
treated with the same cell lysis buffer. After determination of
protein concentrations by the BCA method, protein samples were
separated by SDS-PAGE and transferred to polyvinylidene difluoride
(PVDF) membranes (Millipore Corp., Billerica, MA, USA), followed by
blocking with 5% bovine serum albumin (BSA) for 2 h, and incubation
with the primary antibody for 1 h at room temperature, including
anti-phosphorylated (p)-AKT (cst4051, 1:200), anti-AKT (cst9272,
1:1,800), p-ERK (cst9101, 1:1,000), ERK (cst9102, 1:1,000),
anti-E-cadherin (cst9961, 1:1,000) and anti-vimentin (cst3932,
1:500) (Cell Signaling Technology, Inc., Danvers, MA, USA). After
being washed with TBST for 3 times, the membranes were then
incubated with peroxidase-conjugated secondary antibodies for 1 h.
Band intensity was determined used chemiluminescent reagents
(Pierce; Thermo Fisher Scientific, Waltham, MA, USA) and analyzed
by ImageJ software v1.48 (National Institutes of Health, Bethesda,
MD, USA).
Construction of stably overexpressing
RSK4 in MDA-MB-231 cells
We constructed the lentiviral expression vector of
the RSK4 gene and the MDA-MB-231 cell line which stably
overexpressed the RSK4 gene, following the procedure as previously
described by Roy and Jacobson (16). The RSK4 gene was synthesized by
the gene synthesis method and amplified by PCR, and then cloned
into the pLVEF-1a/GFP-Puro vector to construct the
pLVEF-1a/GFP-Puro-RSK4 lentiviral vector. After restriction enzyme
analysis and sequence identification, the lentiviral vector was
packaged and the titer was detected. The human breast cancer
MDA-MB-231 cells for transfection were cultured in DMEM media
containing 10% FBS, sodium pyruvate and antibiotics. These cells
were transfected with the recombinant lentiviral vector and
cultured selectively by puromycin to acquire stably transfected
cells. MDA-MB-231 cells which expressed GFP were observed by
fluorescence microcopy. After 48 h of transfection, the expression
levels of RSK4 mRNA and protein in the transfected MDA-MB-231 cells
were detected by real-time quantitative PCR and western blot
analysis, respectively. MDA-MB-231 cells (the untreated group,
control), MDA-MB-231 cells transfected with blank vectors (the
vector group) and MDA-MB-231 cells overexpressing RSK4 proteins
(the MDA-MB-231-RSK4 group) were used for further analysis, and
MDA-MB-231 cells transfected with blank pEGFP vectors were applied
as control.
Colony formation assay
For analysis of cell proliferation potential, the
untreated group, vector group and MDA-MB-231-RSK4 cells were
separately seeded at 500 cells/6-cm dish in complete medium. After
growing in the dish for 3 weeks, three groups of cells were fixed
and visualized by staining with 0.1% crystal violet (w/v) in 20 nM
4-morpholinepropanesulfonic acid. Visible colonies of each group
were compared by counting the number of cells in each colony and
three biological replicates were performed.
Cell migration and invasion assays
Breast cancer cell migration and invasion assays
were performed as previously described (17). Briefly, the cell migration assay
was carried out in Boyden chambers without coating of Matrigel.
Migrated breast cancer cells were stained and counted. Cell
invasion analysis was performed in a Boyden chamber with
polyethylene terephthalate filter inserts. Three groups of breast
cancer cells were plated in BSA-DMEM and inserted into the upper
chamber, and the lower chamber was filled with FCS-DMEM. The
invaded cells were then treated with fixation and staining solution
containing 0.1% crystal violet, 1% formalin, and 20% ethanol.
Invaded breast cancer cells were stained and counted.
DNA methylation sequencing
For prediction of DNA methylation in the RSK4 gene,
the DNA methylation in the RSK4 gene was analyzed by using
bisulfite direct sequencing assay, and for primers specific to
bisulfite-treated DNA, the DNA region of the RSK4 gene was analyzed
by using the free web program MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi).
MethPrimer determines the presence and location of CpG islands
within the RSK4 DNA sequence. Direct bisulfite sequencing for
analysis of DNA methylation levels is a technique that was
developed in order to determine CpG site-specific changes.
Determination of DNA methylation changes is fundamental to
understanding control of RSK4 expression within breast cancer cells
and how these changes affect tumorigenesis. Prior to bisulfite
conversion and sequencing, the total DNA samples were extracted
from MDA-MB-231 cells and the quality of extracted DNA was assessed
using the Qubit Platform (Invitrogen Life Technologies, Carlsbad,
CA, USA). By treating DNA samples with bisulfite, the cytosine
residues were converted to uracil while 5-methylcytosine residues
remained unaffected, thus methylated and unmethylated residues in
the RSK4 promoter regions were recognized by sequencing. Following
bisulfite treatment, PCR amplification was performed with
bisulfite-compatible primers which target a specific genomic
region. During the PCR amplification, cytosine bases which are
converted to uracil during bisulfite treatment are converted to
thymine. Both forward and reverse primers were used at the
concentration of 10 µM. A mix of 10 µl of SYBR-Green
Supermix (Bio-Rad) was prepared with 8 µl of nuclease free
water, plus 0.5 µl each of the forward and reverse primers
and 1 µl of bisulfite-treated DNA was added to the mixture,
for a total of 20 µl of reaction system. The PCR reaction
conditions were as follows: 95°C for 5 min; 50 cycles of 95°C for 1
min, 60°C for 1 min and 72°C for 1 min; followed by a cycle of 72°C
for 1 min and termination at 4°C. The PCR products were purified
and 50 ng of the resulting products was prepared for sequencing.
The sequencing results were analyzed using ABI Sequence Scanner
(http://www.appliedbiosystems.com/absite/us/en/home.html).
The methylation levels for each CpG site within the DNA amplicon
were quantified by measuring the ratio between peak height values
of guanine (G) and adenine (A). In addition, to explore the effect
of methylation on RSK4 expression and metastasis of human breast
cancer, MDA-MB-231 cells were treated with the demethylation
reagent 5-Aza-2′-deoxycytidine (5-Aza-CdR). The methylation
sequencing was then performed and qPCR was used to detect the mRNA
expression of RSK4 in the MDA-MB-231 cells.
Nude mouse tumorigenicity assay
For analyzing the RSK4 involvement in tumorigenicity
in severe combined immunodeficient mice, the vector group and
MDA-MB-231-RSK4 cells were collected, and washed 3 times with PBS.
A total of 12 female athymic nude mice, 5–6 weeks old (18–22 g)
were purchased from Beijing Slac Laboratory Animal Co. Ltd.
(Beijing, China), and maintained in high efficiency particulate
air-filtered cages in a pathogen-free facility. Mice were fed
autoclaved diet with 14% protein and 3% fat. MDA-MB-231-RSK4 cells
(2×106) were injected into the mammary fat pad of 6
female nude mice. The vector group cells were injected into the
mammary fat pad of another 6 female nude mice. Mice infected with
the breast cancer cells were kept in laminar flow animal rooms with
stable humidity and temperature. Estradiol supplementation (0.4
mg/kg) was given every week after cell injection. After injection,
tumor appearance in mice was inspected each day by observation and
palpation for 6 weeks, and tumor growth was measured weekly using a
digital caliper (Shanghai Jiang Lai Biotechnology Co., Ltd.,
Shanghai, China). At the end of 6 weeks, all mice were sacrificed
by cervical dislocation, and the necropsy was performed to check
the presence of tumor nodules.
Statistical analysis
SPSS software, version 19.0 (IBM SPSS, Armonk, NY,
USA) was used for the statistical analysis. The results are
presented as mean ± standard deviation values. A paired t-test was
used to analyze the differences between tumor and adjacent
non-tumor tissues. One way analysis of variance was used to analyze
differences between more than three groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
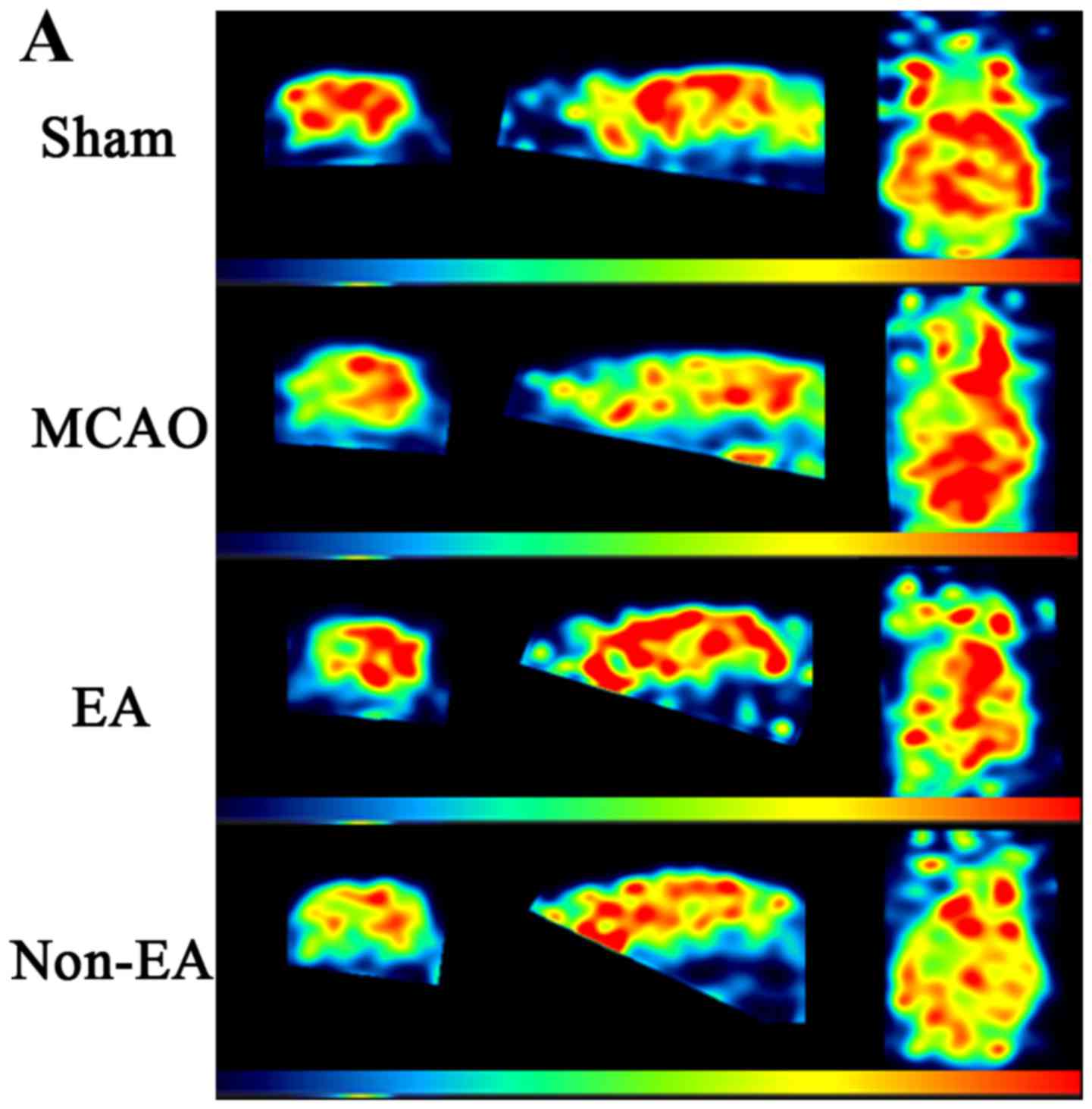
RSK4 expression in breast cancer
tissues
To investigate the expression of RSK4 in tissues,
breast cancer tissues and adjacent benign tissues from 8 breast
cancer patients were collected and subjected to mRNA and protein
level measurement. By real-time PCR and RT-PCR, we found that the
mRNA level of the RSK4 gene in cancerous breast tissues was
significantly decreased in all 8 patients compared with the
corresponding adjacent benign tissues (P<0.05; Fig. 1A and B). Western blot analysis was
performed to determine the RSK4 protein levels between breast
cancer tissues and adjacent benign tissues of the same patients,
and GAPDH was used as an internal control. Consistent with the mRNA
levels, the RSK4 protein levels in breast cancer tissues were also
significantly lower than these levels in the adjacent benign
tissues (Fig. 1C). The expression
level results clearly showed that the expression of RSK4 was
markedly inhibited in the breast cancer tissues.
RSK4 expression in breast cancer cell
lines
For further confirmation of RSK4 expression in
breast cancer, we measured the mRNA and protein levels of RSK4 in 6
breast cancer cell lines, MCF-7, T47D, ZR-75-1, MDA-MB-436, SK-BR-3
and MDA-MB-231, and one normal human epithelial mammary cell line
MCF-10A. As shown in Fig. 2A, the
protein levels of RSK4 in all 6 breast cancer cell lines were lower
than the level in the normal cell line. Among all 6 breast cancer
cell lines, the highly invasive human breast carcinoma cell line
MDA-MB-231 exhibited the most significant decrease in the RSK4
protein level compared with other 5 breast cancer cell lines,
indicating that the expression level of RSK4 may possess a certain
correlation with breast tumor progression (Fig. 2A). Consistence with the protein
level, the result of qPCR also revealed a similar decrease in the
RSK4 mRNA expression in these breast cancer cell lines, compared
with the normal mammary cell line MCF-10A (P<0.05, P<0.01;
Fig. 2B).
Overexpression of RSK4 in MDA-MB-231
cells
To explore the pathological role of RSK4 protein in
breast cancer carcinogenesis and the underlying mechanisms,
MDA-MB-231 cells with overexpression of RSK4 (MDA-MB-231-RSK4) were
prepared using a lentivirus expression system. Untreated MDA-MB-231
cells (untreated) and MDA-MB-231 cells transfected with blank
vectors (vector) were used as controls. The overexpression of RSK4
mRNA and protein was confirmed by western blot analysis (Fig. 3A) and real-time quantitative PCR
(Fig. 3B), respectively. The
MDA-MB-231-RSK4 cells showed high expression of RSK4 mRNA and
protein in comparison with the untreated MDA-MB-231 cells or blank
vector-transfected cells (P<0.05), showing that RSK4 protein was
successfully overexpressed in the MDA-MB-231 cells. Then, we
revealed that the proliferation of MDA-MB-231-RSK4 cells was
significantly inhibited by overexpression of RSK4, as indicated by
the reduced colony number in the MDA-MB-231-RSK4 cells by colony
formation assay (P<0.05; Fig. 3C
and D). Moreover, cell migration and invasion assays showed
that the migration and invasion potentials of the MDA-MB-231-RSK4
cells were also markedly lower than the two control groups
(P<0.05; Fig. 3E–G).
AKT and ERK signaling pathways and
EMT
As described before, previous studies have revealed
the involvement of RSK proteins in AKT and ERK signaling pathway
transduction, and RSK4 was also found to regulate ERK activation
(6,7,15).
To further explore the molecular pathways mediating the regulation
of breast cancer proliferation, migration and invasion by RSK4
protein, we analyzed the expression levels of several AKT and ERK
signaling pathway proteins AKT and ERK, as well as the
phosphorylation of AKT and ERK. Although the protein levels of AKT
and ERK were not significantly changed by overexpressing RSK4 in
the MDA-MB-231 cells, we found that the phosphorylated forms of AKT
and ERK proteins were greatly decreased in the MDA-MB-231-RSK4
cells, clearly showing that RSK4 overexpression in this breast
cancer cell line could inhibit the phosphorylation of major
components of AKT and ERK signalling pathway cascades (Fig. 4). Moreover, E-cadherin and
vimentin have been established as two important markers for EMT,
which is critical for cell migration and cancer invasion. Here, we
found that the expression level of E-cadherin was markedly
increased in the RSK4-overexpressing MDA-MB-231 cells, while the
expression level of vimentin was much lower than the two control
groups, indicating that the role of RSK4 protein in regulating
cancer metastasis may be mediated by EMT (Fig. 4).
Methylation of the RSK4 promoter in
breast cancer cells
To further analyze the molecular mechanisms
mediating RSK4 expression regulation in tumorigenesis, we analyzed
the methylation of the RSK4 promoter in MDA-MB-231 cells (control
cells) and 5-Aza-CdR-treated MDA-MB-231 cells (demethylated cells)
by sequencing. As shown in Fig.
5A, methylation was represented by black circles and white
circles represented an unmethylation status. The sequencing results
showed that a high methylation level of RSK4 was found in the
control cells, and the RSK4 methylation level in demethylated cells
was significantly lower than that in the control cells.
Consistently, the mRNA level of RSK4 in demethylated cells was
markedly higher than that in the control cells (P<0.05; Fig. 5B). In addition, the mRNA level of
RSK4 in the demethylated cells was increased by 4- to 5-fold
(P<0.05; Fig. 5B). Taking
together, these results showed that the RSK4 gene expression in
breast cancer cells was inhibited by DNA methylation in its
promoter area.
RSK4 expression in mouse
tumorigenicity
To further confirm the role of RSK4 in breast cancer
development, the vector group and MDA-MB-231-RSK4 cells were
applied for tumorigenicity assay in female athymic nude mice. The
total body weight of the mice in each group is shown in Fig. 6B. Compared with the mice infected
with the vector group, the tumors formed in the nude mice injected
with MDA-MB-231-RSK4 cells were significantly reduced in weight and
volume from week 3 to 6 (Fig. 6A, C
and D), and showed a time-dependent reduction in tumor growth.
This mouse model showed that overexpression of RSK4 in breast
cancer cells could significantly suppress breast
tumorigenicity.
Discussion
Breast cancer, usually characterized by a painful
lump, change in breast shape, nipple fluid or red scaly patch of
skin on the breast, is still the leading female cancer worldwide
(1). Women with breast cancer may
also suffer from bone pain, breath shortness, swollen lymph nodes
and yellow skin caused by distant metastasis of the breast cancer
(18). Although great progress
has been achieved in reducing breast cancer incidence and death
rate in developed countries, the need to intensively study its
pathological mechanisms and develop more applicable and affordable
early screening and therapy methods is crucial for developing
countries with limited medical resources (18).
Novel cancer biomarkers with high efficiency and
specificity have caused more and more attention in recent years,
considering their potential application in the early screening and
accurate diagnosis as well as personalized therapeutic strategy
design (19). The rational
application of molecular biomarkers may assure that breast cancer
patients are given optimal treatment. Successfully, several
molecular markers such as estrogen receptor, progesterone receptor,
human epidermal growth factor receptor 2, antigen Ki-67 and % S
phase cells, have been applied for classification of heterogeneous
breast cancers (19). Recent
publications have also revealed the potential of several other
molecules such as cyclin E, B-Myb, Twist, and DMP1β as novel
therapeutic targets for breast cancer patients (19). In the present study, we revealed
that RSK4 expression was greatly repressed in breast cancer cells
and tissues, and overexpression of RSK4 in breast cancer cells
inhibited cell migration. Moreover, a mouse model experiment by
overexpressing of RSK4 in breast cancer cells further confirmed its
critical role in regulating breast cancer progression. Combined
with other publications about RSK4 and breast cancer (3,13,14), recent evidence provides an
important research basis for the future application of RSK4 in
breast cancer screening, diagnosis and therapy, although large
prospective clinical studies should be performed to establish its
predictive value in patients with breast cancer.
Comprehensive understanding of breast cancer
progression and breast cell transformation may provide critical
insight for the development of reliable diagnostic and therapeutic
methods for breast cancer treatment. AKT and ERK signaling pathways
have long been established as central mediators in the development
of multiple cancers by regulating cancer cell growth, survival,
proliferation and migration (20,21). Raf/MEK/ERK and PI3K/PDK1/Akt
signaling pathways have been associated with breast cancer
development and it resistance to cytotoxic drugs (22). In view of the significant role of
the AKT and ERK signaling pathways in breast cancer, they have been
extensively applied for development of targeted therapy for breast
cancer patients, and multiple promising therapeutic agents
targeting the AKT and ERK signaling pathways are under evaluation
in clinical trials (23). In the
present study, we found that the involvement of RSK4 in breast
cancer progression is mediated by the AKT and ERK signaling
pathways, which provide meaningful insight into the pathological
mechanisms of breast cancer and provide more choices for the
development of new therapies. In addition, EMT is a cellular
program that is closely associated with lung tumorigenesis and the
majority of female cancer deaths are caused by cell invasion
(24). In the present study, we
found that the expression levels of E-cadherin and vimentin were
markedly altered in RSK4-overexpressing MDA-MB-231 cells, showing
that RSK4 protein may regulate EMT in breast cancer and may be used
as biomarkers for identification of breast tumor subsets with
greater risk occurrence.
Recent research has also pinpointed the increasing
roles of epigenetic processes such as DNA methylation in breast
cancer pathogenicity, prevention and treatment (25). For instance, the methylation of
adenomatous polyposis coli (APC), APC/C activator CDH1 and catenin
β-1 (CTNNB1) genes has been associated with breast cancer
development (26). Also, the
hypomethylation of Alu and LINE-1 promoter regions has been shown
to be linked with the HER-enriched breast cancer subtype (27). Furthermore, the BRCA1 gene is also
subjected to hypermethylation which regulates the gene expression
pattern during breast cancer development (28). Here, we found that the inhibition
of RSK4 expression in breast cancer progression was mediated by the
methylation in its promoter regions, attaching more importance to
the epigenetic mechanisms in breast cancer pathology which may lead
to improved diagnosis and treatment of this disease.
In conclusion, we revealed that RSK4 expression was
markedly suppressed in breast cancer cells and tissues, and
overexpression of RSK4 in breast cancer cells inhibited cell
migration and invasion. A mouse model experiment of overexpression
of RSK4 further confirmed its critical role in regulating breast
cancer progression. The involvement of RSK4 in breast cancer was
mediated by the AKT and ERK signaling pathways and the expression
of RSK4 was altered by DNA methylation in promoter regions. These
results provide important insight into the role of RSK4 in breast
cancer development and may help improve breast cancer prevention,
diagnosis and treatment.
Acknowledgments
The present study was funded by the Guangxi
Scientific and Technology Research Project Foundation
(14124004-1-12) and the National Natural Science Foundation of
China (NSFC81260394).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng H, Zheng R, Zhang S, Zou X and Chen
W: Incidence and mortality of female breast cancer in China, 2009.
Thorac Cancer. 4:400–404. 2013. View Article : Google Scholar
|
|
3
|
Thakur A, Rahman KW, Wu J, Bollig A,
Biliran H, Lin X, Nassar H, Grignon DJ, Sarkar FH and Liao JD:
Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in
breast cancer. Mol Cancer Res. 5:171–181. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yntema HG, van den Helm B, Kissing J, van
Duijnhoven G, Poppelaars F, Chelly J, Moraine C, Fryns JP, Hamel
BC, Heilbronner H, et al: A novel ribosomal S6-kinase (RSK4;
RPS6KA6) is commonly deleted in patients with complex X-linked
mental retardation. Genomics. 62:332–343. 1999. View Article : Google Scholar
|
|
5
|
Kohn M, Hameister H, Vogel M and
Kehrer-Sawatzki H: Expression pattern of the Rsk2, Rsk4 and Pdk1
genes during murine embryogenesis. Gene Expr Patterns. 3:173–177.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Myers AP, Corson LB, Rossant J and Baker
JC: Characterization of mouse Rsk4 as an inhibitor of fibroblast
growth factor-RAS-extracellular signal-regulated kinase signaling.
Mol Cell Biol. 24:4255–4266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dümmler BA, Hauge C, Silber J, Yntema HG,
Kruse LS, Kofoed B, Hemmings BA, Alessi DR and Frödin M: Functional
characterization of human RSK4, a new 90-kDa ribosomal S6 kinase,
reveals constitutive activation in most cell types. J Biol Chem.
280:13304–13314. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niehof M and Borlak J: RSK4 and PAK5 are
novel candidate genes in diabetic rat kidney and brain. Mol
Pharmacol. 67:604–611. 2005. View Article : Google Scholar
|
|
9
|
López-Vicente L, Armengol G, Pons B, Coch
L, Argelaguet E, Lleonart M, Hernández-Losa J, de Torres I and
Ramon y Cajal S: Regulation of replicative and stress-induced
senescence by RSK4, which is downregulated in human tumors. Clin
Cancer Res. 15:4546–4553. 2009. View Article : Google Scholar
|
|
10
|
Dewdney SB, Rimel BJ, Thaker PH, Thompson
DM Jr, Schmidt A, Huettner P, Mutch DG, Gao F and Goodfellow PJ:
Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6
(RSK4) in endometrial cancers. Clin Cancer Res. 17:2120–2129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arechavaleta-Velasco F, Zeferino-Toquero
M, Estrada-Moscoso I, Imani-Razavi FS, Olivares A, Perez-Juarez CE
and Diaz-Cueto L: Ribosomal S6 kinase 4 (RSK4) expression in
ovarian tumors and its regulation by antineoplastic drugs in
ovarian cancer cell lines. Med Oncol. 33:112016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
López-Vicente L, Pons B, Coch L, Teixidó
C, Hernández-Losa J and Armengol G: RSK4 inhibition results in
bypass of stress-induced and oncogene-induced senescence.
Carcinogenesis. 32:470–476. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Q, Jiang Y, Wei W, Ji Y, Gao H and Liu
J: Frequent epigenetic inactivation of RSK4 by promoter methylation
in cancerous and non-cancerous tissues of breast cancer. Med Oncol.
31:7932014. View Article : Google Scholar
|
|
14
|
Zhu J, Li QY, Liu JL, Wei W, Yang HW and
Tang W: RSK4 knockdown promotes proliferation, migration and
metastasis of human breast adenocarcinoma cells. Oncol Rep.
34:3156–3162. 2015.PubMed/NCBI
|
|
15
|
Frödin M and Gammeltoft S: Role and
regulation of 90 kDa ribosomal S6 kinase (RSK) in signal
transduction. Mol Cell Endocrinol. 151:65–77. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roy P and Jacobson K: Overexpression of
profilin reduces the migration of invasive breast cancer cells.
Cell Motil Cytoskeleton. 57:84–95. 2004. View Article : Google Scholar
|
|
17
|
Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD
and Wang LH: Twist transcriptionally up-regulates AKT2 in breast
cancer cells leading to increased migration, invasion, and
resistance to paclitaxel. Cancer Res. 67:1979–1987. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
DeSantis CE, Fedewa SA, Goding Sauer A,
Kramer JL, Smith RA and Jemal A: Breast cancer statistics, 2015:
Convergence of incidence rates between black and white women. CA
Cancer J Clin. 66:31–42. 2016. View Article : Google Scholar
|
|
19
|
Inoue K and Fry EA: Novel molecular
markers for breast cancer. Biomark Cancer. 8:25–42. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sever R and Brugge JS: Signal transduction
in cancer. Cold Spring Harb Perspect Med. Apr 1–2015.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu Z, Ye S, Hu G, Lv M, Tu Z, Zhou K and
Li Q: The RAF-MEK-ERK pathway: Targeting ERK to overcome obstacles
to effective cancer therapy. Future Med Chem. 7:269–289. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Navolanic PM, Steelman LS and McCubrey JA:
EGFR family signaling and its association with breast cancer
development and resistance to chemotherapy (Review). Int J Oncol.
22:237–252. 2003.PubMed/NCBI
|
|
23
|
Alvarez RH, Valero V and Hortobagyi GN:
Emerging targeted therapies for breast cancer. J Clin Oncol.
28:3366–3379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Felipe Lima J, Nofech-Mozes S, Bayani J
and Bartlett JM: EMT in breast carcinoma - A review. J Clin Med.
5:652016. View Article : Google Scholar
|
|
25
|
Basse C and Arock M: The increasing roles
of epigenetics in breast cancer: Implications for pathogenicity,
biomarkers, prevention and treatment. Int J Cancer. 137:2785–2794.
2015. View Article : Google Scholar
|
|
26
|
Hoque MO, Prencipe M, Poeta ML, Barbano R,
Valori VM, Copetti M, Gallo AP, Brait M, Maiello E, Apicella A, et
al: Changes in CpG islands promoter methylation patterns during
ductal breast carcinoma progression. Cancer Epidemiol Biomarkers
Prev. 18:2694–2700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park SY, Seo AN, Jung HY, Gwak JM, Jung N,
Cho NY and Kang GH: Alu and LINE-1 hypomethylation is associated
with HER2 enriched subtype of breast cancer. PLoS One.
9:e1004292014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hedenfalk I, Duggan D, Chen Y, Radmacher
M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M,
Kallioniemi OP, et al: Gene-expression profiles in hereditary
breast cancer. N Engl J Med. 344:539–548. 2001. View Article : Google Scholar : PubMed/NCBI
|