Introduction
Bronchopulmonary dysplasia (BPD), also known as
chronic lung disease (CLD), occurs primarily in premature infants.
BPD has a higher morbidity in premature infants with a lower birth
weight and fetal age (1). In
premature infants with a birth weight <1,250 g, almost 97% also
present with BPD (2). The major
pathological changes associated with BPD are alveolar
simplification, abnormal angiogenesis and various degrees of
pulmonary fibrosis (3), which
result from both injury and repair mechanisms. Alveolar
simplification directly influences the effective alveolar
ventilation area, as well as respiratory function. Alveolar type II
(AT2) epithelial cells play an important role in the formation of
alveoli during pulmonary development. AT2 cells can differentiate
into alveolar type I (AT1) epithelial cells and can promote the
formation of alveolar structure and the air-blood barrier,
promoting normal alveolar development (4). During the normal repair process that
occurs in response to pulmonary epithelial injury, AT2 cells
differentiate into AT1 cells and proliferate to rebuild the
air-blood barrier (5). The normal
differentiation of AT2 cells into AT1 cells is essential for normal
alveolar development and repair function. Previously (6), we provided evidence of
epithelial-mesenchymal transition (EMT) in AT2 cells isolated from
rat lung tissue from BPD model systems; we hypothesized that the
development of EMT may influence both normal development, as well
as the repair processes of alveoli. However, the mechanisms by
which EMT occurs in BPD remain poorly understood. In this study, we
explored the factors which regulate the development of EMT in AT2
cells in the context of BPD.
Runx3 is a runt domain transcription factor; its
expression is closely associated with several diseases and is also
a key regulator of gene expression during lung vasculogenesis
(7–9). The majority of recent studies on
Runx3 have focused on the role of the transcription factor in
cancer metastasis; these studies have found that low levels of
Runx3 can promote EMT (10),
contribute to cancer metastasis (11) and can even be used as an early
predictor of cancer development (12). Increased Runx3 expression in
cancer cells may inhibit the production of cancer stem cells and
reduce the occurrence of tumors (13). In addition, Runx3 exhibits a
spatiotemporal expression during embryonic development (14) and also plays a role in pulmonary
development. Lee et al (15) studied lung tissue from
Runx3−/− mice. They found that, compared with wild-type
mice, Runx3−/− mice exhibited abnormal alveolar
remodeling and alveolar dysplasia 1 day after birth; in addition,
they observed the co-expression of surfactant protein B (SPB), an
alveolar epithelial marker and Clara cell 10 kDa protein (CC10), a
bronchiolar epithelial marker in the bronchiolar epithelial cells
of Runx3−/− mice, indicating the decreased
differentiation of epithelial cells. Recently, Lee et al
(16) confirmed the presence of
abnormal EMT in the lungs of Runx3−/− mice.
BPD is a result of the disruption of the normal
pulmonary development process; Runx3 is an important factor in such
a process and can affect EMT. Our previous study (6) confirmed the presence of EMT in lung
tissues of newborn rats with BPD. In this study, we wished to
determine whether the decreased expression of Runx3 in BPD-affected
lung tissue promotes EMT in AT2 cells, and may thus influence the
normal pulmonary development process and contribute to the
occurrence of BPD. Therefore, we measured Runx3 expression in lung
tissues and isolated AT2 cells from the tissue of rats with BPD,
and observed the effects of abnormal Runx3 expression on the
occurrence of EMT in the RLE-6TN cell line in vitro. We also
analyzed the correlation between Runx3 protein expression in lung
tissues and pulmonary development indicators in vivo.
Materials and methods
Animal models and lung tissue
preparation
Animal models of BMD were established as previously
described (17). Pregnant Wistar
rats with a body weight of 200–220 g were purchased from the
Experimental Animal Center of China Medical University. Each
pregnant Wistar rat was fed independently and gave birth naturally
at 22 days of gestation. Newborn rats were randomly allocated to
either the BPD model group (n=80) or the control group (n=80)
within 12 h after birth. Newborn rats in the BPD model group were
placed in a plexiglass oxygen tank and subjected to 95% oxygen
inhalation for 1 to 21 days; an oxygen analyzer was used to
continuously monitor the oxygen concentration. The tank was opened
for 10 min and nursing rat dams were switched every 24 h between
the hyperoxia and normoxia tanks to avoid oxygen toxicity. Newborn
rats in the control group inhaled fresh air as opposed to
hyperbaric oxygen; however, all other experimental conditions and
control factors were the same as those for the BPD model group.
Eight animals were euthanized and lung tissue was collected at each
time-point (1, 3, 7, 14 and 21 days). The left lungs were fixed in
4% paraformaldehyde for hematoxylin and eosin (H&E) staining
and immunofluorescence assay. The right lungs were preserved in
liquid nitrogen for mRNA detection and western blot analysis. All
animal procedures were evaluated and approved by the Experimental
Animal Ethics Committee of China Medical University, Shenyang,
China.
AT2 cell isolation and purification
Another 8 animals were euthanized and AT2 cells were
isolated at each time-point (1, 3, 7, 14 and 21 days), respectively
in each group. AT2 epithelial cells were isolated from the newborn
rat lungs as previously described (6). The isolated AT2 cells were frozen at
−80°C for real-time PCR and western blot analysis.
RLE-6TN cultures and groups
RLE-6TN cells constitute a cell line derived from
rat AT2 cells that was purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA). The Runx3 expression plasmid
and the double-stranded siRNAs against rat Runx3 were synthesized
by Shanghai Genebank Co. Lipophilic transfection reagent
(Lipofectamine 2000; Invitrogen Life Technologies, Carlsbad, CA,
USA) was used. Human transforming growth factor-β1
(TGF-β1)-mammalian was purchased from PeproTech, Inc. (Rocky Hill,
NJ, USA).
The cells were grown on 6-well plates in Dulbecco's
modified Eagle's medium (DMEM), nutrient mixture F-12 Ham
supplemented with 10% fetal bovine serum (FBS), 40 mmol/l HEPES at
37°C in a humidified 5% CO2 atmosphere (18). The cells were divided into 5
groups according to the different interventional methods as
follows: the control group (untransfected RLE-6TN cell monolayer),
the Runx3 group (transfected with Runx3 overexpression plasmid for
72 h), the siRunx3 group (Runx3-deficient RLE-6TN cell monolayer
transfected with siRNA of Runx3 for 72 h), the TGF-β1 group (2.5
ng/ml TGF-β1 was used to treat the RLE-6TN cell monolayer followed
by culture for 48 h), the Runx3 + TGF-β1 group (transfected with
Runx3 overexpression plasmid for 24 h, and then 2.5 ng/ml TGF-β1
was used followed by culture for the next 48 h).
Generation of Runx3-overexpressing
RLE-6TN cells
The pFlag-control and pFlag-Runx3 expression
plasmids were both purchased from Genechem (Shanghai, China).
Transfection with the pFlag-control and the pFlag-Runx3 plasmids
into the RLE-6TN cells was carried out using Lipofectamine 2000
transfection reagent (Invitrogen Life Technologies) following the
manufacturer's instructions. At 72 h after transfection, western
blot analysis and real-time PCR were used, respectively for the
analysis of protein and mRNA analysis. For immunofluorescence
experiments, the cells were seeded on 22 mm round coverslips.
Silencing by small interfering RNA
(siRNA)
Logarithmically growing RLE-6TN cells were seeded at
a density of 105 cells and transfected with siRNA
against Runx3 using Lipofectamine (Invitrogen Life Technologies)
following the manufacturer's instructions. At 72 h after
transfection, lysates were prepared and analyzed by SDS-PAGE and
immunoblotting, anhd real-time PCR was used for the analysis of
mRNA expression. Immunofluorescence experiments were also carried
out.
Stimulation of RLE-6TN cells with
TGF-β1
Confluent cultures (70%) of RLE-6TN cells were
maintained in serum-free medium for 24 h. The cells were stimulated
with 2.5 ng/ml (100 pmol/l) TGF-β1 for the next 48 h. The collected
cells after stimulation were use for mRNA detection,
immunofluorescence and western blot analysis.
Runx3-overexpressing RLE-6TN cells
stimulated with TGF-β1
Transfection of the pFlag-Runx3 plasmids into the
RLE-6TN cells was carried out as mentioned above. Twenty-four hours
later, the cells were stimulated with 2.5 ng/ml TGF-β1 for the next
48 h. The collected cells were stored at −80°C for mRNA detection
and western blot analysis. For immunofluorescence experiments, the
cells were seeded on 22 mm round coverslips. For ultrastructural
analysis by transmission electron microscopy (TEM), the collected
cells from the 5 different groups were fixed in 2.5%
glutaraldehyde.
Validation of Runx3 plasmid transfection
and siRNA interference effects
In order to validate the effects of Runx3 plasmid
transfection and siRNA interference, we collected RLE-6TN cells at
48 and 72 h post-transfection and measured Runx3 protein and mRNA
expression using western blot analysis and real-time PCR.
Analysis of Runx3 in lung tissues and
isolated AT2 cells
Runx3 protein expression in lung tissues and AT2
cells was analyzed by western blot analysis. Total protein
extracted from the right lung tissue or isolated AT2 cells was
quantified using the BCA protein assay kit (Pierce Biotechnology.
Rockford, IL, USA). Equivalent amounts of protein were separated
using 10% SDS-PAGE gels and transferred onto polyvinylidene
difluoride (PVDF) membranes (Millipore Billerica, MA, USA). The
membranes were incubated for 1 h in 10% normal donkey serum to
block non-specific binding. Next, the membranes were incubated,
respectively with primary antibodies against Runx3 (1:1,000,
ab49117) and β-actin (1:1,000, mouse monoclonal [ACTN05 (C4)] to
actin; ab3280) (both from Abcam, Hong Kong, China) diluted in
phosphate-buffered saline (PBS) incubated overnight at 4°C. The
following day, after being washed in Tris-buffered saline with 1%
Tween-20 (TBST), the membranes were incubated for 2 h in
horseradish peroxidase-conjugated secondary antibodies (cat. nos.
31466 and 31430; Thermo Fisher Scientific, Waltham, MA, USA), then
imaged by enhanced chemiluminescence reagents. ImageJ software was
used to analyze the optical density of protein bands and then
normalized to that of β-actin.
Real-time PCR was used to examine Runx3 mRNA
expression. Total RNA was extracted from right lung lobes or
isolated AT2 cells using TRIzol reagent and frozen at −80°C
(Invitrogen Life Technologies, Camarillo, CA, USA) according to the
manufacturer's instructions. A total of 1 µg RNA from each
sample was reverse-transcribed into cDNA using SuperScript III
according the manufacturer's instructions (SuperScript III;
Invitrogen Life Technologies). Real-time PCR was performed on a
LightCycler (7500 Fast Real-Time PCR system; Applied Biosystems,
Foster City, CA, USA) using appropriate primers designed by Primer
Premier 5.0 software (Premier Biosoft International, Palo Alto, CA,
USA; Table I). The amplification
reaction was carried as follows: 95°C for 30 sec (95°C for 5 sec,
60°C for 34 sec) 40 cycles. The relative level of mRNA expression
was calculated following normalization to β-actin.
 | Table IPrimers design for real-time PCR. |
Table I
Primers design for real-time PCR.
| Names | Primer
sequences | Distribution | AT (°C) | Product size
(bp) | Extension time
(sec) |
|---|
| Runx3 | F:
5′-GCAACGCTTCCGCTGTCA-3′ | NM_130425 | 60 | 215 | 34 |
| R:
5′-GGCTTTGGTCTGGTCCTCTATC-3′ | 368–582 | | | |
| SPC | F:
5′-GCCAGTTTCGCATTCC-3′ | NM_017342 | 60 | 282 | 34 |
| R:
5′-GCTTATAGGCGGTCAGG-3′ | 76–357 | | | |
| α-SMA | F:
5′-CTTGCTAACGGAGGCG-3′ | BC158550 | 60 | 160 | 34 |
| R:
5′-TCCAGAGTCCAGCACAATA-3′ | 366–525 | | | |
| E-cad | F:
5′-ACAGTCAAACGGCATCTAA-3′ | NM_031334 | 60 | 190 | 34 |
| R:
5′-GGTGAAAGCTGGGAAAC-3′ | 388–577 | | | |
| N-cad | F:
5′-GACCCAGAAGATGATGTAAG-3′ | NM_031333.1 | 60 | 171 | 34 |
| R:
5′-CTCAGCGTGGATAGGC-3′ | 2617–2787 | | | |
| β-actin | R:
5′-CGTGCGTGACATTAAAGAG-3′ | NM_031144 | 60 | 132 | 34 |
| F:
5′-TTGCCGATAGTGATGACCT-3′ | 702–833 | | | |
Analysis of surfactant protein C (SPC),
α-smooth muscle actin (α-SMA), E-cadherin (E-cad) and N-cadherin
(N-cad) levels in RLE-6TN cells
The expression levels of proteins involved in EMT
(SPC, SMA, E-cad and N-cad) in RLE-6TN cells were analyzed by
western blot analysis as mentioned above. The antibody
concentrations were as follows: SPC (1:100, goat polyclonal
antibody; sc-7706; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA), α-SMA (1:500, mouse monoclonal to α-SMA; ab7817; Abcam),
E-cad (1:1,000, rabbit polyclonal to E-cad; GTX100443, GeneTex,
Irvine, CA, USA), N-cad (1:1,000, mouse monoclonal to N-cad;
ab98952; Abcam) or β-actin (1:1,000, mouse monoclonal [ACTN05 (C4)]
to actin; ab3280; Abcam). ImageJ software was used to analyze the
optical density of the protein bands and then normalized to that of
β-actin.
The mRNA expression of SPC, α-SMA, E-cad, N-cad in
the RLE-6TN cells was analyzed by real-time PCR as mentioned above.
The primers and amplification conditions are shown in Table I. The relative level of mRNA
expression was calculated following normalization to β-actin.
Double-label immunofluorescence assay for
RLE-6TN cells
The RLE-6TN cells were cultured, respectively on
glass slides in the 5 groups, fixed with 4% paraformaldehyde and
permeabilized with 0.5% Triton X-100, then washed 3 times with PBS.
The sections were subsequently incubated in 5% FBS for 30 min for
antigen blocking. The slides were then incubated with a mixture of
two primary antibodies [anti-SPC (1:100 diluted, goat poly-clonal
antibody; sc-7706; Santa Cruz Biotechnology, Inc.) and α-SMA (1:100
diluted, mouse monoclonal to α-SMA; ab7817; Abcam)] at −4°C
overnight. Normal goat IgG (sc-2028) or normal mouse IgG (sc-2025)
(both from Santa Cruz Biotechnology, Inc.) was used respectively in
place of the primary antibodies in the negative control samples.
The slides were then washed 3 times with PBS and then incubated
with a mixture of two secondary antibodies [anti-mouse antibody
(Alexa Fluor-488, green fluorescence) and anti-goat antibody (Alexa
Fluor-594, red fluorescence)] at 37°C for 60 min. Subsequently, the
sections were further washed with PBS 3 times and then subjected to
DAPI (1:2,000; Sigma-Aldrich, St. Louis, MO, USA) nuclear staining
for 5 min. After washing thoroughly with PBS, images of the
sections were acquired using a fluorescence microscope at x400
magnification.
TEM
The RLE-6TN cells collected, respectively from the 5
different groups were fixed in PBS solution (pH 7.4) containing
2.5% glutaraldehyde for 2 h, washed with PBS 3 times, post-fixed in
1% osmium tetroxide for 2 h, washed with distilled water 3 times,
dehydrated with gradient ethanol and acetone, and immersed in Epon
812 overnight; the lung tissue blocks were then embedded in Epon
812 and cut into ultrathin sections (60 nm). The ultrathin sections
were double-stained with uranyl acetate and aluminum citrate and
cellular morphology was observed under a transmission electron
microscope (Hitachi H-7650; Hitachi Co., Tokyo, Japan).
Pathology and morphometry
The left lung tissues were fixed in 4%
paraformaldehyde for 24 h, dehydrated with gradient ethanol,
vitrified with xylene, embedded in paraffin, cut into sections (5
µm) and stained with H&E (G1120; Solarbio Life Sciences,
Beijing, China). Morphological changes were assessed using an
optical microscope (x40 magnification). Ten fields were randomly
selected for analysis from each section to evaluate the development
of pulmonary alveoli. The radial alveolar count (RAC) developed by
Emery and Mithal (19) was
determined. Scion Image software (Scion Corp., Frederick, MD, USA)
was used to measure alveolar wall thickness (20). Immunofluorescence assay for α-SMA
in lung tissues was carried out according to our previously
described methods (6). The number
of branch points per millimeter squared and surface density of
secondary septa were calculated by computer-assisted image analysis
as previously described (21),
and the percentage of SMA-positive secondary septa was calculated.
Each section was evaluated by two independent pathologists who were
blinded to the experimental design.
Statistical analysis
We used SPSS 11.5 software (SPSS, Inc., Chicago, IL,
USA) for statistical analysis. Values are presented as the means ±
standard deviation (SD); an independent-sample t-test was used to
compare differences between 2 groups. One-way analysis of variance
(ANOVA) was used to determine the significant difference among
multiple groups. Pearson's test was applied for correlation
analyses. Statistical significance was assessed at P<0.05. A
P-value <0.05 was considered to indicate a statistically
significant difference.
Results
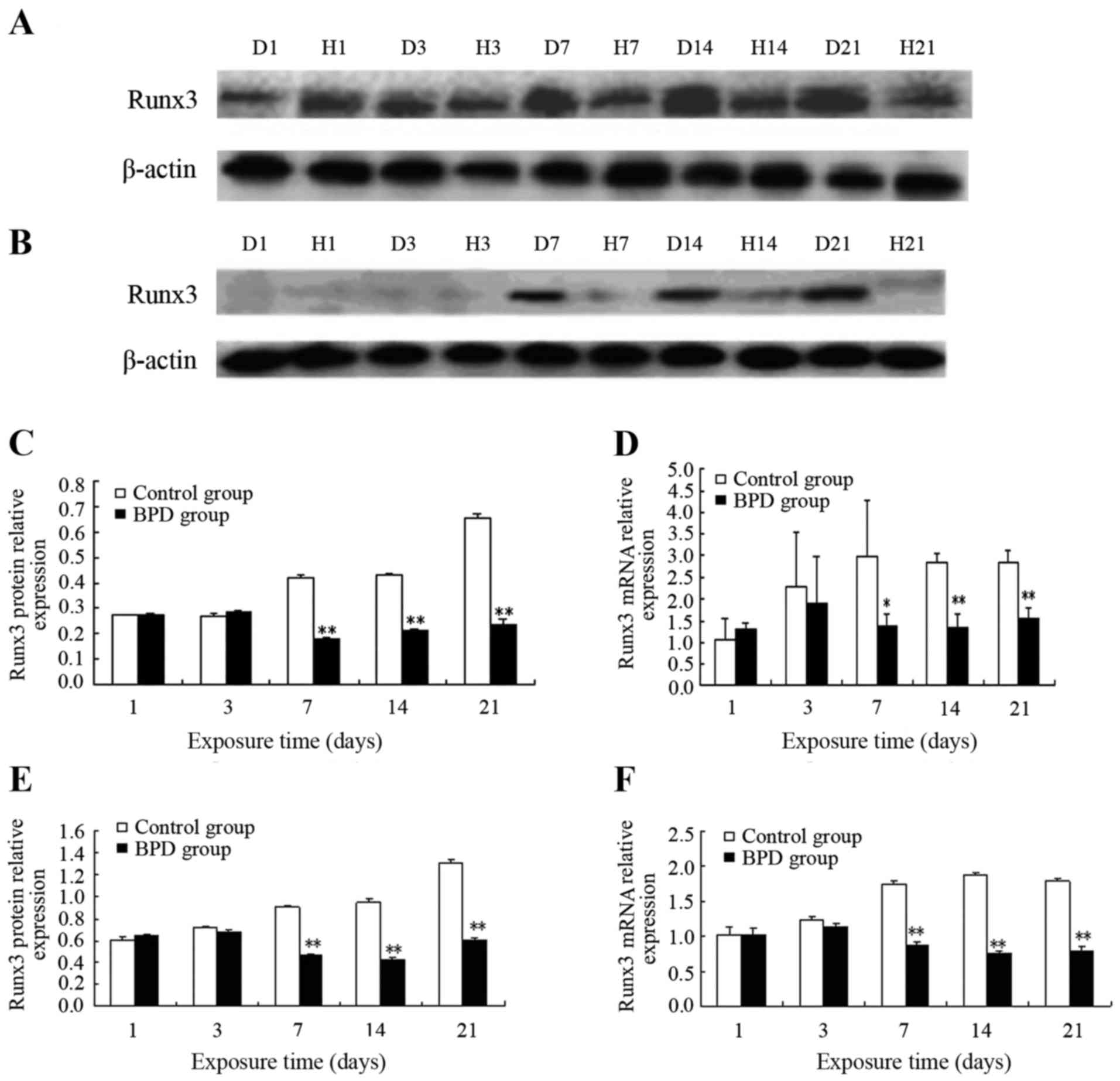
Protein and mRNA expression of Runx3 in
lung tissues and isolated AT2 cells
On days 1 and 3, the protein expression level of
Runx3 in lung tissue did not differ significantly between the 2
groups. On day 7, Runx3 protein expression in the control group
remained high, while Runx3 protein expression progressively
decreased in the BPD group. On days 7, 14 and 21, the protein
expression level of Runx3 in the control group was significantly
higher than that in the BPD group (P<0.01) (Fig. 1A and C). Similarly, no significant
difference in Runx3 mRNA expression was observed between the 2
groups on days 1 or 3, while the Runx3 mRNA expression level was
significantly higher in the control group than in the BPD group on
days 7, 14 and 21 (P<0.01) (Fig.
1D).
In the AT2 cells isolated from primary rats at
different time points, no significant differences in both Runx3
protein and mRNA expression were observed between the 2 groups on
days 1 and 3. On day 7 and at later time points, the Runx3 protein
expression level in the control group was markedly higher than that
in the BPD group (P<0.01) (Fig. 1B
and E). On days 7, 14 and 21, the Runx3 mRNA expression level
in the control group was significantly greater than that in the BPD
group (P<0.01) (Fig. 1F).
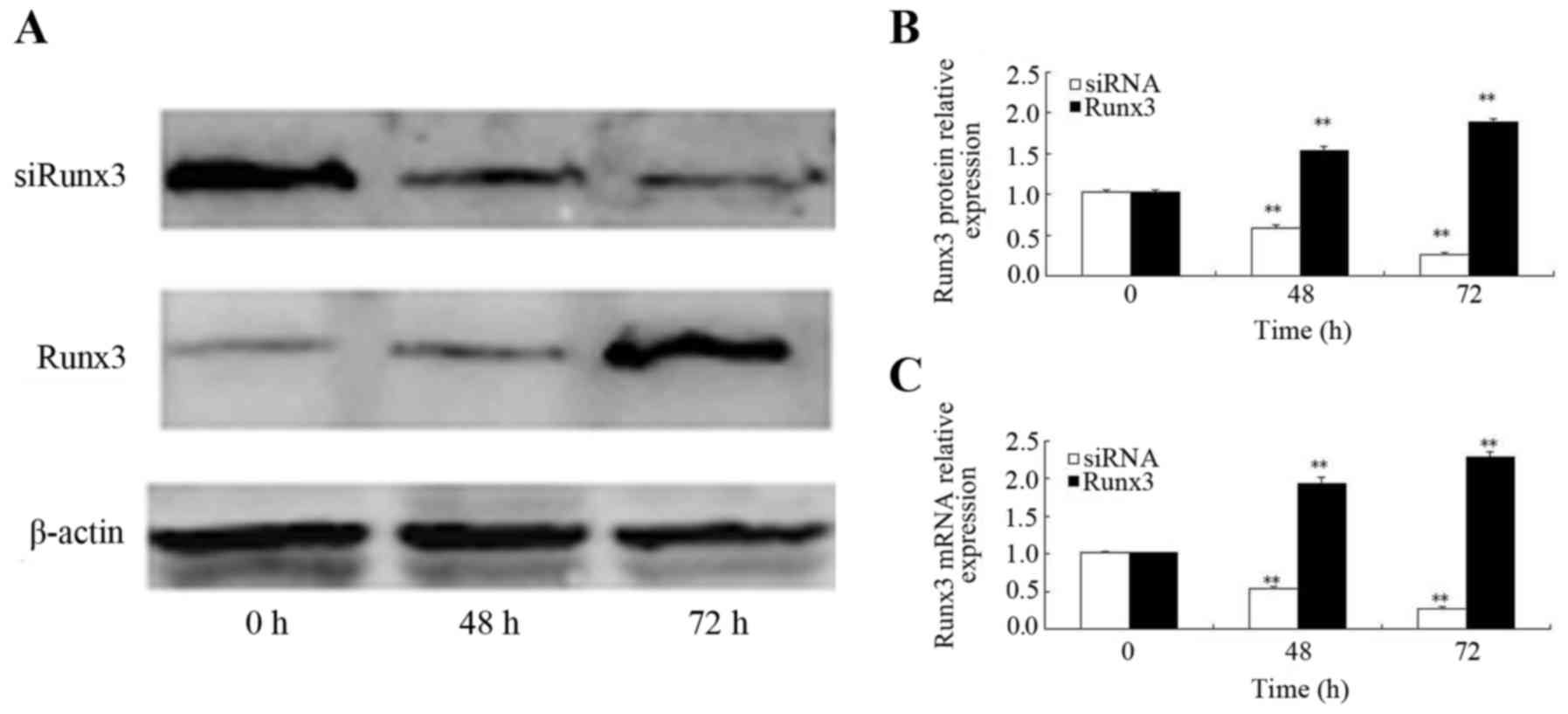
Effects of ectopic Runx3 expression and
siRNA interference
At 48 h following transfection with the Runx3
plasmid, the increased mRNA expression of Runx3 was observed; Runx3
mRNA expression was higher at both 48 and 72 h post-transfection as
compared to time 0 (P<0.01) (Fig.
2B). Similarly, the Runx3 protein expression level increased
beginning at 48 h post-transfection and it was also significantly
higher at 48 and 72 h post-transfection as compared to time 0
(P<0.01) (Fig. 2A and C). At
48 h following transfection of the cells with siRNA, Runx3 mRNA
expression was decreased; Runx3 mRNA expression 24, 48 and 72 h
post-siRNA transfection was lower than that at time 0 (P<0.01)
(Fig. 2B). Similarly, the Runx3
protein expression level began to decrease beginning at 48 h
post-siRNA transfection and it remained lower at 48 and 72 h
post-transfection than at time 0 (P<0.01) (Fig. 2A and C).
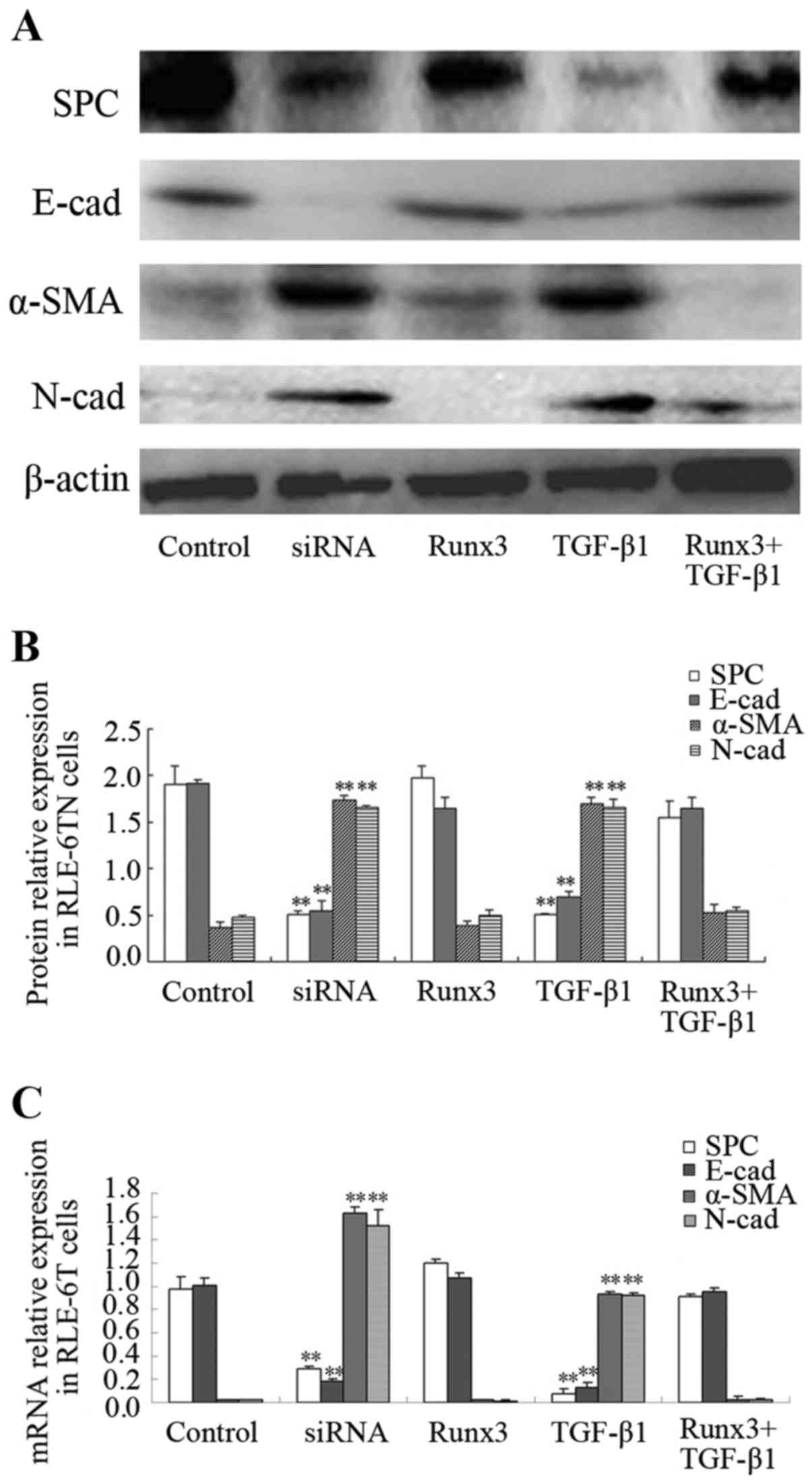
Expression of SPC, SMA, E-cad, N-cad in
RLE-6TN cells
The protein expression of SPC, α-SMA, E-cad and
N-cad in the 5 RLE-6TN cell groups was measured. The protein
expression levels of the mesenchymal cell markers, α-SMA and N-cad,
in the TGF-β1 and siRunx3 groups were significantly higher than
those in the control group (P<0.01; Fig. 3A and B), while the protein
expression levels of the epithelial cell markers, SPC and E-cad,
were significantly lower than those in the control group
(P<0.01; Fig. 3A and B). No
significant differences in either the epithelial or the mesenchymal
cell markers were found between the Runx3 overexpression group or
the Runx3 + TGF-β1 group and the control group (Fig. 3A and B). Similarly, the mRNA
expression levels of α-SMA and N-cad in the TGF-β1 group and
siRunx3 group were significantly higher than those in the control
group (P<0.01; Fig. 3C), while
the mRNA expression levels of SPC and E-cad were significantly
lower than those in control group (P<0.01; Fig. 3C). No significant differences in
either the epithelial cell markers or the mesenchymal cell markers
were found between the Runx3 overexpression group or the Runx3 +
TGF-β1 group and the control group (Fig. 3C).
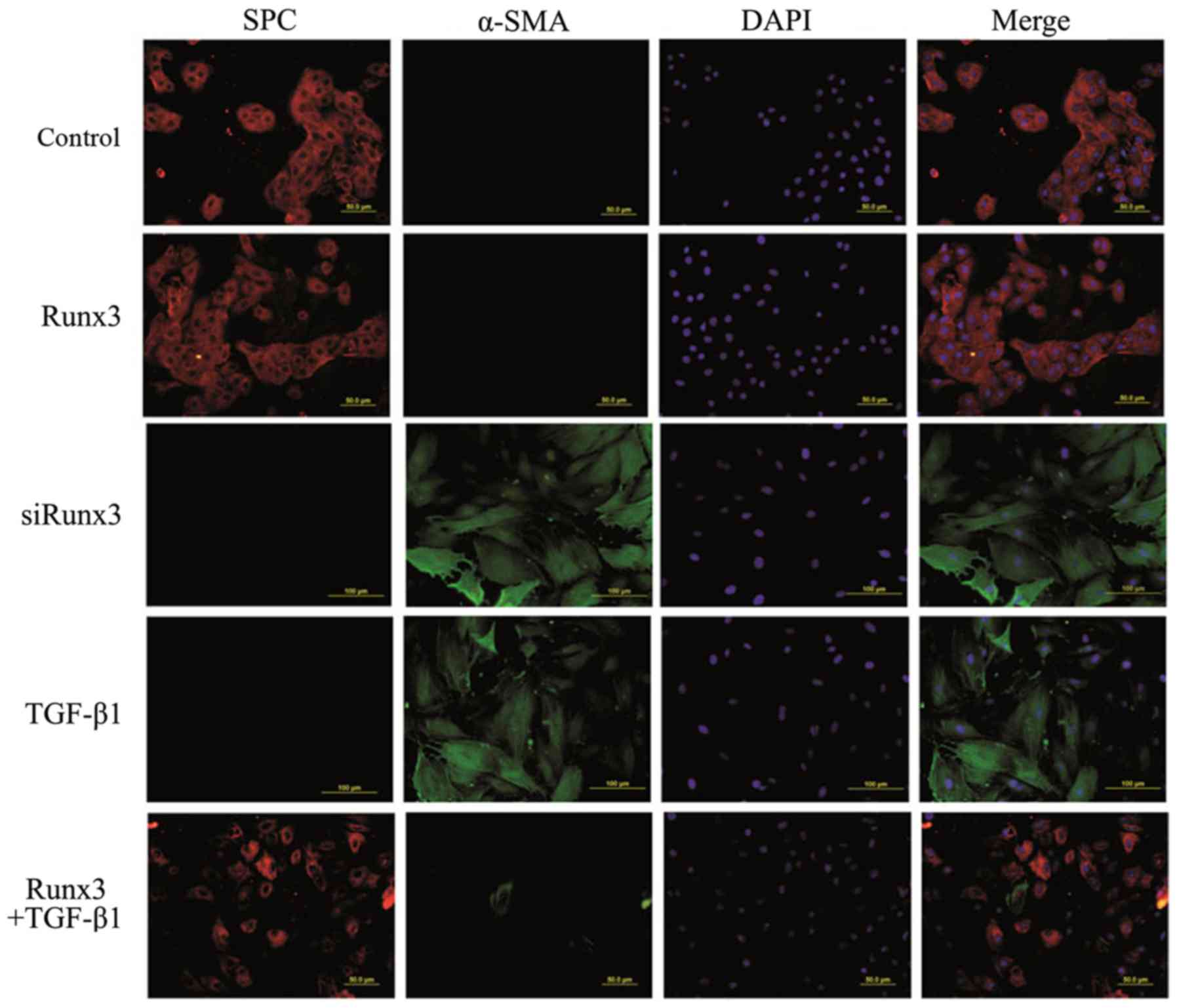
Double-label immunofluorescence
assay
SPC is a specific marker of AT2 cells, while α-SMA
is a marker of mesenchymal cells; their co-expression suggests EMT
occurrence in RLE-6TN cells. Using two-color immunofluorescence
staining we found that in both the control group and the Runx3
group, SPC was expressed in the cytoplasm (red fluorescence), while
the expression of α-SMA was not detectable (green fluorescence). In
the TGF-β1 group and the siRunx3 group, SPC expression was not
observed, yet α-SMA expression was detectable in the cytoplasm.
However, in the Runx3 + TGF-β1 group, a large amount of SPC
expression and a trace amount of α-SMA expression were found
(Fig. 4).
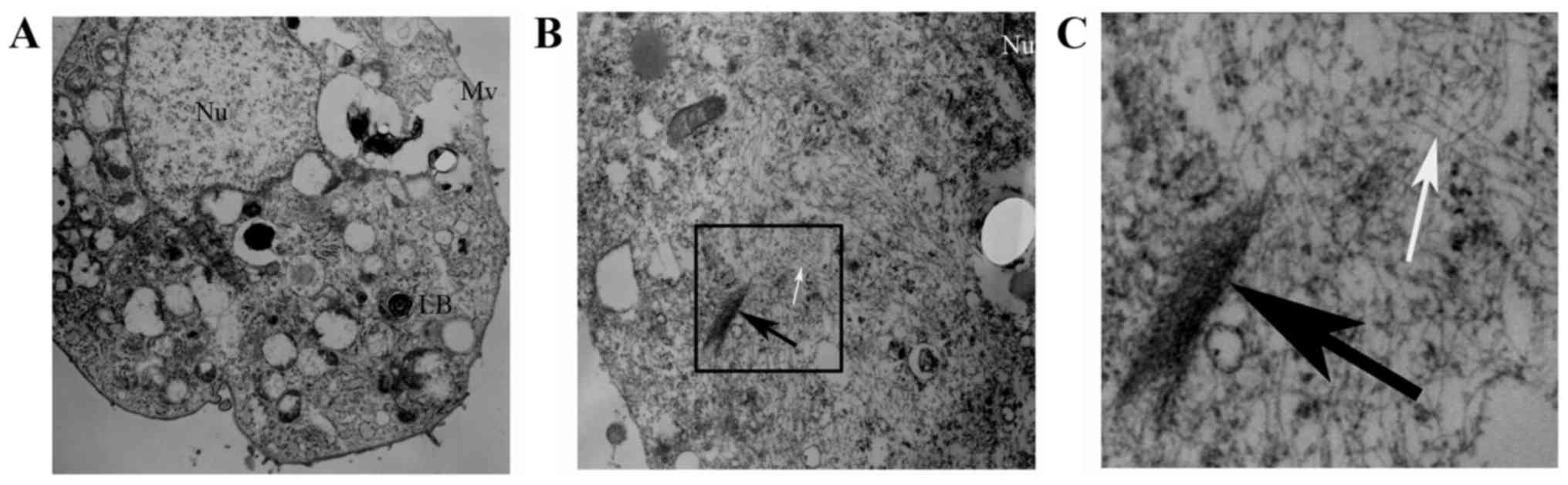
Ultrastructural evidence of EMT in
RLE-6TN cells
We observed the ultrastructure changes of the 5
RLE-6TN groups by TEM. The normal ultrastructure of RLE-6TN cells
(microvilli on cell surface and lamellar bodies in the cells) was
observed in the control group, the Runx3 group and the Runx3 +
TGF-β1 group (Fig. 5A). However,
in the TGF-β1 group and the siRunx3 group, there were a large
number of sparse actin microfilaments and some fasciculated actin
microfilaments in the cytoplasm of the RLE-6TN cells (Fig. 5B and C).
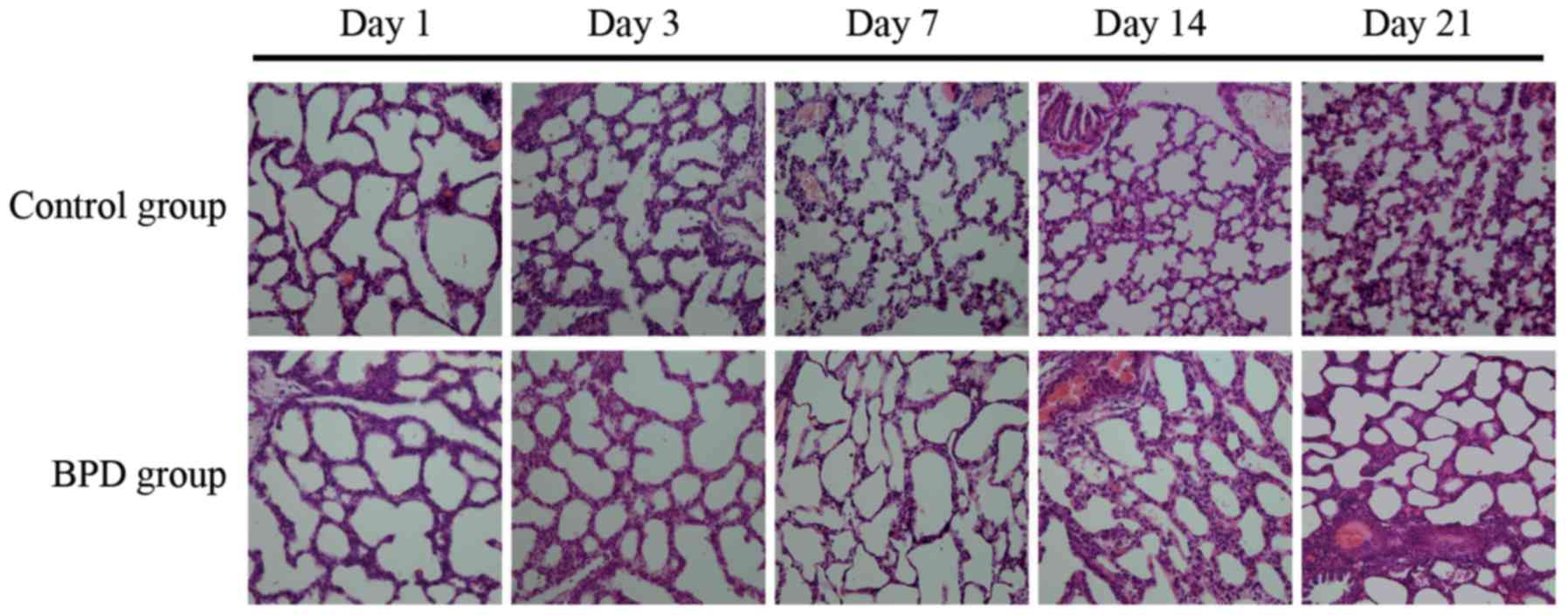
Comparison of pulmonary development
indicators
The sections were stained with H&E.
Morphological changes were assessed using an optical microscope
(x40 magnification; Fig. 6). On
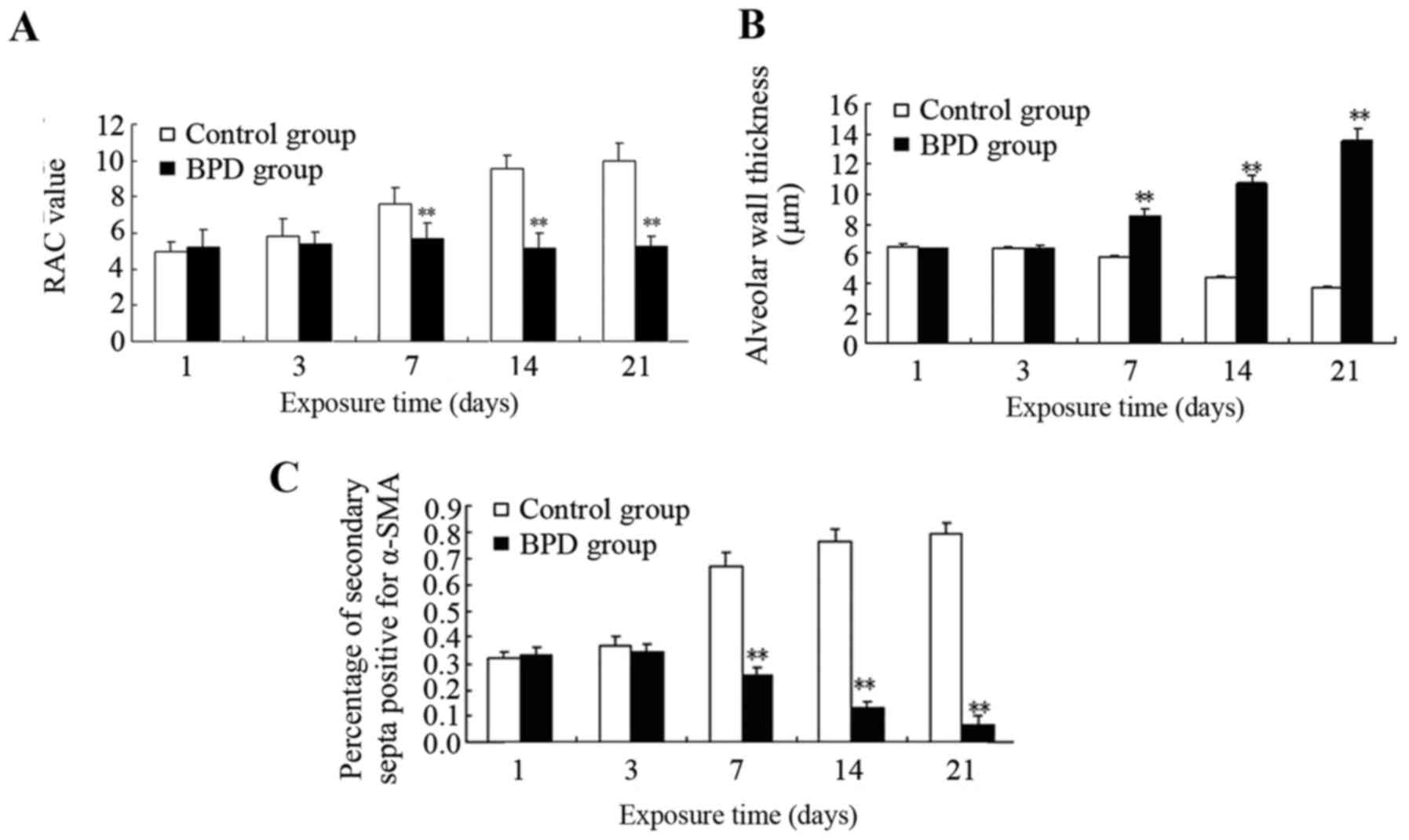
days 1 and 3, there was no significant difference in the RAC
between the 2 groups; however, on days 7, 14 and 21, the RAC of the
control group was significantly greater than that of the BPD group
(P<0.01; Fig. 7A). On days 7,
14 and 21, the alveolar wall thickness in the BPD group was
significantly greater than that in control group (P<0.01;
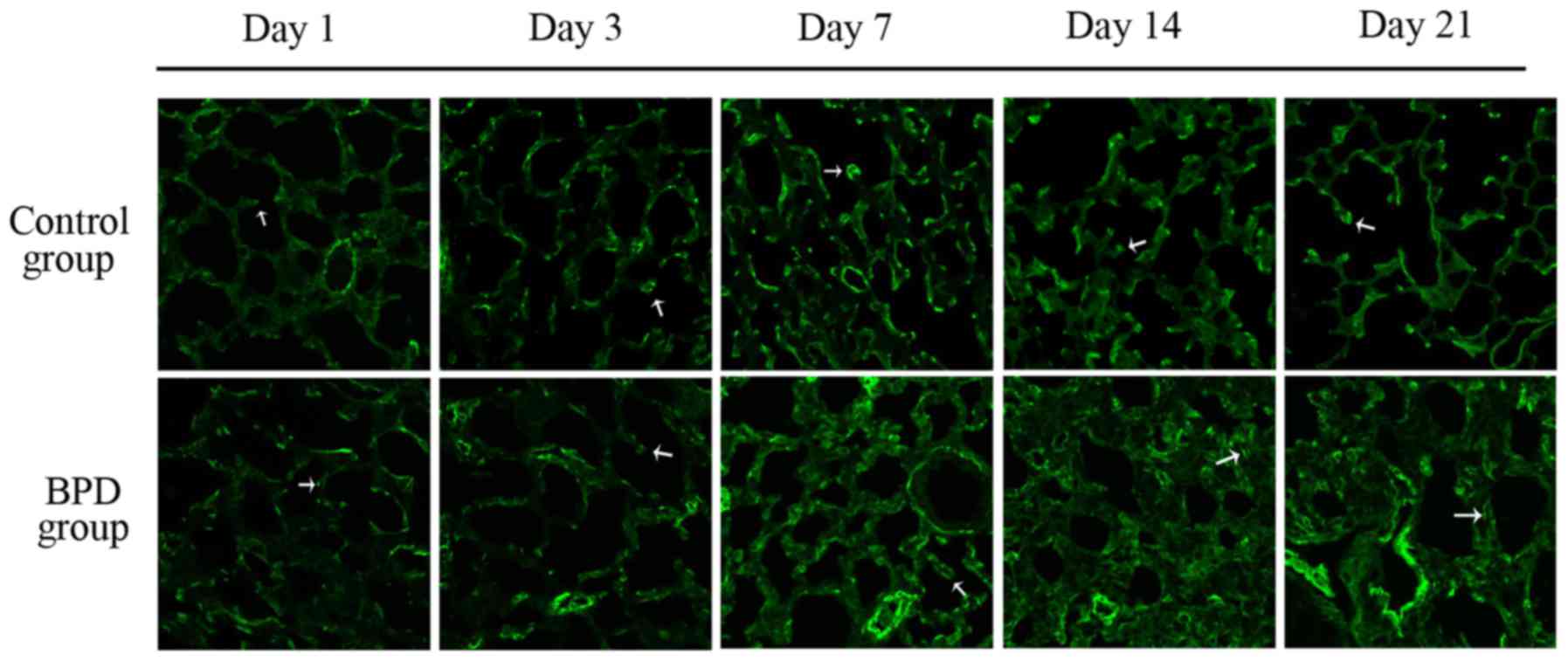
Fig. 7B). On days 1 and 3, α-SMA
expression in the lung tissues did not differ significantly between
the 2 groups; α-SMA was primarily expressed at the top of secondary
septa in the control group samples, but was expressed in the
pulmonary mesenchyma in the BPD samples beginning on day 7. On days
7, 14 and 21 the percentage of α-SMA-positive secondary septa in
the control group was markedly higher than that in the BPD group
(Figs. 7C and 8).
Analysis of the correlation between Runx3
protein expression in lung tissue and pulmonary development
indicators
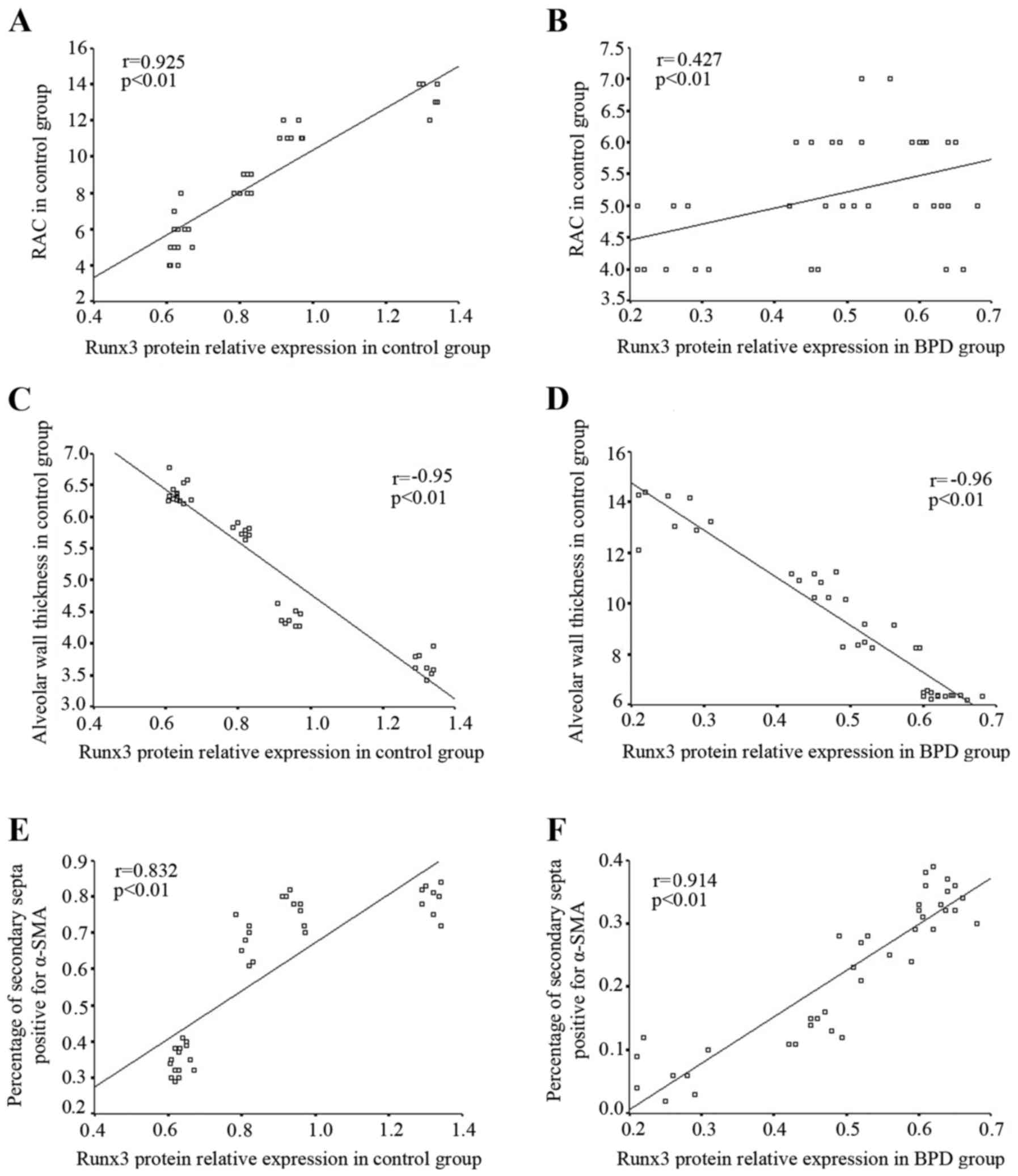
In the control group, the RAC gradually increased in
direct correlation with the increase in Runx3 protein expression;
Spearman correlation analysis revealed that Runx3 protein
expression positively correlated with RAC (r=0.925, P<0.01;
Fig. 9A). In the BPD group, Runx3
protein expression and RAC both decreased gradually over time;
Runx3 protein expression positively correlated with RAC (r=0.427,
P<0.01; Fig. 9B). In the
control group, the alveolar wall became thinner with time and
Spearman correlation analysis indicated that Runx3 protein
expression negatively correlated with alveolar wall thickness
(r=−0.95, P<0.01; Fig. 9C); by
contrast, in the BPD group, the alveolar wall became thicker over
time although Runx3 protein expression remained negatively
correlated with alveolar wall thickness (r=−0.96, P<0.01;
Fig. 9D). In the control group,
Runx3 protein expression positively correlated with the percentage
of α-SMA-positive secondary septa (r=−0.832, P<0.01; Fig. 9E), while in the BPD group, both
Runx3 protein expression and the percentage of α-SMA-positive
secondary septa in lung tissue gradually decreased over time.
Spearman correlation analysis indicated that Runx3 protein
expression positively correlated with the percentage of
α-SMA-positive secondary septa within the BPD group (r=−0.914,
P<0.01; Fig. 9F).
Discussion
In mammals, the Runx family of transcription factors
includes three genes: Runx1, 2 and 3. Each member contains a highly
conserved runt domain and their gene products are similar in
structure; however, each member has different biological functions
and demonstrates tissue-specific expression. All Runx family
members play an important role in the normal development of the
body, as well as the tumor formation (22). Runx1 participates in
hematopoiesis; for example, the development of leukemia is often
associated with a change in the expression of Runx1 (23). Runx2 is associated with
osteoporosis; changes in Runx2 expression are associated with
cleidocranial dysplasia (24) and
osteosarcoma (25). Runx3 is the
smallest member of the Runx family and acts to inhibit tumor
formation (26). Human Runx3 is
located at chromosome 1p36.1 (27). Human Runx3 is 67 kb long and
contains two promoters (P1 and P2) and 6 exons. Runx3 gene
transcription is mainly controlled by P2 which is located at the
front of exon 2 and surrounded by an obvious CpG island with a GC
content of ~64% (28).
The Runx3 gene plays a regulatory role in pulmonary
development (16), but also
affects the occurrence of EMT (29). At E15.5d, the protein expression
of Runx3 has been shown to be low in wild-type fetal rats, while at
E17.5d, Runx3 protein expression in bronchioles and alveoli
increases gradually until the maturation phase (15). In this study, we demonstrated that
Runx3 expression in control newborn rat lung tissue was
significantly increased between 7 and 21 days after birth; by
contrast, Runx3 expression in newborn rat tissue from the BPD group
was significantly lower than that in the control group. These data
indicate that Runx3 expression in lung tissues was downregulated in
the BPD group, perhaps due the abnormal pulmonary development
present in this group. The suppressed Runx3 expression may also
hinder normal pulmonary development of newborn rats with BPD. In
order to further study Runx3 expression in AT2 cells, we isolated
AT2 cells from lung tissue in two rat groups with different day
ages and then measured the mRNA and protein expression of Runx3 in
AT2 cells. We found that the protein and mRNA expression levels of
Runx3 in BPD group were markedly lower than those in the control
group on days 7, 14 and 21. Previously, we confirmed that EMT
occurred in AT2 cells within the BPD group beginning at day 7
(6). Consistent with the above
timeline, a decrease in Runx3 expression in AT2 cells within the
BPD group also appeared beginning at day 7. Taking these data
together with the finding in our previous study that Runx3 can
inhibit the EMT process, we hypothesized that a low level of Runx3
in AT2 cells of BPD lung tissues may be associated with the
occurrence of EMT.
EMT is a common biological phenomenon defined as the
differentiation of epithelial cells into mesenchymal cells
(30). During the development of
EMT, the levels of epithelial cell markers (e.g., E-cadherin and
occludin) gradually decrease (31), while those of mesenchymal cell
markers (e.g., N-cadherin and α-SMA) (32,33) progressively increase. In order to
verify the regulatory effect of Runx3 on EMT in AT2 cells, we
purchased a rat alveolar type II epithelial cell line (RLE-6TN)
(ATCC) for Runx3 plasmid transfection and siRNA interference to
observe the effects of changes in Runx3 expression on EMT. In
addition, we stimulated the RLE-6TN cell line with TGF-β1 to induce
EMT and overexpressed Runx3 in RLE-6TN cells in combination with
TGF-β1 to observe the effects of Runx3 on TGF-β1-induced EMT.
TGF-β1 is a common stimulator used to induce EMT in epithelial
cells (34). We also used it to
induce EMT in RLE-6TN cells, which was characterized by a decrease
in the expression of epithelial cell markers (SPC and E-cad) and an
increase in the expression of mesenchymal cell markers (N-cad and
α-SMA). In the present study, we found the decreased expression of
SPC (an AT2 marker) and E-cad (an epithelial cell marker) and the
increased expression of N-cad and α-SMA (mesenchymal cell markers)
in Runx3-silent RLE-6TN cells, indicating the development of EMT.
Thus, we can conclude that a decrease in Runx3 expression can
directly promote the occurrence of EMT. It has been shown that a
high level of TGF-β1 is present in BPD lung tissue (35). The pathological changes present in
the lungs of transgenic mice overexpressing TGF-β1 are similar to
those observed in BPD, which suggests that high levels of TGF-β1
can contribute to the occurrence of BPD (36). We hypothesize that high levels of
TGF-β1 may be related to EMT occurrence in AT2 cells from
BPD-affected lung tissue. EMT was directly induced using TGF-β1 in
RLE-6TN cells cultured under normal conditions, while it was not
induced by stimulating Runx3-overexpressing RLE-6TN cells with
TGF-β1; specifically, epithelial cell markers were still highly
expressed in the Runx3 + TGF-β1 group. This indicates that a high
level of Runx3 can suppress TGF-β1-induced EMT.
Epithelial cells and mesenchymal cells have
different morphologies and subcellular structures. Specific changes
in subcellular structure are found during EMT in renal tubular
epithelial cells (37,38); based on these findings, we
hypothesized that there may also be a similar change in AT2 cells
experiencing EMT. Using TEM, we observed normal ultrastructure of
AT2 cells within the control group, the Runx3 overexpression group,
and the Runx3 + TGF-β1 group (microvilli on cell surface and
characteristic lamellar bodies in the cytoplasm). By contrast, we
observed a larger number of sparse, fasciculated actin
microfilaments, but no characteristic lamellar bodies in AT2 cells
within the Runx3 gene knockdown group and TGF-β1 stimulation group;
these data indicate that AT2 cells completely transform into
mesenchymal cells via EMT. The above-mentioned findings were
confirmed at the ultrastructure level as well. Therefore, we
conclude that a low level of Runx3 can directly promote the
occurrence of EMT, while a high level of Runx3 has an inhibitory
effect on EMT.
Runx3 can directly regulate the EMT process in AT2
cells; furthermore, EMT in AT2 cells of lung tissues may influence
the normal development of alveoli. In order to evaluate the
association between Runx3 and alveolar development, we performed a
correlation analysis of Runx3 protein expression and pulmonary
development indicators at the level of lung tissue. RAC, alveolar
wall thickness (39), and the
percentage of α-SMA-positive secondary septa (20) are the most important indicators of
alveolar development. Correlation analysis revealed that Runx3
expression positively correlated with RAC and the percentage of
α-SMA-positive secondary septa, but negatively correlated with
alveolar wall thickness. This study also demonstrated that in the
lung tissues of the normal control group, α-SMA was mainly
expressed at the top of secondary septa and the percentage of
α-SMA-positive secondary septa was increased significantly from day
7 to 21 after birth. In our previous study, we found that α-SMA
protein expression was sharply increased in the BPD group beginning
at day 7 after birth, which was significantly greater than that in
the control group (6). In this
study, using immunofluorescence staining, we found that α-SMA was
distributed mainly in pulmonary mesenchyma and rarely in the top of
secondary septa in BPD tissues, which is similar to the α-SMA
expression results obtained in a premature baboon model of BPD
(40). These findings suggest
that the correct localization of myofibroblasts is essential for
normal pulmonary development and that increased myofibroblasts in
the mesenchyma may promote the remodeling of lung tissues in BPD
models.
Our study confirmed that Runx3 expression was
decreased in the lung tissues of rats with BPD in vivo, that
Runx3 expression closely correlated with pulmonary development
indicators in vivo, and that Runx3 can promote EMT in AT2
cells in vitro. Therefore, we infer that a low level of
Runx3 in lung tissues may contribute to EMT occurrence in AT2 cells
and thus influence the normal pulmonary development and promote the
occurrence of BPD. The following factors are considered to be
associated with the decreased expression of Runx3: methylation of
its promoter region (41),
histone modification (42),
micro-RNA regulation (43),
cytoplasmic allotopic expression (44,45) and hemizygote absence (46). Further investigation into the
mechanisms by which Runx3 deficiency promtes BPD is necessary and
may suggest therapeutic options for the treatment of neonatal
BPD.
In conclusion, this study demonstrated that Runx3
expression was decreased in BPD-affected lung tissue, and the lower
Runx3, at least in part, impaired normal lung development and
contributed to BPD by inducing EMT in AT2 cells. However, the
mechanisms underlying the decreased expression of Runx3 in BPD
warrant further investigation.
Abbreviations:
|
BPD
|
bronchopulmonary dysplasia
|
|
EMT
|
epithelial-mesenchymal transition
|
|
AT2 cells
|
alveolar type II epithelial cells
|
|
RAC
|
radial alveolar count
|
|
α-SMA
|
α-smooth muscle actin
|
|
CLD
|
chronic lung disease
|
|
AT1 cells
|
alveolar type I epithelial cells
|
|
SPC
|
surfactant protein C
|
|
E-cad
|
E-cadherin
|
|
N-cad
|
N-cadherin
|
|
TEM
|
transmission electron microscope
|
Acknowledgments
We would like to thank Mr. Fugui Zhang (Central
Laboratory of China Medical University) for providing instructions
for the transmission electron microscope and Mr. Zhihong Zong
(Central Laboratory of China Medical University) for providing
technical support for real-time PCR. This study was funded by the
Natural Science Foundation of China (nos. 81471489 and
81571479).
References
|
1
|
Fanaroff AA, Stoll BJ, Wright LL, Carlo
WA, Ehrenkranz RA, Stark AR, Bauer CR, Donovan EF, Korones SB,
Laptook AR, et al: Trends in neonatal morbidity and mortality for
very low birth-weight infants. Am J Obstet Gynecol. 196:147.e1–e8.
2007. View Article : Google Scholar
|
|
2
|
Walsh MC, Szefler S, Davis J, Allen M, Van
Marter L, Abman S, Blackmon L and Jobe A: Summary proceedings from
the bronchopulmonary dysplasia group. Pediatrics. 117(Suppl 1):
S52–S56. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Husain AN, Siddiqui NH and Stocker JT:
Pathology of arrested acinar development in postsurfactant
bronchopulmonary dysplasia. Hum Pathol. 29:710–717. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Huang C, Reddy Chintagari N,
Bhaskaran M, Weng T, Guo Y, Xiao X and Liu L: miR-375 regulates rat
alveolar epithelial cell trans-differentiation by inhibiting
Wnt/β-catenin pathway. Nucleic Acids Res. 41:3833–3844. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bitterman PB, Polunovsky VA and Ingbar DH:
Repair after acute lung injury. Chest. 105(Suppl 3): 118S–121S.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang H, Fu J, Xue X, Yao L, Qiao L, Hou A,
Jin L and Xing Y: Epithelial-mesenchymal transitions in
bronchopulmonary dysplasia of newborn rats. Pediatr Pulmonol.
49:1112–1123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Inoue K, Shiga T and Ito Y: Runx
transcription factors in neuronal development. Neural Dev.
3:202008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ito Y: RUNX genes in development and
cancer: regulation of viral gene expression and the discovery of
RUNX family genes. Adv Cancer Res. 99:33–76. 2008. View Article : Google Scholar
|
|
9
|
Lee JM, Kwon HJ, Lai WF and Jung HS:
Requirement of Runx3 in pulmonary vasculogenesis. Cell Tissue Res.
356:445–449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Z, Chen L, Zhang X, Xu X, Xing H,
Zhang Y, Li W, Yu H, Zeng J and Jia J: RUNX3 regulates vimentin
expression via miR-30a during epithelial-mesenchymal transition in
gastric cancer cells. J Cell Mol Med. 18:610–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mei PJ, Bai J, Liu H, Li C, Wu YP, Yu ZQ
and Zheng JN: RUNX3 expression is lost in glioma and its
restoration causes drastic suppression of tumor invasion and
migration. J Cancer Res Clin Oncol. 137:1823–1830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YS, Lee JW, Jang JW, Chi XZ, Kim JH,
Li YH, Kim MK, Kim DM, Choi BS, Kim EG, et al: Runx3 inactivation
is a crucial early event in the development of lung adenocarcinoma.
Cancer Cell. 24:603–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nishina S, Shiraha H, Nakanishi Y, Tanaka
S, Matsubara M, Takaoka N, Uemura M, Horiguchi S, Kataoka J,
Iwamuro M, et al: Restored expression of the tumor suppressor gene
RUNX3 reduces cancer stem cells in hepatocellular carcinoma by
suppressing Jagged1-Notch signaling. Oncol Rep. 26:523–531.
2011.PubMed/NCBI
|
|
14
|
Levanon D, Brenner O, Negreanu V, Bettoun
D, Woolf E, Eilam R, Lotem J, Gat U, Otto F, Speck N, et al:
Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1
(Aml1) indicates non-redundant functions during mouse
embryogenesis. Mech Dev. 109:413–417. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee KS, Lee YS, Lee JM, Ito K, Cinghu S,
Kim JH, Jang JW, Li YH, Goh YM, Chi XZ, et al: Runx3 is required
for the differentiation of lung epithelial cells and suppression of
lung cancer. Oncogene. 29:3349–3361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JM, Shin JO, Cho KW, Hosoya A, Cho SW,
Lee YS, Ryoo HM, Bae SC and Jung HS: Runx3 is a crucial regulator
of alveolar differentiation and lung tumorigenesis in mice.
Differentiation. 81:261–268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
You K, Xu X, Fu J, Xu S, Yue X, Yu Z and
Xue X: Hyperoxia disrupts pulmonary epithelial barrier in newborn
rats via the deterioration of occludin and ZO-1. Respir Res.
13:362012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Willis BC, Liebler JM, Luby-Phelps K,
Nicholson AG, Crandall ED, du Bois RM and Borok Z: Induction of
epithelial-mesenchymal transition in alveolar epithelial cells by
transforming growth factor-beta1: potential role in idiopathic
pulmonary fibrosis. Am J Pathol. 166:1321–1332. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cooney TP and Thurlbeck WM: The radial
alveolar count method of Emery and Mithal: a reappraisal 1 -
postnatal lung growth. Thorax. 37:572–579. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Emanuel RL, Torday JS, Mu Q, Asokananthan
N, Sikorski KA and Sunday ME: Bombesin-like peptides and receptors
in normal fetal baboon lung: roles in lung growth and maturation.
Am J Physiol. 277:L1003–L1017. 1999.PubMed/NCBI
|
|
21
|
Cullen A, Van Marter LJ, Allred EN, Moore
M, Parad RB and Sunday ME: Urine bombesin-like peptide elevation
precedes clinical evidence of bronchopulmonary dysplasia. Am J
Respir Crit Care Med. 165:1093–1097. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ito Y: Oncogenic potential of the RUNX
gene family: 'overview'. Oncogene. 23:4198–4208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Speck NA and Gilliland DG: Core-binding
factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2:502–513.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mundlos S, Otto F, Mundlos C, Mulliken JB,
Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et
al: Mutations involving the transcription factor CBFA1 cause
cleidocranial dysplasia. Cell. 89:773–779. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thomas DM, Johnson SA, Sims NA, Trivett
MK, Slavin JL, Rubin BP, Waring P, McArthur GA, Walkley CR,
Holloway AJ, et al: Terminal osteoblast differentiation, mediated
by runx2 and p27KIP1, is disrupted in osteosarcoma. J
Cell Biol. 167:925–934. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bae SC and Choi JK: Tumor suppressor
activity of RUNX3. Oncogene. 23:4336–4340. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kato N, Tamura G, Fukase M, Shibuya H and
Motoyama T: Hypermethylation of the RUNX3 gene promotor in
testicular yolk sac tumor of infants. Am J Pathol. 163:387–391.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bone KR, Gruper Y, Goldenberg D, Levanon D
and Groner Y: Translation regulation of Runx3. Blood Cells Mol Dis.
45:112–116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tanaka S, Shiraha H, Nakanishi Y, Nishina
S, Matsubara M, Horiguchi S, Takaoka N, Iwamuro M, Kataoka J,
Kuwaki K, et al: Runt-related transcription factor 3 reverses
epithelial-mesenchymal transition in hepatocellular carcinoma. Int
J Cancer. 131:2537–2546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thomson S, Petti F, Sujka-Kwok I, Mercado
P, Bean J, Monaghan M, Seymour SL, Argast GM, Epstein DM and Haley
JD: A systems view of epithelial-mesenchymal transition signaling
states. Clin Exp Metastasis. 28:137–155. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lemieux E, Bergeron S, Durand V, Asselin
C, Saucier C and Rivard N: Constitutively active MEK1 is sufficient
to induce epithelial-to-mesenchymal transition in intestinal
epithelial cells and to promote tumor invasion and metastasis. Int
J Cancer. 125:1575–1586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gheldof A and Berx G: Cadherins and
epithelial-to-mesenchymal transition. Prog Mol Biol Transl Sci.
116:317–336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou B, Buckley ST, Patel V, Liu Y, Luo J,
Krishnaveni MS, Ivan M, DeMaio L, Kim KJ, Ehrhardt C, et al:
Troglitazone attenuates TGF-β1-induced EMT in alveolar epithelial
cells via a PPARγ-independent mechanism. PLoS One. 7:e388272012.
View Article : Google Scholar
|
|
34
|
Kojima T, Takano K, Yamamoto T, Murata M,
Son S, Imamura M, Yamaguchi H, Osanai M, Chiba H, Himi T, et al:
Transforming growth factor-beta induces epithelial to mesenchymal
transition by down-regulation of claudin-1 expression and the fence
function in adult rat hepatocytes. Liver Int. 28:534–545. 2008.
View Article : Google Scholar
|
|
35
|
Nakanishi H, Sugiura T, Streisand JB,
Lonning SM and Roberts JD Jr: TGF-beta-neutralizing antibodies
improve pulmonary alveologenesis and vasculogenesis in the injured
newborn lung. Am J Physiol Lung Cell Mol Physiol. 293:L151–L161.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vicencio AG, Lee CG, Cho SJ, Eickelberg O,
Chuu Y, Haddad GG and Elias JA: Conditional overexpression of
bioactive transforming growth factor-beta1 in neonatal mouse lung:
a new model for bronchopulmonary dysplasia? Am J Respir Cell Mol
Biol. 31:650–656. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fan JM, Ng YY, Hill PA, Nikolic-Paterson
DJ, Mu W, Atkins RC and Lan HY: Transforming growth factor-beta
regulates tubular epithelial-myofibroblast transdifferentiation in
vitro. Kidney Int. 56:1455–1467. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ng YY, Huang TP, Yang WC, Chen ZP, Yang
AH, Mu W, Nikolic-Paterson DJ, Atkins RC and Lan HY: Tubular
epithelial-myofibroblast transdifferentiation in progressive
tubulointerstitial fibrosis in 5/6 nephrectomized rats. Kidney Int.
54:864–876. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Joss-Moore L, Carroll T, Yang Y, Fitzhugh
M, Metcalfe D, Oman J, Hale M, Dong L, Wang ZM, Yu X, et al:
Intrauterine growth restriction transiently delays alveolar
formation and disrupts retinoic acid receptor expression in the
lung of female rat pups. Pediatr Res. 73:612–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Subramaniam M, Bausch C, Twomey A,
Andreeva S, Yoder BA, Chang L, Crapo JD, Pierce RA, Cuttitta F and
Sunday ME: Bombesin-like peptides modulate alveolarization and
angiogenesis in bronchopulmonary dysplasia. Am J Respir Crit Care
Med. 176:902–912. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang Y, Tong D, Lou G, Zhang Y and Geng
J: Expression of RUNX3 gene, methylation status and
clinicopathological significance in breast cancer and breast cancer
cell lines. Pathobiology. 75:244–251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee TI, Jenner RG, Boyer LA, Guenther MG,
Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K,
et al: Control of developmental regulators by Polycomb in human
embryonic stem cells. Cell. 125:301–313. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lai KW, Koh KX, Loh M, Tada K, Subramaniam
MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, et
al: Singapore Gastric Cancer Consortium: MicroRNA-130b regulates
the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer.
46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada
K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al: RUNX3, a
novel tumor suppressor, is frequently inactivated in gastric cancer
by protein mislocalization. Cancer Res. 65:7743–7750. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Soong R, Shah N, Peh BK, Chong PY, Ng SS,
Zeps N, Joseph D, Salto-Tellez M, Iacopetta B and Ito Y: The
expression of RUNX3 in colorectal cancer is associated with disease
stage and patient outcome. Br J Cancer. 100:676–679. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yanada M, Yaoi T, Shimada J, Sakakura C,
Nishimura M, Ito K, Terauchi K, Nishiyama K, Itoh K and Fushiki S:
Frequent hemizygous deletion at 1p36 and hypermethylation
downregulate RUNX3 expression in human lung cancer cell lines.
Oncol Rep. 14:817–822. 2005.PubMed/NCBI
|